Abstract
Nucleophosmin 1 (NPM1) is an oligomeric, nucleolar phosphoprotein that functions as a molecular chaperone for both proteins and nucleic acids. NPM1 is mutated in approximately one-third of patients with AML. The mutant NPM1c+ contains a 4-base insert that results in extra C-terminal residues encoding a nuclear export signal, which causes NPM1c+ to be localized in the cytoplasm. Here, we determined the effects of targeting NPM1 in cultured and primary AML cells. Treatment with siRNA to NPM1 induced p53 and p21, decreased the percentage of cells in S-phase of the cell cycle, as well as induced differentiation of the AML OCI-AML3 cells that express both NPMc+ and unmutated NPM1. Notably, knockdown of NPM1 by shRNA abolished lethal AML phenotype induced by OCI-AML3 cells in NOD/SCID mice. Knockdown of NPM1 also sensitized OCI-AML3 to all-trans retinoic acid (ATRA) and cytarabine. Inhibition of NPM1 oligomerization by NSC348884 induced apoptosis and sensitized OCI-AML3 and primary AML cells expressing NPM1c+ to ATRA. This effect was significantly less in AML cells coexpressing FLT3-ITD, or in AML or normal CD34+ progenitor cells expressing wild-type NPM1. Thus, attenuating levels or oligomerization of NPM1 selectively induces apoptosis and sensitizes NPM1c+ expressing AML cells to treatment with ATRA and cytarabine.
Introduction
Nucleophosmin (NPM1 or B23.1) is a ubiquitously expressed, nucleolar phosphoprotein that functions as a molecular chaperone, shuttling between the nucleolus and the cytoplasm.1-3 NPM1 plays multiple roles in cell growth and proliferation by participating in diverse biologic processes, including ribosome biogenesis and transport, centrosome duplication, DNA repair, transcriptional regulation and histone chaperoning.4-7 Intracellular NPM1 is predominantly oligomeric and binds to other proteins, including the tumor suppressor proteins p14ARF and p53.1,8-10 Multifunctional characteristic of NPM1 appears to be dictated not only by its sub-cellular localization and its binding partners, but is also influenced by the various post translational modifications in NPM1, including acetylation, phosphorylation, poly-ubiquitination and sumoylation.11-14 Wild-type (WT) NPM1 contains distinct structural domains that account for its ability to act as a multifunctional protein.1,15 NPM1 has an N-terminal conserved, hydrophobic, oligomerization domain (residues, 1-110), which is common to all isoforms of NPM1 and critical for its chaperone activity.1-3 Recently, NSC348884 was identified as a small molecule inhibitor that disrupts NPM1 dimer/oligomer formation, inducing apoptosis of cancer cells.16 Oncogenic fusion proteins created by chromosomal translocation involving NPM1 gene, or mutations in NPM1 are observed in leukemia and lymphoma.17 Notably, NPM-ALK fusion protein is found in CD30+ anaplastic large-cell lymphoma,18 while leukemia related NPM1 fusion proteins include NPM-MLF1 and NPM-RARα.17,19,20 These chimeric fusion proteins contain the N-terminal NPM1 oligomerization domain and a C-terminal fragment of the other protein.17 NPM1 gene is also mutated in one third of adult acute myeloid leukemia (AML), especially those with the normal karyotype.21 NPM1 mutations are heterozygous and, in the majority, localized to exon 12 of the gene.21,22 Approximately 50 different types of mutations have been found, all creating the cytoplasm-dislocated mutant (Mt) NPM1 (NPM1c+) protein.21,22 The most common is the type-A mutation, accounting for 75% of cases, which consists of TCTG tetra-nucleotide tandem duplication at position 956-959 of the NPM1 coding sequence.22-24 This mutation causes the loss of tryptophans 288 and 290 (or 290 alone) from the carboxy-terminus and the creation of an additional leucine-rich nuclear export motif in the NPM1 protein, which causes the aberrant cytoplasmic dislocation of NPM1c+.22-24 Knockout of NPM1 in mice is embryonically lethal because of defects in hematopoiesis, and mice hypomorphic for NPM1 develop a condition similar to myelodysplastic syndrome.24 Overexpression of cytoplasmic Mt NPM1 in the mouse model conferred a proliferative advantage in the myeloid lineage.25 In addition, expression of the cytoplasmic Mt NPM1 in zebrafish resulted in the expansion of hematopoietic cells including erythromyeloid progenitor cells.26
AML with Mt NPM1 shows distinctive biologic and clinical features. These include unique gene expression profiles, high association with FLT3-ITDs, low CD34 expression and multi-lineage involvement.2,22 Generally, AML with Mt NPM1 exhibit good response to induction therapy.21,22 However, the overall outcome is poorer when FLT3-ITD is coexpressed with Mt NPM1.21,22 AMLs with Mt NPM1 overexpress HOXA9 and Meis1, which are important regulators of hematopoiesis expressed early in embryogenesis, and are generally down regulated during differentiation.27-29 Deregulated coexpression of HOXA9 and Meis1 has been implicated in leukemogenesis.28,29 Other biologic activities of Mt NPM1 also appear to contribute to the leukemia biology.30,31 In the present studies, using cultured AML cells, we show that down regulation of NPM1 by siRNA induces p53, p21 and C/EBPα, attenuates HOXA9 and Meis1 mRNA and protein expression, as well as induces morphologic differentiation and diminishes clonogenic survival of AML cells. Knockdown of NPM1 dramatically inhibited leukemia initiation by cultured AML cells in NOD/SCID mice. In addition, importantly, treatment with NPM1 siRNA significantly sensitized Mt NPM1c+ expressing AML cells to all-trans retinoic acid (ATRA) and Ara-C. Our studies also demonstrate that by disrupting oligomerization of NPM1, treatment with NSC348884 induced apoptosis of cultured and primary AML with Mt NPM1 but not those AML cells that coexpressed FLT3-ITD. NSC348884 also sensitized AML cells with Mt NPM1 to apoptosis induced by ATRA.
