Abstract
IDH1 SNP rs11554137 was recently reported in association with poor prognosis in normal karyotype adult acute myeloid leukemia (AML). We aimed to determine the prevalence, clinical associations, and prognostic significance of SNP rs11554137 in unselected pediatric and adult AML patients. Diagnostic marrow specimens from 527 AML patients treated on the pediatric trial Children's Oncology Group-AAML03P1 (N = 253) or adult SWOG trials (N = 274) were analyzed for the presence of the SNP. SNP rs11554137 was present in 11% of all patients. SNP status had no prognostic impact on survival in pediatric patients. In adult AML, overall survival for SNP-positive patients was 10% versus 18% for SNP-negative patients (P = .44). Among the 142 adults who achieved complete remission, 5-year relapse-free survival was significantly worse for SNP-positive patients (0% vs 25%, P = .0014). However, among adults with normal cytogenetics, FLT3/ITD was present in 90% of SNP-positive patients versus 59% of SNP-negative patients (P = .0053). In multivariate analysis, adjusting for the effects of age, cytogenetics, and FLT3/ITD, the independent prognostic effect of SNP positivity was not statistically significant (hazard ratio = 1.72, P = .18). The clinical profile of SNP-positive patients suggests that SNP rs11554137 may have biologic effects that bear further investigation. The clinical trials in this study are registered at http://www.clinicaltrials.gov as #NCT000707174 and #NCT00899171.
Introduction
Single nucleotide polymorphisms (SNPs) represent the most common form of genetic diversity within a species and account for much of the variation in genetic traits between persons.1 Such traits include disease susceptibility and response to therapy. The nucleotide substitution in a synonymous SNP does not result in a change in the amino acid sequence encoded. Thus, such “silent” SNPs were originally thought to be nonfunctional and of little consequence to human health and disease, serving only as surrogate biomarkers for other functional genetic variations with which they might be inherited in linkage disequilibrium. However, synonymous SNPs have recently been shown to directly impact gene function through various translational or post-translational mechanisms, such as altering miRNA binding, protein folding, the spliceosome, mRNA stability, or expression in general.2 Further, such “silent” SNPs in the mutational hotspots of acute myeloid leukemia (AML)–associated genes have recently been reported to be of prognostic significance, as first reported in adult AML in the WT1 gene by the German AML SHG cooperative group.3 We corroborated the favorable prognostic profile of WT1 SNP rs16754 in pediatric AML.4
Isocitrate dehydrogenase 1 is a citric acid cycle enzyme encoded by the IDH1 gene. Missense mutations at the arginine 132 codon in exon 4 of IDH1 are a common finding in adult malignant glial tumors and are associated with favorable prognosis in that setting.5 After the discovery by Mardis et al of an IDH1R132 mutation via massively parallel DNA sequencing analysis of an AML patient genome,6 we recently reported IDH1R132 mutations in 4.4% of adult AML patients but did not detect the mutation in pediatric AML.7 These mutations, as well as similar mutations in the related IDH2 gene, have been shown to generate the putative oncogenic metabolite 2-hydoxyglutarate, and may contribute to leukemogenesis via the induction of DNA hypermethylation.8 Our initial study of the IDH1 gene in AML also detected germline polymorphisms in IDH1 exon 4.7 The most common of these was the SNP rs11554137, representing a GGC to GGT transversion at the glycine residue at codon position 105. Although the functional effects of harboring this SNP are unclear, the German AML SHG also recently reported SNP rs11554137 in 12% of cytogenetically normal adult AML patients and notably found that presence of the IDH1 SNP correlated with decreased OS and RFS in these AML patients.9 We sought to investigate the incidence of IDH1 SNP rs11554137 in AML patients enrolled in Children's Oncology Group (COG) or SWOG trials and to determine the clinical associations and prognostic significance of harboring the SNP in these unselected pediatric and adult AML patients.
Methods
Patient samples
Pediatric AML patients enrolled in the COG trial AAML03P1 as well as adult patients with de novo AML enrolled in any of 3 SWOG trials (SWOG-9031, SWOG-9333, and SWOG-9500) were eligible for this study. COG-AAML03P1 enrolled de novo AML patients 0 to 21 years of age; SWOG-9500 enrolled “younger” adult AML patients (18-55 years), whereas SWOG-9031 and SWOG-9333 enrolled “older” adult AML patients (> 55 years). Patients with acute promyelocytic leukemia (M3 AML) were excluded, as were SWOG-9333 patients randomized to induction chemotherapy with mitoxantrone and etoposide, as somewhat poorer outcomes were reported for patients on that treatment arm.10 Details of these clinical trials have been published.10-13 Genomic DNA extracted from diagnostic marrow specimens was available from 257 COG and 274 SWOG patients. Consent was obtained from all study participants in accordance with the Declaration of Helsinki. Institutional review board approval was obtained from the Fred Hutchinson Cancer Research Center before mutation analysis, and this study was approved by the Myeloid Disease Biology Committees of the COG and SWOG.
