Abstract
Hydroxyurea is the standard therapy of chronic myelomonocytic leukemia (CMML) presenting with advanced myeloproliferative and/or myelodysplastic features. Response to hypomethylating agents has been reported in heterogeneous series of CMML. We conducted a phase 2 trial of decitabine (DAC) in 39 patients with advanced CMML defined according to a previous trial. Median number of DAC cycles was 10 (range, 1-24). Overall response rate was 38% with 4 complete responses (10%), 8 marrow responses (21%), and 3 stable diseases with hematologic improvement (8%). Eighteen patients (46%) demonstrated stable disease without hematologic improvement, and 6 (15%) progressed to acute leukemia. With a median follow-up of 23 months, overall survival was 48% at 2 years. Mutations in ASXL1, TET2, AML1, NRAS, KRAS, CBL, FLT3, and janus kinase 2 (JAK2) genes, and hypermethylation of the promoter of the tumor suppressor gene TIF1γ, did not predict response or survival on DAC therapy. Lower CJUN and CMYB gene expression levels independently predicted improved overall survival. This trial confirmed DAC efficacy in approximately 40% of CMML patients with advanced myeloproliferative or myelodysplastic features and suggested that CJUN and CMYB expression could be potential biomarkers in this setting. This trial is registered at EudraCT (eudract.ema.europa.eu) as #2008-000470-21 and www.clinicaltrials.gov as #NCT01098084.
Introduction
Chronic myelomonocytic leukemia (CMML), which is the most frequent myelodysplastic/myeloproliferative disorder,1 is characterized by the accumulation of monocytes and a variable proportion of immature dysplastic granulocytes in the PB and the BM.2 Clonal cytogenetic abnormalities are detected in ∼ 40%,3 and a copy-neutral uniparental disomy in leukemic cells of ∼ 50% of the patients.4 Mutations in ASXL1, TET2, and RUNX1 genes are detected in 25% to 50% of patients, RAS and CBL gene mutations in 10% to 25% of patients, and mutations in JAK2, FLT3, LNK, UTX, EZH2, IDH1, and IDH2 in < 10% of patients.5 Epigenetic changes could also play a role in the disease pathogenesis.6 For example, transcription-intermediary factor-1γ gene (TIF1γ), whose disruption in myeloid cells induces an age-dependent CMML phenotype in the mouse, is down-regulated in leukemic cells of ∼ 35% of CMML patients because of the gene promoter hypermethylation.7
The prognosis of this disease of the elderly is quite variable, with an approximately 2.5-year median survival.3 BM and PB blast percentages have major prognostic value and distinguish CMML-1, with < 10% BM and < 5% PB blasts, from CMML-2 with ≥ 10% BM and/or ≥ 5% PB blasts.1 Other well-documented prognostic factors include white blood cell (WBC) count, splenomegaly (SMG), extramedullary disease (EMD), cytopenias, and cytogenetic abnormalities.8-11 ASXL1 and EZH2 mutations negatively affect the disease outcome, whereas the prognostic influence of TET2 mutation is more controversial.12-16
The short survival of patients with poor prognostic factors is related to the limited availability of effective treatments.3 The only potentially curative therapeutic option is allogeneic stem cell transplantation, however excluded in most patients by age and comorbidities.17 In the only randomized trial conducted specifically in CMML, to our knowledge, hydroxyurea appeared more efficient than oral etoposide18 but still associated with short survival; whereas in other reports, the response rate of CMML to low-dose cytarabine,19 oral topotecan,20 and intensive chemotherapy21 remained low. The hypomethylating agents, 5-azacitidine (AZA) and 5-aza-2′-deoxycytidine (decitabine [DAC]) received Federal Drug Administration approval for treatment of myelodysplastic syndrome, including CMML, AZA being also approved in the European Union for CMML with marrow blasts > 10%. Pooled AZA and DAC published studies suggest an overall response rate of 39% to 45% and an overall survival (OS) benefit for responders.22-26 However, CMML patient populations included in those series were generally heterogeneous.27
We conducted a phase 2 trial of DAC in CMML patients with features of advanced disease defined with previously used criteria,18 exploring in particular biologic parameters predicting drug efficacy.
Methods
Clinical trial design
Between November 2008 and June 2009, the Groupe Francophone des Myélodysplasies (GFM) activated a prospective phase 2 clinical trial of DAC in advanced CMML. Inclusion criteria were as follows: (1) age 18 years or older; (2) diagnosis of CMML, based on World Health Organization 2008 criteria, except that patients with CMML features and 20% to 29% marrow blasts could also be included; (3) the following poor prognostic criteria, based on our previous experience18 (ie, if WBC < 13 g/L, having International Prognostic Scoring System intermediate-2 or high risk; if WBC ≥ 13 g/L, having 2 of the following criteria: marrow blasts ≥ 5%, hemoglobin (Hb) < 10g/dL, platelets < 100 g/L, abnormal cytogenetics, SMG > 5 cm below costal margin, and EMD); and (4) patients must have signed an informed consent form, in accordance with the Declaration of Helsinki. Patients with proliferative CMML treated with hydroxyurea were allowed to continue hydroxyurea to maintain WBC < 15 g/L.
The main objective of the trial was response to DAC. Secondary objectives included response duration, OS, and biologic parameters that could affect efficacy of DAC. The sample size (n = 41) was determined using Flemming single-stage design to detect an overall response rate > 35% with 80% power and a level of α = 5% (2-sided test). The trial was approved by the ethical committee of Dijon and registered at eudract.ema.europa.eu as EudraCT #2008–000470–21.