Methods
Cell culture
HL-60 and OCI-AML3 cells were obtained and cultured as previously described.32,33 U937 cells were grown in RPMI-1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin. OCI-AML2 cells (known to express WT p53 and WT NPM1) were kindly provided by Mark Minden (Ontario Cancer Institute/Princess Margaret Hospital, Ontario, Canada) and cultured in α-MEM with 20% heat inactivated FBS. Cells were passaged 2-3 times per week. Logarithmically growing cells were used for all experiments. OCI-AML3 cells have been previously demonstrated to express NPM1c+.34
Reagents and antibodies
Nucleofector kits were purchased from Lonza. ATRA, Ara-C, monoclonal anti-NPM1 (detects both Mt NPM1 and WT NPM1) and anti–β-actin antibodies were purchased from Sigma-Aldrich. HOXA9 and Meis1 antibodies were purchased from Upstate. Antibody for p21 was purchased from Thermo Scientific Lab Vision. Monoclonal EZH2 was purchased from Cell Signaling Technology. Anti-Fibrillarin antibody was purchased from Abcam. Anti-PARP, anti-C/EBP-α, and anti-FLT3 were purchased from Santa Cruz Biotechnology. Monoclonal anti-p53, monoclonal FITC-conjugated anti-CD11b and FITC-conjugated isotype control antibody and FITC-conjugated annexin V were obtained from BD Biosciences. A polyclonal antibody to mut-NPM1 was raised in the current study by immunizing rabbits with the peptide sequence H2N-CLAVEEVSLRK-COOH conjugated to keyhole limpet hemocyanin (KLH) peptide (Sigma-Genosys). Small molecule inhibitor NSC348884 was purchased from Axon Medchem. All the oligonucleotide primers and NPM1 siRNA duplexes were purchased from Integrated DNA Technologies.
Primary AML blasts
Primary AML samples and normal CD34+ mononuclear cells were obtained with informed consent in accordance with the Declaration of Helsinki. Peripheral blood or bone marrow aspirate samples were collected and mononuclear cells were separated using Lymphoprep, washed once with complete RPMI 1640 medium, before their use in experiments.32,33 Banked, de-linked, and de-identified donor peripheral blood CD34+ mononuclear cells procured for recipients who had since died were purified by immunomagnetic beads conjugated with anti-CD34 antibody (StemCell Technologies) before use in cell viability assays.32,35
RNA isolation, RT-PCR, and quantitative PCR
Mutant NPM1 (mut-NPM1) was detected in cultured and primary AML cells using primers listed in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article) by qPCR. Total RNA was isolated using RNAqueous Kit (Ambion) and reverse transcribed with High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems) according to manufacturer's instructions. The resulting cDNAs were used for semi-quantitative detection of wild-type NPM1 or mutant NPM1. The amplified PCR products were separated on a 2% agarose gel and imaged using a UV transilluminator (Kodak Gel Logic 200 Imaging System). Mutant and wild type NPM1 were also detected by real time quantitative PCR. To screen for NPM1 mutations in primary AML cells from patients, quantitative PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) and a StepOnePlus Real Time PCR system (Applied Biosystems). Mut-NPM1 mRNA expression was normalized to GAPDH and is represented as arbitrary units. OCI-AML3 cDNA was used as a positive control and HL-60 was used as a negative control for the amplification.