Molecular genotyping
Genetic material was extracted from diagnostic marrow specimens using the AllPrep DNA/RNA Mini Kit via the QIAcube automated system (QIAGEN). The coding region of IDH1 exon 4, containing codon G105, was amplified via PCR as previously described.7 Thermocycler conditions were 94°C for 5 minutes; 35 cycles at 94°C for 30 seconds, 58°C for 45 seconds, and 72°C for 1 minute; and a final extension step at 72°C for 7 minutes. Purified PCR products were sequenced using the BigDye Terminator sequencing reaction and run on an ABI 3730xl DNA analyzer (Applied Biosystems).
IDH1 mRNA expression analysis
Reverse transcription (RT) was performed on 1 μg total RNA, extracted via the QIAcube system as in “Molecular genotyping” (QIAGEN), per standard RT protocol using Invitrogen reagents. mRNA expression analysis was performed via quantitative RT-PCR on a StepOnePlus instrument using TaqMan master mix and primer/probe sets for IDH1 and the control gene β-glucuronidase (Applied Biosystems). The ΔΔCt method was used to determine relative IDH1 expression levels, normalized to pooled normal marrow controls.
Statistical methods
Data regarding clinical characteristics and treatment outcomes were collected and evaluated according to the standard practices of the COG and SWOG for their respective studies. Overall survival (OS) was calculated as time from study entry until death from any cause, with the observation censored at the date of last contact for patients last known to be alive. The Kaplan-Meier method was used to estimate OS. Relapse-free survival (RFS) was defined as the time from complete remission (CR) until relapse or death from any cause. Estimates of relapse risk (RR) were obtained by the method of cumulative incidence that accounts for competing events. RR was defined as the time from end of course 1 for patients in CR to relapse or death because of progressive disease where deaths from nonprogressive disease were considered competing events. The significance of predictor variables was tested with the log-rank statistic for OS and with Gray's statistic for RR. The significance of observed differences in proportions was tested by the χ2 test and Fisher exact test when data were sparse. The Mann-Whitney test was used to determine the significance between differences in medians. Cox proportional hazard models were used to estimate hazard ratios (HRs) for univariate and multivariate analyses for OS and RFS.
Results
Patient population
Cryopreserved diagnostic specimens were available from 253 (74%) of the 340 eligible pediatric patients enrolled in COG-AAML03P1 and 274 (56%) of the 487 eligible adult patients enrolled in the 3 SWOG studies. To ensure that the patients included in this study were representative, we compared demographics, laboratory and clinical characteristics, and outcome for those patients with and without available specimens. There were no significant differences in the pediatric (COG) cohort. SWOG patients with diagnostic specimens available had significantly higher (P < .001) median diagnostic WBC count (29.8 × 109/L vs 4.6 × 109/L), peripheral blood blast percentage (42% vs 12%), and marrow blast percentage (71% vs 59%) than the 213 patients without available samples. Outcome measures did not differ significantly between patients with and without available specimens.
Pediatric AML
In the pediatric (COG) cohort (median age, 9.8 years), SNP rs11554137 was present in 27 of 253 (11%; 95% confidence interval [CI], 7%-15%) patients. Of the 27 SNP-positive patients, 1 (3.7%) patient was homozygous for SNP rs11554137. Demographic, laboratory, and clinical characteristics of patients with and without IDH1 SNP rs11554137 were compared (Table 1). SNP-positive pediatric patients did not differ significantly from SNP-negative patients in terms of presenting clinical characteristics, including racial distribution. Age distribution was similar in patients with and without the SNP, except in patients 0-2 years of age, who accounted for 44% of SNP-positive patients versus 23% of SNP-negative patients (P = .013).