Patients received DAC (Janssen Cilag) 20 mg/m2 per day intravenously for 5 days every 28 days for at least 3 cycles. Responders were to continue treatment until progression. In case of grade 4 toxicity, according to National Cancer Institute toxicity criteria (Common Terminology Criteria for Adverse Events, Version 3.0), except for neutropenia and/or thrombocytopenia, cycles could be delayed up to 49 days. In case of response, subsequent cycles were to be repeated every 28 to 49 days. Complete blood count was monitored weekly during the treatment period. The use of granulocytic colony-stimulating factor and erythropoietin was allowed in case of febrile neutropenia or grade 3 or 4 neutropenia and/or red blood cell transfusion-dependent (RBC-TD) anemia. Grade 3 and 4 neutropenia was defined by an absolute neutrophil count between 0.5 and 1 g/L and < 0.5 g/L, respectively. Grade 3 and 4 thrombocytopenia was defined by a platelet count between 25 and 50 g/L and < 25 g/L, respectively. RBC transfusion thresholds were Hb level < 8 g/dL, or 9 to 10 g/dL in case of severe infection, underlying cardiac or pulmonary disease, or severe symptoms of anemia. Platelet transfusion thresholds were a platelet count < 20 g/L or higher in case of fever, rapid platelet decrease, mucositis, and concomitant coagulopathy. BM aspirate and blood samples were collected systematically for biologic studies before the first cycle, and blood samples were collected every 3 cycles until progression. Patients signed an independent informed consent for these associated studies.
Assessment of response
Responses, including complete remission (CR), partial response, marrow CR, and stable disease (SD) with hematologic improvement (HI) were defined according to International Working Group (IWG) 2006 criteria and evaluated every 3 cycles. Patients with SD without HI were allowed to continue DAC until progression. In accordance with IWG 2006 criteria, complete cytogenetic response was defined by the disappearance of all chromosomal abnormalities without appearance of new ones, and partial cytogenetic response by at least a 50% reduction of the number of mitoses with any chromosomal abnormality.
Cell sorting and flow cytometry
Mononuclear cells were selected from blood samples collected on ethylenediaminetetraacetic acid by Fycoll Hypaque, washed with ice-cold phosphate-buffered saline, and incubated at 4°C for 1 hour in 100 μL of phosphate-buffered saline containing 0.1% BSA with a combination of PE-conjugated mouse anti-CD14 and FITC-conjugated mouse anti-CD24 antibodies. Negative controls were obtained by substitution of the monoclonal antibody by allophycocyanin- and PE-conjugated mouse IgG1 control antibodies (BD Biosciences PharMingen). After phosphate-buffered saline wash, cells were fixed in 2% paraformaldehyde and analyzed with a LSRII (LSRII; BD Biosciences) by analyzing a total of 10 000 events per sample using FlowJo 9.2 software. CD14+CD24− cells were considered as monocytes and CD14−CD24+ cells as immature granulocytes.2 We also used the AutoMacs system (Miltenyi Biotec) to enrich CD14+ population from healthy donor and CMML PB samples as described.2
Gene mutation analysis
DNA was extracted from CD14+ sorted monocytes using commercial kits (Norgen Biotek) and submitted to whole genome amplification (GenomePlex, Sigma-Aldrich). FLT3 internal tandem duplications (FLT3-ITD) were detected by PCR and fragment analysis using a fluorescently labeled forward primer. PCR products were subjected to capillary electrophoresis on denaturing polyacrylamide gel and analyzed by the CEQ 8000 Genetic Analysis System (Beckman Coulter). Data were processed using Genetic Analysis System Software (Beckman Coulter). FLT3 tyrosine kinase domain mutations (FLT3D835/I836) were screened by PCR and EcoRV restriction enzyme digestion, with subsequent direct sequencing for samples showing an abnormal profile. The screening of NRAS and KRAS mutations was performed by melting curve analysis on the LightCycler 480 instrument (Roche Diagnostics). Data were analyzed using the LightCycler, Version 1.5 software (Roche Diagnostics). All suspected mutations were confirmed by direct sequencing. JAK2V617F mutation analysis was performed by TaqMan single nucleotide polymorphism genotyping assay using the JAK2 MutaScreen kit (Ipsogen). Real-time PCR assays were performed on an ABI PRISM 7900HT (Applied Biosystems). Screening for mutations in TET2 exon 3 to exon 11, c-CBL exons 8 and 9, RUNX1 exon 3 to exon 8, and ASXL1 exon 12 was performed by bidirectional direct sequencing, as described elsewhere.12,13,28 Seqscape (Applied Biosystems) was used to detect sequence variations. Gene abnormalities were numbered according to EMBL nucleotide sequence database. Patients with TET2 nonsense or frameshift variations were considered as TET2 mutated, whereas patients with no or missense variations were considered as wild-type as described.14 The most common ASXL1 variant c.1934dupG;p.Gly646TrpfsX12 was considered as a mutation.29 Previously annotated single nucleotide polymorphisms (http://www.hapmap.org) were not considered pathogenic.