RNA interference
For NPM1 knockdown by siRNA, a duplex composed of sequences 5′-GAAGCAGAGGCAAUGAAUUACGA-3′ and 5′-CUUCGUAAUUCAUUGCCUCUGCUUCAA-3‘ was used. Oligos were annealed by boiling for 5 minutes and slowly cooling to room temperature. HL-60 and OCI-AML3 cells were nucleofected with 100nM siRNA duplex using Nucleofector Kit V (Lonza) according to the manufacturer's protocol. Cells were incubated for 24-96 hours after nucleofection before performing experiments. To obtain knockdown of NPM1 in OCI-AML2 cells or in OCI-AML3 cells for in vivo studies, nontargeting (sh-NT) and NPM1-targeting (sh-NPM1) lentiviral shRNAs were obtained from Sigma-Aldrich. Lentiviral particles were generated according to the manufacturer's protocol. Cells were transduced with the lentivirus particles for 48 hours and selected with puromycin.
Cell fractionation and immunoblot analyses
After the designated treatments, cells were harvested and nuclear and cytoplasmic fractions were prepared using NE-PER extraction kit (Pierce), according to the manufacturer's protocol. Fifty micrograms of protein from whole cell lysates or sub-cellular fractions were used for immunoblot analyses of NPM1, mut-NPM1, HOXA9, Meis1, and FLT3 as previously described.32,33,35 Immunoblot analyses were performed at least twice and horizontal scanning densitometry was performed on representative blots as previously described.32,33,35 For native PAGE, cell lysates were not heat denatured and electrophoresis was performed in the absence of SDS.
Cell-cycle analyses
Cell viability and apoptosis
After designated treatments, the percentages of nonviable cells were determined by trypan blue dye uptake in a hemocytometer and are reported as a percentage of untreated cells. For flow cytometric determination of apoptotic cells, cells were stained with annexin V (BD Biosciences Pharmingen) and propidium iodide, as previously described.32,33,35 Alternatively, cells were cytospun onto glass slides and evaluated for morphologic features of apoptosis (ie, cell shrinking, membrane blebbing, fragmentation of nuclei, and formation of apoptotic bodies) as previously described.32,33,35
Differentiation of AML cells
For evaluating AML cell differentiation, cells were stained with mouse anti-CD11b (clone ICRF44) conjugated to Alexa Fluor 488 (BD Biosciences Pharmingen). For 2-color analysis, cells were also stained with mouse NPM1 antibody (Sigma-Aldrich) conjugated to DyLight 647 (Pierce). After staining, cells were washed and analyzed by flow cytometry. For assessment of morphologic differentiation, HL-60 and OCI-AML3 cells were treated as indicated for 96 hours. After this, cells were cytospun and stained with Wright-Giemsa stain. Cellular morphology was observed by light microscopy and percent differentiation was estimated by counting at least 200 cells per condition according to previously defined criteria for differentiation.32,33
Colony-forming assay
After the indicated treatments, cells were harvested, washed with 1× PBS, counted on a hemocytometer, and 500 cells per condition were plated in complete Methocult medium (StemCell Technologies) and cultured for 7 to 10 days at 37°C in a humidified, 5% CO2 incubator. Colony growth was measured as a percentage of control colony growth as previously described.33
Assessment of DNA fragmentation
For assessment of DNA fragmentation, control and NSC348884-treated cells were pelleted and lysed in a buffer containing 10mM Tris·HCl, pH 7.4, 10mM NaCl, 10mM EDTA, 100 μg/mL proteinase K, and 0.5% SDS and incubated for 1 hour at 50°C. Briefly, DNA was phenol extracted; ethanol precipitated and treated with Rnase A. Resulting DNA was resolved on 1.5% agarose gel and visualized by a UV transilluminator.36
Lentiviral transfection of FLT3-ITD into OCI-AML3 cells
Human full-length FLT3-ITD cDNA was isolated from MV4-11 cells by PCR, sub-cloned into pLVX-puro vector (Clontech) using the BamHI and XbaI restriction sites. The construct was validated by DNA sequencing. The production and harvest of high-titer lentivirus was performed using lenti-X HT packaging system (Clontech) as recommended by the manufacturer. OCI-AML3 cells were transiently infected with empty vector or pLVX-FLT3-ITD lentivirus particles. Transfected cells were treated with NSC348884 and/or ATRA.
In vivo model of AML
Female nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice were sub-lethally irradiated (250 rads) using a cesium source. The following day, 10 million OCI-AML3 cells transduced with a nontargeted shRNA or NPM1 shRNA were injected into the tail vein of 5 mice, and the mice were monitored daily for evidence of leukemia. Mice were humanely killed when they displayed hunched appearance and/or had lost > 15% of their body weight or exhibited hind limb paralysis. The spleen from the killed mice was measured to determine whether the mice had evidence of splenic enlargement. The survival of mice from the tail vein model is represented with a Kaplan-Meier survival plot. NOD/SCID mice, human OCI-AML3 cell xenograft studies were sanctioned by protocol approved by the Kansas University Medical Center Institutional Animal Care and Use Committee.