Characteristics of adult and pediatric AML patients by SNP status
| Characteristic . | Pediatric AML . | Characteristic . | Adult AML . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IDH1 SNP+ (n = 27) . | IDH1 SNP− (n = 226) . | P . | IDH1 SNP+ (n = 30) . | IDH1 SNP− (n = 244) . | P . | ||||||
| N . | % . | N . | % . | N . | % . | N . | % . | ||||
| Sex | Sex | ||||||||||
| Male | 17 | 63 | 123 | 54 | .399 | Male | 11 | 37 | 137 | 56 | .052 |
| Female | 10 | 37 | 103 | 46 | Female | 19 | 63 | 107 | 44 | ||
| Age, y | Age, y | ||||||||||
| Median (range) | 5.8 | (0.15-17.2) | 10.4 | (0.07-21.6) | .046* | Median (range) | 63 | (22-76) | 63 | (18-88) | .59 |
| 0-2 y | 12 | 44 | 51 | 23 | 18-55 y | 11 | 37 | 71 | 29 | ||
| 3-10 y | 6 | 22 | 69 | 31 | 56-67 y | 11 | 37 | 83 | 34 | ||
| > 11 y | 9 | 33 | 106 | 47 | > 68 y | 8 | 27 | 90 | 37 | ||
| Race | .55 | Race | .0039* | ||||||||
| White | 18 | 69 | 157 | 77 | White | 20 | 67 | 217 | 89 | ||
| Black | 6 | 23 | 30 | 15 | Black | 9 | 30 | 17 | 7 | ||
| Asian | 2 | 8 | 12 | 6 | Asian | 0 | 0 | 7 | 3 | ||
| Other | 0 | 0 | 6 | 3 | Other | 0 | 0 | 1 | 0 | ||
| Unknown | 1 | 21 | Unknown | 1 | 3 | 2 | 1.00 | ||||
| Ethnicity | .19 | Ethnicity | .21 | ||||||||
| Hispanic | 2 | 8 | 40 | 18 | Hispanic | 2 | 7 | 6 | 2 | ||
| Non-Hispanic | 23 | 92 | 177 | 82 | Non-Hispanic | 28 | 93 | 238 | 98 | ||
| Unknown | 2 | 9 | Unknown | 0 | 0 | ||||||
| WBC, × 109/L, median (range) | 33.2 | (0.98-495) | 19.6 | (0.8-409) | .240 | WBC, × 109/L, median (range) | 26.5 | (2.7-298) | 29.8 | (0.7-274) | .82 |
| BM blasts, %, median (range) | 72 | (22-97) | 68 | (0-100) | .596 | BM blasts, %, median (range) | 80 | (8-97) | 70 | (0-99) | .025* |
| Cytogenetics | Cytogenetics | ||||||||||
| Normal | 3 | 13 | 44 | 21 | .426 | Normal | 11 | 46 | 76 | 42 | .83 |
| t(8;21) | 2 | 8 | 27 | 13 | .747 | CBF† | 0 | 0 | 22 | 12 | .083 |
| Abnormal 16 | 5 | 21 | 26 | 13 | .336 | −7/7q− | 1 | 4 | 15‡ | 8 | .7 |
| Abnormal 11 | 3 | 13 | 46 | 22 | .274 | +8 | 1 | 4 | 27 | 15 | .21 |
| t(6;9)(p23;q34) | 0 | 0 | 7 | 3 | 1.000 | Other | 11 | 46 | 40 | 22 | .022* |
| −7/7q− | 2 | 8 | 7 | 3 | .236 | No data | 6 | 65 | |||
| −5/5q− | 0 | 0 | 3 | 1 | 1.000 | ||||||
| +8 | 1 | 4 | 19 | 9 | .703 | FLT3/ITD | |||||
| Other | 8 | 33 | 29 | 14 | .033* | Present | 13 | 48 | 75 | 33 | .140 |
| No data | 3 | 18 | Absent | 14 | 52 | 151 | 67 | ||||
| 3 | 18 | ||||||||||
| FLT3/ITD | FLT3/ITD by karyotype | ||||||||||
| Present | 4 | 15 | 25 | 11 | .527 | Normal | |||||
| Absent | 23 | 85 | 201 | 89 | Present | 9 | 90 | 29 | 41 | .0053* | |
| Unknown | 0 | 0 | Absent | 1 | 10 | 41 | 59 | ||||
| Abnormal | |||||||||||
| Present | 1 | 9 | 12 | 13 | 1.00 | ||||||
| Absent | 10 | 91 | 83 | 86 | |||||||
| NPM1 mutation | NPM1 mutation | ||||||||||
| Present | 0 | 0 | 10 | 5 | .604 | Present | 9 | 35 | 70 | 32 | .823 |
| Absent | 23 | 100 | 191 | 95 | Absent | 17 | 65 | 151 | 68 | ||
| Unknown | 4 | 25 | Unknown | 4 | 23 | ||||||
| CEBPA mutation | CEBPA mutation | ||||||||||
| Yes | 2 | 7 | 10 | 5 | .629 | Yes | 0 | 0 | 6 | 3 | 1.000 |
| No | 25 | 93 | 206 | 95 | No | 28 | 100 | 217 | 97 | ||
| Unknown | 0 | 10 | Unknown | 2 | 21 | ||||||