Gene expression analysis
Gene expression analysis of healthy donor (n = 6) and patients (n = 36) sorted PB CD14+ cells was performed with Agilent 4 × 44 Human Gene Expression arrays (Agilent Technologies). After single color hybridization and array scanning, data were normalized by the quantile method. An unsupervised cluster was computed using Euclidian distance and Ward method for clusterisation., and validated by bootstrapping. Supervised analysis of genes differentially expressed between CMML and controls was performed using the moderated t test from LIMMA package. The expression profile of the set of significant differentially expressed genes was used to compute a supervised cluster using the same parameters as for unsupervised cluster. For validation of selected gene expression, RNA was isolated with Trizol (Invitrogen), reverse transcribed by SuperScriptII reverse transcriptase (Invitrogen) with random hexamers (Invitrogen), and real-time quantitative PCR was performed with AmpliTaq Gold polymerase in an Applied Biosystem 7500 thermocycler using the standard Power SyBr Green detection protocol as outlined by the manufacturer (Applied Biosystems). Briefly, 12 ng of total complementary DNA, 50nM of each primer, and 1× SyBr Green mix were used in a total volume of 20 μL. Primer sequences will be given on request. Real-time quantitative PCR expression levels of CJUN and CMYB were expressed relative to L32 control RNA expression, and the resulting arbitrary units were analyzed as continuous variables. Only for visual display of OS curves was gene expression dichotomized as “high ” versus “low, ” using the median expression of each gene as cut-off value.
Statistical analyses
The closing date of the study was February 1, 2011. Baseline characteristics were compared between responders (patients achieving CR, partial response, mCR, and SD with HI) and nonresponders (all other patients) by nonparametric tests (Fisher exact test for qualitative variables, Wilcoxon rank-sum test for quantitative variables). All variables with P < .05 in univariate analysis were included in a multivariate logistic regression for response. Censored endpoints were estimated by the nonparametric Kaplan-Meier method. OS was defined as the time between treatment onset and death or last contact. Regarding disease free survival, death and progression to acute myeloid leukemia (AML) were considered as competing events. Univariate analyses were performed with the log-rank test and Cox model for dichotomic and continuous variables, respectively, and multivariate analyses were performed by a Cox model, after accounting for interactions. The proportional hazard hypothesis was verified by visual display of the Schönfeld residuals, and a limited backward selection was performed to retain significant parameters with P < .05. All tests were 2-tailed. Statistical analysis was performed on STATA Version 10 and R Version 2.10.1 software packages.
Results
Baseline patient characteristics
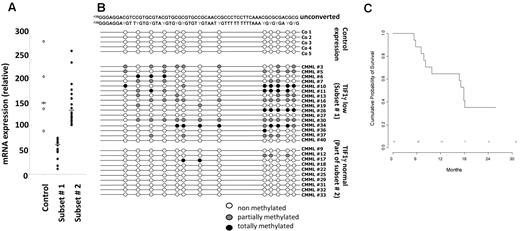
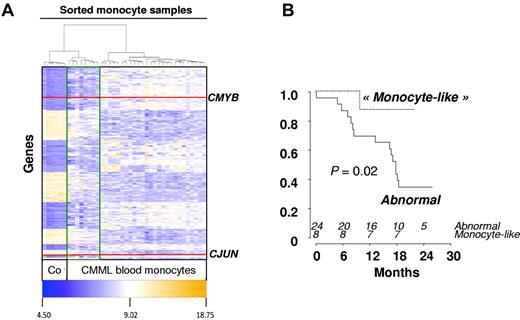
Forty-one patients from 16 GFM centers were included, of whom 39 completed at least 1 cycle and were considered as evaluable for response. The remaining 2 patients died of septic shock, before starting treatment, and during the first DAC cycle, respectively. Baseline characteristics of the 39 treated patients are summarized in Table 1. Of the 7 patients with WBC < 13 g/L, 5 had International Prognostic Scoring System intermediate-2 and 2 had International Prognostic Scoring System high. Eight patients had EMD, including skin infiltration in 5 cases and lymphadenopathy in 3 cases. Only one patient had > 20% BM blasts (29%) at inclusion, and this patient died of sepsis before onset of DAC. Therefore, restricting our analyses to World Health Organization-defined CMML (marrow blasts < 20%) did not affect the conclusions of this trial. Abnormal karyotype included trisomy 8 and monosomy 7 in 7 and 1 case, respectively. Twenty-two patients (56%) were RBC-TD at baseline. At least one gene mutation was identified in 90% of the patients. Genetic analysis of sorted CD14+ cells was available in 38 patients (Table 1). TIF1γ gene expression assessed by real-time quantitative PCR was decreased in 16 of 38 (50%) studied patients compared with control monocytes at baseline (Figure 1A). Promoter hypermethylation was identified in all the 16 patients with low TIF1γ while missing in the 10 patients with normal TIF1γ mRNA level studied (Figure 1B). Supervised clustering of gene expression analyzed in sorted monocytes of 32 CMML blood samples before DAC treatment identified a disease signature with differential expression of 1803 genes compared with healthy control monocytes (fold-change > 2, P < .05; Figure 2). The overexpression of 6 of these genes (ie, ERG, CMYB, arginase-1 [ARG1], metalloproteinase-9 [MMP9], CJUN, and α-defensin 1-3 [HNP1-3]) was confirmed by real-time quantitative PCR analysis of CMML compared with healthy control monocytes (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In addition, an unsupervised gene expression analysis identified a group of 8 CMML samples with a “normal monocyte-like” signature closer to control samples than to the 24 other CMML cases (supplemental Figure 2). A supervised analysis of expression profiles in these subgroups is highlighted in Figure 2. The 8 patients with a “normal monocyte-like” signature were significantly younger (median age 66 vs 74 years; P = .01) and less proliferative (median WBC 13.0 vs 22.8 g/L; P = .01) than other CMML patients but were otherwise comparable. Notably, there was no mutation pattern associated with this subgroup (supplemental Table 1).
Characteristics of patients at baseline
| Characteristic (n = 39) . | Value . |
|---|---|
| Median age, y (range) | 71 (54-88) |
| Sex, M/F, n | 30/9 |
| CMML-1/2, n | 17/22 |
| Abnormal karyotype, n (%) | 18 (46) |
| Splenomegaly > 5 cm, n (%) | 15 (38) |
| Extramedullary disease, n (%) | 8 (21) |
| Prior treatment with hydroxyurea, n (%) | 16 (41) |
| Median WBC, g/L (range) | 20.9 (4.1-147.3) |
| Median peripheral blood monocytes, g/L (range) | 4.8 (1.0-95.7) |
| Median Proportion of immature granulocytes, % (range) | 35 (0-84) |
| Median Hb level, g/dL (range) | 9.3 (1-14) |
| Median bone marrow blasts, % (range) | 10 (0-29) |
| Median platelet number, g/L (range) | 81 (12-560) |
| ASXL1, n mutated/n studied, % | 19/38 (50) |
| TET2, n mutated/n studied, % | 13/38 (34) |
| AML1, n mutated/n studied, % | 10/38 (26) |
| NRAS, n mutated/n studied, % | 6/38 (16) |
| KRAS, n mutated/n studied, % | 5/38 (13) |
| CBL, n mutated/n studied, % | 5/38 (13) |
| FLT3, n mutated/n studied, % | 3/38 (8) |
| JAK2, n mutated/n studied, % | 1/38 (3) |
| No mutation in the 8 screened genes, % | 4/38 (10) |
| Characteristic (n = 39) . | Value . |
|---|---|
| Median age, y (range) | 71 (54-88) |
| Sex, M/F, n | 30/9 |
| CMML-1/2, n | 17/22 |
| Abnormal karyotype, n (%) | 18 (46) |
| Splenomegaly > 5 cm, n (%) | 15 (38) |
| Extramedullary disease, n (%) | 8 (21) |
| Prior treatment with hydroxyurea, n (%) | 16 (41) |
| Median WBC, g/L (range) | 20.9 (4.1-147.3) |
| Median peripheral blood monocytes, g/L (range) | 4.8 (1.0-95.7) |
| Median Proportion of immature granulocytes, % (range) | 35 (0-84) |
| Median Hb level, g/dL (range) | 9.3 (1-14) |
| Median bone marrow blasts, % (range) | 10 (0-29) |
| Median platelet number, g/L (range) | 81 (12-560) |
| ASXL1, n mutated/n studied, % | 19/38 (50) |
| TET2, n mutated/n studied, % | 13/38 (34) |
| AML1, n mutated/n studied, % | 10/38 (26) |
| NRAS, n mutated/n studied, % | 6/38 (16) |
| KRAS, n mutated/n studied, % | 5/38 (13) |
| CBL, n mutated/n studied, % | 5/38 (13) |
| FLT3, n mutated/n studied, % | 3/38 (8) |
| JAK2, n mutated/n studied, % | 1/38 (3) |
| No mutation in the 8 screened genes, % | 4/38 (10) |
TFI1γ gene expression in CMML monocytes at inclusion. (A) Quantitative PCR analysis of TIF1γ gene expression allowed to define 2 subsets of patients with low (subset 1) or normal/high (subset 2) TIF1γ gene expression compared with healthy monocytes. Subset 1 indicates gene expression lower than mean ± 2 SD of that measured in normal samples; and Subset 2, others. (B) Methylation pattern of the TIF1γ promoter in a series of 5 healthy donors, the 16 patients with low TIF1γ mRNA level (subset 1), and 10 of the 16 patients with normal TIF1γ mRNA level. (C) Kaplan-Meier survival estimates comparing the 2 subsets of patients, based on TIF1γ mRNA level.
TFI1γ gene expression in CMML monocytes at inclusion. (A) Quantitative PCR analysis of TIF1γ gene expression allowed to define 2 subsets of patients with low (subset 1) or normal/high (subset 2) TIF1γ gene expression compared with healthy monocytes. Subset 1 indicates gene expression lower than mean ± 2 SD of that measured in normal samples; and Subset 2, others. (B) Methylation pattern of the TIF1γ promoter in a series of 5 healthy donors, the 16 patients with low TIF1γ mRNA level (subset 1), and 10 of the 16 patients with normal TIF1γ mRNA level. (C) Kaplan-Meier survival estimates comparing the 2 subsets of patients, based on TIF1γ mRNA level.
Gene expression in sorted CMML monocytes. (A) Heatmap of 1803 genes differentially expressed between healthy donor (n = 6) and CMML-sorted monocytes (n = 36; fold-change > 2; P < .05) results from a supervised analysis based on Euclidian distances. Red lines indicate CMYB and CJUN genes. Unsupervised analysis had previously identified 2 groups of CMML patients: those with a “normal monocyte-like” (highlighted with a green line, n = 8) and those with an “abnormal” (the others, n = 24) signature. (B) Kaplan-Meier survival estimates of OS according to the “normal monocyte-like” compared with the “abnormal” signature.
Gene expression in sorted CMML monocytes. (A) Heatmap of 1803 genes differentially expressed between healthy donor (n = 6) and CMML-sorted monocytes (n = 36; fold-change > 2; P < .05) results from a supervised analysis based on Euclidian distances. Red lines indicate CMYB and CJUN genes. Unsupervised analysis had previously identified 2 groups of CMML patients: those with a “normal monocyte-like” (highlighted with a green line, n = 8) and those with an “abnormal” (the others, n = 24) signature. (B) Kaplan-Meier survival estimates of OS according to the “normal monocyte-like” compared with the “abnormal” signature.