| Characteristic . | Pediatric AML . | Characteristic . | Adult AML . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IDH1 SNP+ (n = 27) . | IDH1 SNP− (n = 226) . | P . | IDH1 SNP+ (n = 30) . | IDH1 SNP− (n = 244) . | P . | ||||||
| N . | % . | N . | % . | N . | % . | N . | % . | ||||
| Sex | Sex | ||||||||||
| Male | 17 | 63 | 123 | 54 | .399 | Male | 11 | 37 | 137 | 56 | .052 |
| Female | 10 | 37 | 103 | 46 | Female | 19 | 63 | 107 | 44 | ||
| Age, y | Age, y | ||||||||||
| Median (range) | 5.8 | (0.15-17.2) | 10.4 | (0.07-21.6) | .046* | Median (range) | 63 | (22-76) | 63 | (18-88) | .59 |
| 0-2 y | 12 | 44 | 51 | 23 | 18-55 y | 11 | 37 | 71 | 29 | ||
| 3-10 y | 6 | 22 | 69 | 31 | 56-67 y | 11 | 37 | 83 | 34 | ||
| > 11 y | 9 | 33 | 106 | 47 | > 68 y | 8 | 27 | 90 | 37 | ||
| Race | .55 | Race | .0039* | ||||||||
| White | 18 | 69 | 157 | 77 | White | 20 | 67 | 217 | 89 | ||
| Black | 6 | 23 | 30 | 15 | Black | 9 | 30 | 17 | 7 | ||
| Asian | 2 | 8 | 12 | 6 | Asian | 0 | 0 | 7 | 3 | ||
| Other | 0 | 0 | 6 | 3 | Other | 0 | 0 | 1 | 0 | ||
| Unknown | 1 | 21 | Unknown | 1 | 3 | 2 | 1.00 | ||||
| Ethnicity | .19 | Ethnicity | .21 | ||||||||
| Hispanic | 2 | 8 | 40 | 18 | Hispanic | 2 | 7 | 6 | 2 | ||
| Non-Hispanic | 23 | 92 | 177 | 82 | Non-Hispanic | 28 | 93 | 238 | 98 | ||
| Unknown | 2 | 9 | Unknown | 0 | 0 | ||||||
| WBC, × 109/L, median (range) | 33.2 | (0.98-495) | 19.6 | (0.8-409) | .240 | WBC, × 109/L, median (range) | 26.5 | (2.7-298) | 29.8 | (0.7-274) | .82 |
| BM blasts, %, median (range) | 72 | (22-97) | 68 | (0-100) | .596 | BM blasts, %, median (range) | 80 | (8-97) | 70 | (0-99) | .025* |
| Cytogenetics | Cytogenetics | ||||||||||
| Normal | 3 | 13 | 44 | 21 | .426 | Normal | 11 | 46 | 76 | 42 | .83 |
| t(8;21) | 2 | 8 | 27 | 13 | .747 | CBF† | 0 | 0 | 22 | 12 | .083 |
| Abnormal 16 | 5 | 21 | 26 | 13 | .336 | −7/7q− | 1 | 4 | 15‡ | 8 | .7 |
| Abnormal 11 | 3 | 13 | 46 | 22 | .274 | +8 | 1 | 4 | 27 | 15 | .21 |
| t(6;9)(p23;q34) | 0 | 0 | 7 | 3 | 1.000 | Other | 11 | 46 | 40 | 22 | .022* |
| −7/7q− | 2 | 8 | 7 | 3 | .236 | No data | 6 | 65 | |||
| −5/5q− | 0 | 0 | 3 | 1 | 1.000 | ||||||
| +8 | 1 | 4 | 19 | 9 | .703 | FLT3/ITD | |||||
| Other | 8 | 33 | 29 | 14 | .033* | Present | 13 | 48 | 75 | 33 | .140 |
| No data | 3 | 18 | Absent | 14 | 52 | 151 | 67 | ||||
| 3 | 18 | ||||||||||
| FLT3/ITD | FLT3/ITD by karyotype | ||||||||||
| Present | 4 | 15 | 25 | 11 | .527 | Normal | |||||
| Absent | 23 | 85 | 201 | 89 | Present | 9 | 90 | 29 | 41 | .0053* | |
| Unknown | 0 | 0 | Absent | 1 | 10 | 41 | 59 | ||||
| Abnormal | |||||||||||
| Present | 1 | 9 | 12 | 13 | 1.00 | ||||||
| Absent | 10 | 91 | 83 | 86 | |||||||
| NPM1 mutation | NPM1 mutation | ||||||||||
| Present | 0 | 0 | 10 | 5 | .604 | Present | 9 | 35 | 70 | 32 | .823 |
| Absent | 23 | 100 | 191 | 95 | Absent | 17 | 65 | 151 | 68 | ||
| Unknown | 4 | 25 | Unknown | 4 | 23 | ||||||
| CEBPA mutation | CEBPA mutation | ||||||||||
| Yes | 2 | 7 | 10 | 5 | .629 | Yes | 0 | 0 | 6 | 3 | 1.000 |
| No | 25 | 93 | 206 | 95 | No | 28 | 100 | 217 | 97 | ||
| Unknown | 0 | 10 | Unknown | 2 | 21 | ||||||
Significant P value.
CBF abnormality includes t(8;21), inv(16), and t(16;16). Three patients with +8 also have CBF abnormalities, and another 3 have −7/7q−.
Two patients with −7/7q− also have +8; another 2 patients with −7/7q− also have t(8;21); and a fifth patient with −7 also has der(8)t(8;21)(q12;p11) as part of a complex karyotype.
Clinical outcome data were compared for pediatric patients with and without IDH1 SNP rs11554137. SNP-positive and SNP-negative patients had similar CR rates after one course of induction (81% vs 82%, P = 1.00). OS at 3 years from study entry for SNP-positive patients was 75% ± 18% versus 66% ± 6% for those without the SNP (HR = 0.76, P = .641). The corresponding RFS from CR was 59% ± 23% and 57% ± 7% for those with and without SNP rs11554137 (P = .923). For patients who achieved an initial remission (n = 200), those with and without the IDH1 SNP had similar rates of relapse (31% ± 21% vs 34% ± 7%; P = .759) and treatment-related mortality (10% ± 14% vs 8% ± 4%; P = .390) from the end of course 1. Thus, IDH1 SNP status had no prognostic impact on survival in the pediatric cohort. These findings were recapitulated when analysis was restricted to standard-risk pediatric patients. The standard-risk group of pediatric patients was defined as lacking either low-risk (CEBPA mutation, NPMc mutation, or core-binding factor AML [CBF AML: inv(16), t(16;16), or t(8;21)] or high-risk (−7, −5/del 5q, or FLT3/ITD with high allelic ratio) features. Standard risk SNP-positive and SNP-negative patients had similar rates of 3-year OS (69% ± 26% vs 62% ± 9%%, P = .862), RFS (56 ± 33% vs 53% ± 11, P = .971), and RR (33% ± 31% vs 42% ± 12%, P = .652).
Adult AML
In the adult (SWOG) cohort (median age, 63 years), the IDH1 SNP was present in 30 of 274 patients (11%; 95% CI, 8%-15%). Of the 30 SNP-positive patients, 1 (3.3%) patient was homozygous for SNP rs11554137. Notably, none of the patients with SNP rs11554137 had concomitant IDH1R132 mutations. Demographic, laboratory, and clinical characteristics of patients with and without the IDH1 SNP were compared (Table 1). The prevalence of SNP rs11554137 in adult AML varied significantly between racial groups (P = .0046), mostly because of a high proportion of blacks (30%) among SNP-positive patients. The IDH1 SNP was not found in adult patients with t(8,21) or inv(16)/t(16,16) (CBF AML). CEBPA mutations occurred at similar frequencies in IDH1 SNP-positive patients compared with SNP-negative patients (0% vs 3%, P = 1.00). Likewise, the incidence of NPM1 mutations did not differ between SNP-positive and SNP-negative patients (35% vs 32%, P = .823).
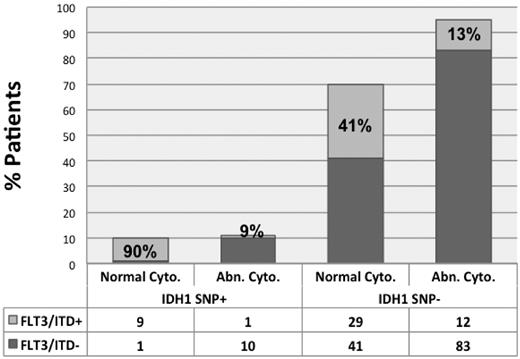
For the 253 patients in whom FLT3 status was known, FLT3/ITD occurred more commonly in SNP-positive patients (13 of 27 = 48%) compared with SNP-negative patients (75 of 226 = 33%), but this difference was not statistically significant (P = .14). However, within the normal karyotype subset, FLT3/ITD was present in 9 of 10 (90%) SNP-positive patients versus 41% of wild-type patients (P = .0053, Table 1). Conversely, only 9% of SNP-positive patients with abnormal karyotype harbored FLT3/ITD (Figure 1).
Distribution of FLT3/ITD within cytogenetic groups by IDH1 SNP status in adult AML.FLT3/ITD was present in 90% of SNP-positive adult AML patients with normal karyotype; conversely, FLT3/ITD was present in only 9% of SNP-positive adult AML patients with cytogenetic abnormalities.
Distribution of FLT3/ITD within cytogenetic groups by IDH1 SNP status in adult AML.FLT3/ITD was present in 90% of SNP-positive adult AML patients with normal karyotype; conversely, FLT3/ITD was present in only 9% of SNP-positive adult AML patients with cytogenetic abnormalities.