Response to DAC and survival
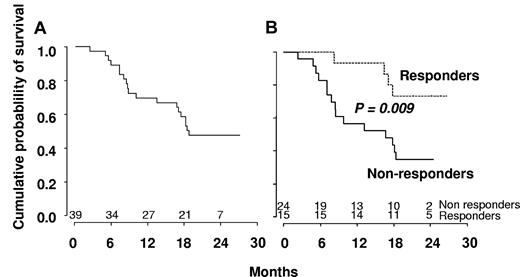
The median number of cycles received was 10 (range, 1-24). Fifteen patients (38%) responded, and their characteristics are summarized in Table 2. These responses included 4 CR (10%), 8 marrow CR (21%) associated in 3 cases with hematologic improvement (HI, including 1 patient with HI-platelets [HI-P] only and 2 patients with HI-P and HI-erythroid [HI-E]) and 3 SD with HI (8%, including 2 HI-P and 1 HI-P + HI-E). Eighteen patients (46%) demonstrated SD without HI, and 6 (15%) progressed to AML. Eight of the 22 RBC-TD patients (36%) became RBC transfusion independent (RBC-TI). Three of the 10 patients with platelet count < 50 g/L reached platelet count > 100 g/L. The 4 patients who reached CR had a normal karyotype at baseline. Six patients received erythropoietin (5 at inclusion and 1 after cycle 3), including 1 patient with marrow CR and HI for whom erythropoietin could be stopped after 9 cycles with Hb > 12 g/dL. A cytogenetic response was obtained in 4 patients (3 CR and 1 partial response), all with trisomy 8, one of whom reached SD without HI and the 3 other marrow CR. SMG disappeared in 6 of 15 (40%) and EMD in 6 of 8 (75%) patients. Hydroxyurea could be stopped in 12 of 16 patients (75%) receiving the drug at inclusion. Median number of cycles to achieve best response was 3 cycles. Eight of the 15 responders achieved best response after 3 cycles of DAC, 4 patients after 6 cycles, and 3 patients after 9 cycles. All responders continued DAC until relapse. Among responders, median peripheral monocyte count decreased from 4.8 to 0.3 g/L after 3 cycles of DAC. Seven of the 15 responders relapsed after 5.5 to 14 months, whereas 6 were still responding after 13 to 21 months (Table 2). One of the 2 remaining responders received allo-SCT after 3 months in CR and was censored for response duration at that point. This patient was still alive at 14.5 months from best response. The last responder with marrow CR died of infection 4 months from best response. Median disease-free survival was 18 months among responders. With a median follow-up of 23 months, OS was 48% at 2 years and median survival of all patients was 555 days (Figure 3A). At the closing date of the study, 20 patients were alive, including 11 of the responders. Eight of these responders were still receiving treatment.
Characteristics of the responding patients
| Patient no. . | WHO . | WBC, g/L . | Hb, g/dL . | Platelet, g/L . | BM blasts, % . | Karyotype . | SMG . | EMD . | Hydroxyurea . | Response . | Cytogenetic response . | Duration of response, months . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CMML-2 | 4.9 | 13.3 | 29 | 7 | 46, XY | Yes | Yes | Yes | CR | NA | 18* |
| 2 | CMML-2 | 22.8 | 8.9 | 34 | 19 | 46, XX | No | No | Yes | CR | NA | 21* |
| 3 | CMML-1 | 13.7 | 9.9 | 81 | 3 | 46, XX | Yes | Yes | Yes | CR | NA | 6 |
| 4 | CMML-2 | 26.9 | 9.7 | 51 | 19 | 46, XY,del20q | NA† | No | Yes | CRm | None | 15* |
| 5 | CMML-1 | 77.5 | 7.3 | 76 | 6 | 46, XY | Yes | No | No | HI | NA | 8 |
| 6 | CMML-2 | 6.6 | 9.3 | 52 | 13 | complex | No | No | No | CRm + HI | None | 9 |
| 7 | CMML-2 | 29.1 | 12.9 | 88 | 20 | 47, XY,+8 | No | Yes | No | CRm | CR | 14 |
| 8 | CMML-1 | 29.6 | 10.1 | 66 | 2 | 47, XX,del5q,+8 | No | No | No | HI | ND | 8 |
| 9 | CMML-2 | 21.5 | 6.7 | 36 | 12 | 46, XY | No | No | Yes | CRm | NA | 13* |
| 10 | CMML-2 | 13.7 | 10.9 | 238 | 20 | 47, XY,+8 | No | Yes | No | CRm | PR | 13 |
| 11 | CMML-2 | 16.3 | 9.5 | 61 | 15 | 47, XY,+Y | Yes | No | No | CRm + HI | None | 19* |
| 12 | CMML-2 | 22.3 | 8.3 | 67 | 18 | 47, XY,+8 | Yes | No | No | CRm | CR | 4 |
| 13 | CMML-2 | 14.1 | 8.3 | 22 | 16 | 46, XY | No | No | No | CRm + HI | NA | 5,5 |
| 14 | CMML-2 | 12.4 | 9.1 | 73 | 11 | 46, XY | No | No | Yes | HI | NA | 17.5* |
| 15 | CMML-2 | 8.8 | 6.5 | 118 | 7 | 46, XX | No | No | Yes | CR | NA | 14.5* |
| Patient no. . | WHO . | WBC, g/L . | Hb, g/dL . | Platelet, g/L . | BM blasts, % . | Karyotype . | SMG . | EMD . | Hydroxyurea . | Response . | Cytogenetic response . | Duration of response, months . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CMML-2 | 4.9 | 13.3 | 29 | 7 | 46, XY | Yes | Yes | Yes | CR | NA | 18* |