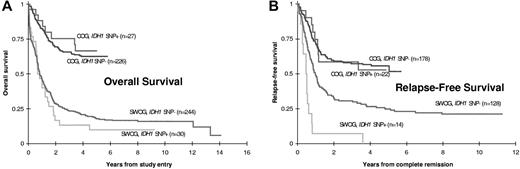
CR rates were similar for adult patients with and without SNP rs11554137 (47% vs 52%, P = .57). SNP-positive patients had somewhat poorer OS (Figure 2; 10% vs 18% at 5 years, HR = 1.17, 95% CI, 0.79-1.76), although this difference was not statistically significant (P = .44). Among the 142 patients who achieved CR, however, RFS was significantly worse for SNP-positive patients (0% vs 25% at 5 years, HR = 2.89, 95% CI, 1.62-5.15, P = .0014). Of the 14 SNP-positive patients who achieved CR, 13 relapsed (all but 1 within 10 months) and the 14th patient died of sepsis in remission after 61 days.
Prognostic significance of IDH1 SNP rs11554137 in adult and pediatric AML patients. Kaplan-Meier estimates of (A) OS and (B) RFS for adult (SWOG) and pediatric (COG) patients with or without SNP rs11554137. RFS was significantly worse in SNP-positive adult patients (P = .0014).
Prognostic significance of IDH1 SNP rs11554137 in adult and pediatric AML patients. Kaplan-Meier estimates of (A) OS and (B) RFS for adult (SWOG) and pediatric (COG) patients with or without SNP rs11554137. RFS was significantly worse in SNP-positive adult patients (P = .0014).
Because of the wide age distribution of adult patients included in this study, we performed subset outcome analysis of the SWOG patients within the following age categories: age 18-55 years (all patients on SWOG-9500), age 56-67 years, and age 68 or greater years. The groups 56-67 years and more than 68 years were defined to have approximately equal numbers of patients; these 2 groups encompassed all patients enrolled in SWOG-9031 and SWOG-9333. No association was detected between SNP status and age, and no significant interaction was found between age and the effect of SNP status on OS/RFS (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
In univariate analyses of the 142 adult patients who achieved CR, RFS decreased significantly with increasing age (P = .0014). RFS was not significantly heterogeneous among the 4 cytogenetic groups (P = .059), although large differences were observed with 5-year RFS of 40%, 29%, 16%, and 0% for the CBF, normal, other, and −7/7q− groups, respectively.
Multivariate regression analyses of treatment outcomes in the adult patients were performed to assess whether the association of SNP status with outcomes might be because of confounding effects of the following factors: age, sex, race, performance status (0 or 1 vs 2 or 3), FLT3/ITD (present vs absent), NPM mutation (present vs absent), marrow blast percentage, WBC, peripheral blast percentage, and cytogenetics (normal without nonclonal abnormalities vs CBF vs −7/7q− vs all others). Analyses that included cytogenetic data were limited to the 203 patients with evaluable karyotypes; for these analyses, 3 patients with +8 and −7/7q− were assigned to the −7/7q− category, and 3 others with +8 and t(8;21) were assigned to the CBF category. As expected, OS decreased significantly with increasing age (P < .0001), was significantly lower for patients with performance status 2 or 3 (P = .0093), and varied significantly among the 4 cytogenetic categories (P < .0001), with 5-year OS of 37%, 24%, 13%, and 0% for the CBF, normal, other, and −7/7q− groups, respectively. After adjusting for the effects of age, performance status, and cytogenetics in multivariate proportional hazards regression, SNP status was not significantly associated with OS (HR = 1.17, 95% CI, 0.73-1.87, P = .51).
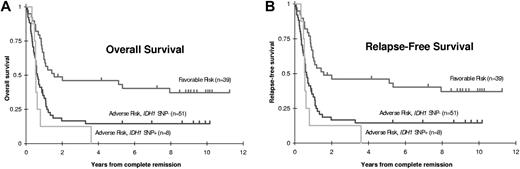
For RFS, in multivariate proportional hazards analysis, after adjusting for the effects of age and cytogenetics, SNP status did retain a significant independent prognostic effect (HR = 2.82, 95% CI, 1.41-5.68, P = .0036). However, when FLT3/ITD status was added to the multivariate analysis, the independent prognostic effect of SNP positivity was markedly reduced and no longer statistically significant (HR = 1.72, P = .18). For the 186 SWOG patients for whom both cytogenetic status and FLT3 status were known, patients were divided into 4 risk categories: CBF AML, normal cytogenetics with negative FLT3/ITD, normal cytogenetics with positive FLT3/ITD, and any non-CBF clonal cytogenetic abnormality. These categorizations allowed classification into 2 risk groups: patients with normal cytogenetics negative for FLT3/ITD had similar outcomes to those with CBF AML, and together compose the “favorable risk” group, whereas patients with normal cytogenetics positive for FLT3/ITD had similar outcomes to those with non-CBF cytogenetic abnormalities, and together compose the “adverse risk” group (supplemental Figure 1). Notably, only one of the SNP-positive patients with known cytogenetics and FLT3 status fell into the “favorable risk” group, whereas the remaining 20 SNP-positive patients were categorized in the “adverse risk” group (Table 2). After allowing for the effects of risk group (favorable vs adverse) and NPM mutation status, there was no evidence that the presence of the SNP had any independent prognostic effect within the adverse risk group (Figure 3; P = .75 for CR; P = .70 for refractory disease (RD), HR = 0.90, 95% CI, 0.51-1.58, P = .70 for OS; and HR = 1.81, 95% CI, 0.79-4.12, P = .16 for RFS).