| 2 | CMML-2 | 22.8 | 8.9 | 34 | 19 | 46, XX | No | No | Yes | CR | NA | 21* |
| 3 | CMML-1 | 13.7 | 9.9 | 81 | 3 | 46, XX | Yes | Yes | Yes | CR | NA | 6 |
| 4 | CMML-2 | 26.9 | 9.7 | 51 | 19 | 46, XY,del20q | NA† | No | Yes | CRm | None | 15* |
| 5 | CMML-1 | 77.5 | 7.3 | 76 | 6 | 46, XY | Yes | No | No | HI | NA | 8 |
| 6 | CMML-2 | 6.6 | 9.3 | 52 | 13 | complex | No | No | No | CRm + HI | None | 9 |
| 7 | CMML-2 | 29.1 | 12.9 | 88 | 20 | 47, XY,+8 | No | Yes | No | CRm | CR | 14 |
| 8 | CMML-1 | 29.6 | 10.1 | 66 | 2 | 47, XX,del5q,+8 | No | No | No | HI | ND | 8 |
| 9 | CMML-2 | 21.5 | 6.7 | 36 | 12 | 46, XY | No | No | Yes | CRm | NA | 13* |
| 10 | CMML-2 | 13.7 | 10.9 | 238 | 20 | 47, XY,+8 | No | Yes | No | CRm | PR | 13 |
| 11 | CMML-2 | 16.3 | 9.5 | 61 | 15 | 47, XY,+Y | Yes | No | No | CRm + HI | None | 19* |
| 12 | CMML-2 | 22.3 | 8.3 | 67 | 18 | 47, XY,+8 | Yes | No | No | CRm | CR | 4 |
| 13 | CMML-2 | 14.1 | 8.3 | 22 | 16 | 46, XY | No | No | No | CRm + HI | NA | 5,5 |
| 14 | CMML-2 | 12.4 | 9.1 | 73 | 11 | 46, XY | No | No | Yes | HI | NA | 17.5* |
| 15 | CMML-2 | 8.8 | 6.5 | 118 | 7 | 46, XX | No | No | Yes | CR | NA | 14.5* |
WHO indicates World Health Organization; NA, not available; CRm, bone marrow response; and PR, partial response.
The response persisted, and the treatment was still ongoing at the time of analysis.
Splenectomy.
Kaplan-Meier survival estimates. (A) The whole population included in the trial. (B) Responders versus nonresponders to DAC according to IWG criteria.
Kaplan-Meier survival estimates. (A) The whole population included in the trial. (B) Responders versus nonresponders to DAC according to IWG criteria.
Tolerance of DAC
DAC was administered in an outpatient manner, dose reductions of DAC were not allowed, but interval between cycles could be delayed up to 49 days. The main side effects were cytopenias: 14 patients (36%) developed grade 3 or 4 thrombocytopenia and 13 (33%) experienced grade 3 or 4 neutropenia. Severe infection occurred in 8 patients (2 documented septicemia, 3 pneumonia, 1 febrile colitis, 1 intra-abdominal abscess, and 1 cellulites), febrile neutropenia in 5, and hemorrhgic complications related to thrombocytopenia in 6 (1 intra-alveolar hemorrhage, 3 epistaxis, 1 hemorrhagic cystitis, and 1 spleen hematoma). Cycles were delayed because of cytopenias in 22 (56%) patients. Other grade 3 or 4 events included atrial fibrillation (n = 1), respiratory distress (n = 2), deep venous thrombosis (n = 1), pulmonary embolism (n = 1), and cerebral stroke (n = 1). Twenty-four (62%) patients were hospitalized during DAC therapy. Twenty-one (52%) patients died of disease progression (n = 16), sepsis (n = 2), hemorrhage (n = 1), and respiratory failure (n = 1).
Prognostic factors of response
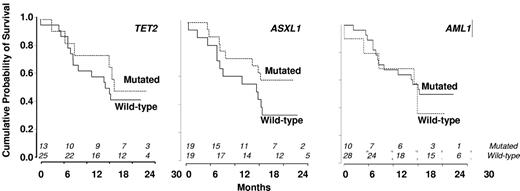
In univariate analysis, WBC, Hb and platelet levels, EMD, proportion of immature granulocytes identified by flow cytometry (Table 1), and cytogenetics did not affect response achievement. Excluding marrow CR (as CMML-1 with BM blasts < 5% could not be evaluated for that response), 17.6% CMML-1 and 18.8% CMML-2 responded (P = not significant). No gene mutation was significantly associated with IWG response. In particular, 7 of 13 patients with mutated TET2 (54%) achieved response, compared with 7 of 25 (28%) patients with WT TET2 (P = .17). In addition, 10 of 13 (77%) TET2 mutated compared with 11 of 25 (44%) TET2 WT patients achieved monocytes ≤ 1.0 g/L at 3 cycles (P = .09). TIF1γ promoter hypermethylation was not predictive of response (not shown). Supervised analysis of gene expression profiles failed to identify a signature associated with IWG response.
We then focused on CJUN expression whose level was assessed by real-time quantitative PCR and analyzed as a continuous variable. Although increased compared with control monocytes (Wilcoxon rank-sum test: P = .0006), CJUN expression was significantly lower in monocytes from responding compared with those from nonresponding patients (Wilcoxon rank-sum test: P = .008, supplemental Figure 3).