Distribution of adult AML patients in risk categories by IDH1 SNP status
| . | IDH1 SNP+ (n = 21) . | IDH1 SNP− (n = 165) . | ||
|---|---|---|---|---|
| N . | % . | N . | % . | |
| Favorable risk | ||||
| CBF AML | 0 | 0 | 20 | 12 |
| CN, FLT3− | 1 | 5 | 41 | 25 |
| Adverse risk | ||||
| CN, FLT3+ | 9 | 43 | 29 | 18 |
| Any other cytogenetics | 11 | 52 | 75 | 45 |
| . | IDH1 SNP+ (n = 21) . | IDH1 SNP− (n = 165) . | ||
|---|---|---|---|---|
| N . | % . | N . | % . | |
| Favorable risk | ||||
| CBF AML | 0 | 0 | 20 | 12 |
| CN, FLT3− | 1 | 5 | 41 | 25 |
| Adverse risk | ||||
| CN, FLT3+ | 9 | 43 | 29 | 18 |
| Any other cytogenetics | 11 | 52 | 75 | 45 |
OS and RFS stratified by risk group and IDH1 SNP status in 186 adult AML patients. Kaplan-Meier estimates of (A) OS and (B) RFS are shown by risk group and, within the adverse risk group, IDH1 SNP status. Tick marks indicate censored observations. Among adult patients with adverse risk based on karyotype or FLT3/ITD, the IDH1 SNP was not significantly associated with OS (P = .94) or RFS (P = .28).
OS and RFS stratified by risk group and IDH1 SNP status in 186 adult AML patients. Kaplan-Meier estimates of (A) OS and (B) RFS are shown by risk group and, within the adverse risk group, IDH1 SNP status. Tick marks indicate censored observations. Among adult patients with adverse risk based on karyotype or FLT3/ITD, the IDH1 SNP was not significantly associated with OS (P = .94) or RFS (P = .28).
IDH1 expression
As SNP rs11554137 has been reported in correlation with elevated IDH1 expression,9 we measured IDH1 expression via quantitative RT-PCR in 200 unselected patients for whom IDH1 SNP status was known. IDH1 mRNA expression level for each sample was normalized to pooled normal marrow controls and ranged between 0.02 and 2.71 times normal marrow in SNP-negative patients (n = 178), compared with a range between 0.07 and 2.51 times normal marrow in SNP-positive patients (n = 22). Median IDH1 expression was similar between the 2 groups (supplemental Figure 2): 0.24 times normal marrow for SNP-negative patients and 0.25 times normal marrow for SNP positive-patients (P = .8954). IDH1 SNP status had no correlation with mRNA expression in our study.
Discussion
In this study, we identified the IDH1 SNP rs11554137 in 11% of both pediatric and adult AML patients. Although the presence of the SNP was not prognostic in pediatric AML patients, SNP positivity in adult AML patients was a significant risk factor for decreased RFS in univariate analysis, but not multivariate analysis, as the correlation with poor outcome for SNP-positive patients was confounded by other adverse risk factors. Notably, the majority of SNP-positive adult AML patients had abnormal cytogenetics, and none of the SNP-positive adult patients had favorable-risk CBF AML. Within the normal cytogenetic subset, 90% of SNP-positive adult patients had FLT3/ITD.
SNP rs11554137 lies in exon 4 near the R132 mutational hotspot, and IDH1R132 mutations were mutually exclusive of the IDH1 SNP. Function-altering mutations of IDH1R132 produce the oncoprotein 2-hydoxyglutarate and also correlate with abnormalities of DNA methylation8 ; any possible metabolic or epigenetic consequences of the SNP rs11554137 have yet to be studied and bear further investigation. Although we previously reported the absence of IDH1R132 mutations in pediatric AML,7 the 11% prevalence of the IDH1 SNP in our study was nearly identical in both pediatric and adult AML, and similar to the incidence of 11.7% in healthy volunteers reported by Wagner et al.9 However, within our study's infant/toddler cohort (age 0-2 years), the SNP was present in 12 of 63 patients (19%). It is possible that the presence of the SNP confers a slight predisposition to AML through an as yet undefined mechanism requiring subsequent genetic events, and that this germline predisposition may be associated with a shorter latency to leukemia in certain cases.
We found that, although IDH1 SNP rs11554137 was not prognostic in pediatric AML, the SNP nonetheless correlated with poor outcomes (significantly worse RFS and a trend toward worse OS) in adult AML. However, this correlation resulted from the fact that nearly all SNP-positive patients had unfavorable risk features by virtue of their karyotype or FLT3/ITD status. We have previously reported in pediatric AML a lack of independent prognostic significance for molecular markers, which have been shown to be prognostic in adults, as in the case of WT1 mutations,14 as well as cKit mutations in CBF AML.15 Differences in disease biology may partially explain the discrepancy between pediatric and adult AML; for example, FLT3/ITD is significantly more common in adult AML compared with pediatric AML, although the prevalence of the IDH1 SNP remains approximately the same. In addition, differences in treatment schema between pediatric and adult AML may also be a contributing factor, as the prognostic value of any biomarker may be therapy-dependent.
The correlation of SNP positivity with clinical outcome in adult AML in our study also differed slightly from the results for German patients treated on AML SHG studies as reported by Wagner et al,9 who found that the IDH1 SNP was an independent predictor of inferior OS, and also correlated with inferior RFS in a univariate model. Their study was restricted to cytogenetically normal adult AML. Although FLT3/ITD was also more common in SNP-positive patients in their cytogenetically normal cohort (40% vs 31%), the difference was not as pronounced as in our study (90% vs 41%). In addition, there was a preponderance of older AML patients in our study, as SWOG-9031 and SWOG-9333, which were limited to patients > 55 years of age, accounted for 70% of the adults analyzed. The median age of adult AML patients in our study was 63 years, compared with only 47 years in the German study. As all survival outcomes diminish with increasing age, it is possible that outcome differences between SNP-positive and SNP-negative patients in our study were obscured by the disproportionate representation of older patients.
Mechanisms by which a “silent” SNP may alter gene function, such as the ability of a synonymous SNP to affect mRNA stability or expression, are currently under investigation. Another such mechanism is the alteration of the kinetics of protein translation, which may be affected by the frequency of usage of a particular codon. SNP rs11554137 represents a GGC to GGT transversion, resulting in the replacement of a common (GGC, 22.2 per thousand) codon by a rare (GGT, 10.8 per thousand) codon encoding glycine (frequencies obtained from the Codon Usage Database, http://www.kazusa.or.jp/codon/). The replacement of a commonly used codon with a rarely used one via a synonymous SNP may slow down the rate of protein translation, resulting in altered protein folding and, ultimately, decreased protein function. This has been demonstrated in the case of the C3435T SNP, which also encodes a GGC to GGT transversion, in the MDR1 gene.16 Although much remains to be elucidated regarding the biology of synonymous SNPs in AML, it is clear that certain SNPs in AML-associated genes are not clinically silent; synonymous SNPs represent another category of molecular alteration that should be investigated in AML for the purposes of disease classification and prognostication.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and families who consented to the use of biologic specimens in these trials and the AML Reference Laboratories of the COG and SWOG for providing diagnostic specimens.
This work was supported by the National Institutes of Health (grants K12 HD043376, K23 CA92405, CA18029, CA32102, CA114563, R01 CA114563-01, R21 CA102624, R21 CA10262-01, U10 CA032102-30, CA38926, CA20319, CA27057, and CA12213 and Children's Oncology Group Chair's grant NIH U10 CA98543), the Conquer Cancer Foundation of American Society of Clinical Oncology, and the Mary Claire Satterly Foundation.
National Institutes of Health
Authorship
Contribution: P.A.H. designed research, performed research, analyzed data, and wrote the manuscript; K.J.K. and T.A.A. served as senior statisticians, performed statistical analyses, and edited the manuscript; R.B.G. performed statistical analyses and edited the manuscript; K.L.M., J.K., R.Z., and R.E.R. performed research and edited the manuscript; S.C.R., B.A.H., V.O., C.A.H., J.L.F., A.S.G., S.H.P., J.E.A., J.E.G., G.H.R., C.L.W., I.D.B., J.P.R., F.R.A., and D.L.S. analyzed data and edited the manuscript; and S.M. designed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Phoenix A. Ho, Fred Hutchinson Cancer Research Center, D2-373, 1100 Fairview Avenue N, Seattle, WA 98109; e-mail: pho@fhcrc.org.