Disease-free and OS
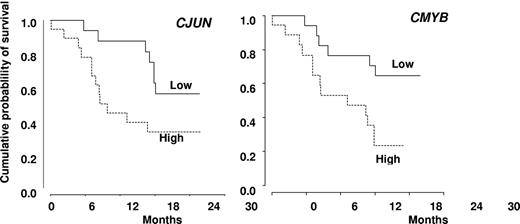
In univariate analysis, World Health Organization, abnormal cytogenetics, Hb, and platelet levels did not affect OS. Increased WBC was associated with shorter OS with a hazard ratio of 1.21 for each 10-g/L increment (95% CI, 1.07-1.36, P = .002, Cox model). Median survival was 503 days in nonresponders and not reached in patients who achieved hematologic response (log-rank test: P = .009, Figure 3B). Among the 18 patients with SD without HI after 3 or more cycles of DAC, 8 had a decrease in PB monocytes to < 1 g/L. Their 2-year survival estimate was 63% (95% CI, 29%-96%), compared with 22% (95% CI, 0%-49%) in the 10 with persistent monocytosis > 1.0 g/L (log-rank test: P = .03). TIF1γ promoter hypermethylation per se was not predictive of survival (Figure 1C). No gene mutation was found to significantly impact OS (Figure 4). Notably, 2-year OS was 50% (95% CI, 22%-78%) in patients with mutated TET2 and 44% (95% CI, 25%-64%) in those with WT TET2 (log-rank test: P = .63). The (A?) gene signature close to that of healthy donor monocytes identified in 8 patients was associated with better OS on DAC therapy, with a 2-year OS of 87.5% (95% CI, 64.6%-100%) compared with 34.8% (95% CI, 15.4%-54.3%) in other CMML patients (P = .02, Figure 2B). When CJUN and CMYB expression levels were expressed as continuous variables, higher CJUN (hazard ratio [HR] = 1.77; 95% CI, 1.13-2.76 for 10 000-arbitrary unit increments, P = .01) and higher CMYB (HR = 1.57; 95% CI, 1.16-2.12 for 50-arbitrary unit increments, P = .0033) expression were both associated with shorter OS (Figure 5). In a multivariate Cox model, including WBC, CJUN and CMYB expression levels as continuous variables, and “normal monocyte-like” signature (as category variable), higher CMYB expression (HR = 1.63; 95% CI, 1.13-2.33, P = .008) and higher CJUN expression (HR = 2.04; 95% CI, 1.27-3.28, P = .003) independently predicted inferior OS. WBC and “normal monocyte-like” signature had no independent impact in this model.
Kaplan-Meier survival estimates according to the absence or presence of mutations. (A) TET2 gene. (B) ASXL1 gene. (C) RUNX1 gene.
Kaplan-Meier survival estimates according to the absence or presence of mutations. (A) TET2 gene. (B) ASXL1 gene. (C) RUNX1 gene.
Kaplan-Meier survival estimates. According to the expression level of CJUN (A) and CMYB (B) genes, expressed as dichotomic variables using the median expression level of each gene in CMML samples as a cut-off value.
Kaplan-Meier survival estimates. According to the expression level of CJUN (A) and CMYB (B) genes, expressed as dichotomic variables using the median expression level of each gene in CMML samples as a cut-off value.
Discussion
The present trial indicates that DAC, administered using a 5-day intravenous schedule that proved superior to other schedules in myelodysplastic syndrome,22 induces a response in ∼ 40% of CMML with prospectively defined features of advanced myeloproliferative and/or myelodysplastic disease. It also identifies CJUN and CMYB gene expression levels as possible molecular predictors of response to DAC, whereas DAC efficacy was not affected by mutations in ASXL1, TET2, RUNX1, KRAS, NRAS, CBL, FLT3, and JAK2 genes. In addition, the methylation status of TIF1γ gene promoter did not predict response to DAC or survival, although we have shown recently that reexpression of TIF1γ in PB monocytes could be a biomarker of the response to DAC.7
Epigenetic therapy with low doses of hypomethylating agents has become the standard of care in high-risk myelodysplastic syndromes.25,30 The 38% overall response rate to DAC observed in the present series of high-risk CMML confirms results of pooled analysis of multicenter trials,24 and previous small size studies with DAC22,23 and AZA26 in this specific disease. In addition, the OS rate of 48% at 2 years in our cohort compares favorably with the median OS obtained in previous series.22-26 In particular, in our previous randomized trial, which included CMML patients based on exactly the same features of advanced disease, hydroxyurea give a significantly better survival than oral etoposide with a survival of 20 and 9 months, respectively.18 However, responses obtained with hydroxyurea were only minor responses that included normalization of WBC count and decrease of SMG and EMD. OS was significantly longer in responders, and myelosuppresssion with associated infections was the main side effects of a treatment feasible in an outpatient setting.
Until recently, the most common molecular abnormalities in CMML were NRAS and KRAS mutations observed in approximately one-third of the patients,31 and JAK2V617F was identified in a minority of them.32 In the last 2 years, additional gene mutations or deletions were identified, although none is specific of the disease.5 It remains unclear how these mutations account for the disease heterogeneity and impact patient survival. For example, we observed a trend for shorter survival of TET2 mutated CMML-1 patients,14 whereas others identified a better outcome for CMML patients who carried TET2 mutations.13 Here, we show, in a series of high-risk patients treated homogeneously, that TET2 mutations do not affect survival of CMML treated with DAC. Similarly, neither mutations in RUNX1 nor mutations in ASXL1, both associated with disease progression in other series,12,33 were predictive of response to DAC. This observation remained true when the most frequently reported mutation in ASXL1 (c.1934dupG;p.Gly646TrpfsX12), which has been challenged as a PCR artifact, was excluded.29 Interestingly, mutations in genes involved in the control of gene expression (TET2, ASXL1, and AML1) were frequently combined, whereas those affecting signaling pathways (NRAS, KRAS, CBL, JAK2, and FLT3) were mutually exclusive. Altogether, a mutation was identified in at least one of the 8 studied genes in 90% of the patients. Analysis of the mutant allele burden in CMML patients demonstrated that clinical response to DAC could be dissociated from the clearance of a mutated allele.34
Unique DNA methylation patterns have been associated with different cancers and participate in leukemogenesis (eg, by silencing tumor suppressor genes through promoter methylation and histone deacetylase recruitment). Methylation of the promoter of p15INK4B gene, whose deletion in myeloid cells generates a CMML-like phenotype in mice,33 is a common epigenetic abnormality in myeloid leukemias.35 Such a methylation was detected in approximately 15% of patients with advanced myelodysplastic syndrome or CMML and was at least temporally decreased on DAC therapy.22 Similarly, in 50% of the high-risk CMML entered in the present trial, we identified a methylation-driven decrease in the expression of TIF1γ gene whose deletion in myeloid cells generates also a CMML-like phenotype in mice.7 Nevertheless, the TIF1γ gene expression level was not predictive of the response to DAC. Gene expression was proposed to be an interesting tool in optimizing demethylating therapy in the clinic,36,37 and TIF1γ increased expression in monocytes could be one of the markers of DAC efficacy after 3 to 6 cycles.7 However, the mode of action of hypomethylating agents remains uncertain,38 and we cannot rule out that DAC selects the cells with a normal level of TIF1γ mRNA level rather than promoting gene promoter demethylation. In 25% of the studied CMML monocytes, gene expression appeared close to that observed in healthy donor monocytes, and this signature was associated with a significantly better survival on DAC therapy in univariate analysis. On the other hand, part of the gene signature that characterized the whole CMML population was confirmed by real-time quantitative PCR, and the prognostic value of the studied genes was explored. The expression of CJUN, which promotes aberrant monocyte differentiation in specific settings39 and cooperates with RAS for cell transformation,40 and the expression of CMYB, whose deregulation has been implicated in leukemias,41 were increased in CMML monocytes. Interestingly, in the setting of the limited sample size of this phase 2 trial, the more these genes were expressed, the lower the response rate or the shorter the survival on DAC therapy. In the CMML mouse model induced by deletion of p15INK4b gene, retrovirus integrations that provided cooperative genetic mutations resulting in myeloid leukemia commonly occurred near c-myb gene whose transcript was up-regulated in leukemia cells,24 suggesting a cooperation of increased CMYB gene with epigenetic extinction of suppressor genes in the disease progression.
DAC treatment was continued in responders until disease progression, a strategy that was shown to improve the quality of the response in previous studies.42,43 Nevertheless, some patients failed to respond to DAC, and those who relapsed after response had short survival. The mechanisms of resistance to DAC are poorly understood, although recent studies pointed to DNMT3B gene amplification44 and deficient incorporation into DNA (eg, through acquired mutations in the deoxycytidine kinase [DCK] gene).45 Improved efficacy of demethylating drugs could be obtained by combination with other drugs, such as histone deacetylase inhibitors46 and etanercept.47 In addition, drugs targeting overexpressed CMYB could potentially be useful as leukemic cells demonstrate greater sensitivity to decreased CMYB levels than their normal counterparts.48 Further studies are required to confirm the overexpression of CJUN and CMYB in CMML and establish their pathogenic role, and independent validation of their prognostic value in the setting of treatment with hypomethylating agents is warranted. Finally although CMML is rare, specific trials are important in this disease to identify drugs able to better control both its “dysplastic” aspects (leading to ineffective hematopoiesis and ultimately AML) and its “proliferative” features.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Margot Morabito, Cindy Racoeur, and Marion Ciudad who collected samples and sorted cells, Fathia Chermat (GFM administrative officer), and Vladimir Lazar (genomic platform at Institut Gustave Roussy). Janssen-Cilag (Paris, France) provided the drug and a scientific grant.
This work was supported by the Ligue Nationale Contre le Cancer (Label), Agence Nationale de la Recherche, Institut National du Cancer, Association Laurette Fuguain, and Fondation de France. The Unité Mixte de Recherche 1009 equipment was supported by the Association pour la Recherche sur le Cancer.
Authorship
Contribution: T.B., R.I., and L.A. analyzed the data and performed statistical analyses; A.R. and C.P. performed gene sequencing; B.d.R., F.D., K.L., K.B., N.V., A.T., C.R., B.R., B.J., A.V., I.L., and L.S. enrolled patients; G.M. and C.O. performed gene expression analysis; T.B., R.I., and C.G. collected data; N.D. performed biologic analyses; P.F., M.F., and E.S. designed the study; and E.S. with T.B., R.I., and P.F. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Eric Solary, Inserm UMR 1009, Institut Gustave Roussy, 114, Rue Edouard Vaillant, 94805 Villejuif cedex, France; e-mail: eric.solary@igr.fr.
References
Author notes
T.B., and R.I. contributed equally to this study as first authors.