Statistical analyses
Significant differences between values obtained in a population of leukemia cells treated with different experimental conditions were determined using the Student t test. P values < .05 were assigned significance.
Results
Effects of WT and Mt NPM1 knockdown on the cell cycle progression and colony growth of AML cells
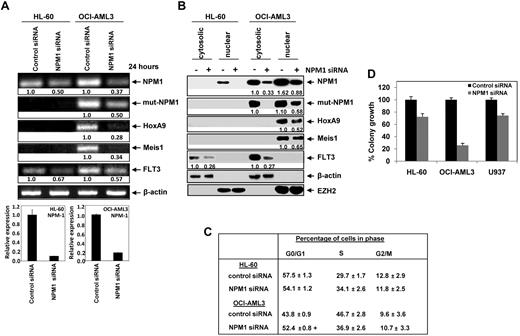
We first confirmed that, while HL-60 cells express WT NPM1 mRNA, as well as express WT NPM1 protein located mostly in the nucleus and nucleoli (supplemental Figures 1A-C). In contrast, OCI-AML3 cells express Mt NPM1 mRNA (supplemental Figure 1A,B), as well as express Mt NPM1 protein. The latter was aberrantly located in the cytoplasm and in the nucleus, as detected by immunoblot analysis of nucleus and cytoplasmic fractions (supplemental Figure 1C), as well as by using immunofluorescent microscopy after staining of the cells with the antibody specific for the Mt NPM1 (supplemental Figure 1D). Next, we transfected OCI-AML3 and HL-60 cells with the siRNA to NPM1, or with control siRNA. As shown in Figure 1A, exposure to NPM1 siRNA depleted the expression of both the Mt and WT NPM1 mRNA in OCI-AML3, and attenuated WT NPM1 mRNA in HL-60 cells, as determined by qPCR analyses. NPM1 mRNA knockdown was also associated with the depletion of HOXA9 and Meis1 mRNA in OCI-AML3 cells (Figure 1A). Consistent with previous reports that have highlighted Meis1-mediated induction of FLT3, down regulation of Meis1 mRNA after NPM1 knockdown was also associated with a significant decline in the FLT3 mRNA levels in OCI-AML3 cells.37 In contrast, although HL60 cells did not express appreciable levels of the HOXA9 and Meis1 mRNA levels, NPM1 knockdown was also associated with a significant decline in the FLT3 mRNA levels in HL-60 cells (Figure 1A). After carrying out the nuclear and cytoplasmic sub-cellular fractionation of HL-60 and OCI-AML3, immunoblot analysis of NPM1 was performed using a polyclonal antibody (raised against Mt-NPM1). In OCI-AML3 cells, Mt NPM1 was depleted by treatment with siRNA in both the nuclear and cytoplasmic fractions. Immunoblot analysis of NPM1 was also performed using a monoclonal antibody that detects both the Mt and WT NPM1. Again, treatment with siRNA to NPM1 led to depletion of Mt and WT NPM1 in both the cytoplasm and nucleus of OCI-AML3 cells, but depleted WT NPM1 in the nucleus of HL-60 cells (Figure 1B). The expressions of β-actin (cytosolic) and EZH2 (nuclear) served as the loading controls, and helped in assessing the purity of the subcellular fractions. The depletion of WT and Mt NPM1 was associated with attenuation of HOXA9 and Meis1 proteins in the nucleus of OCI-AML3 cells, and was associated with attenuation of FLT3 levels (Figure 1B). This is consistent with reports indicating that Meis1 induces FLT3 expression.37 NPM1 siRNA treatment also lowered FLT3 levels in HL60 cells, apparently by a Meis1 independent mechanism (Figure 1B). We also determined the effects of the NPM1 siRNA, and of the protein-expression changes that ensue, on the cell cycle status and colony growth of OCI-AML and HL60 cells. Figure 1C demonstrates that siRNA-mediated depletion of Mt and WT NPM1 in OCI-AML3 cells was associated with a significant increase in the percentage of cells in G1 and decrease in the percentage of cells in the S phase of the cell cycle. This was also associated with marked inhibition of the colony growth of OCI-AML3 cells in the semi-solid medium (P < .004; Figure 1D). While it had no significant effect on the cell cycle status, NPM1 siRNA-mediated attenuation of WT NPM1 also inhibited the colony growth of HL-60 and U937 cells (Figure 1C,D). NPM1 siRNA treatment for 48 hours induced apoptosis in 4.5 ± 0.7% of HL-60 and 4.0 ± 0.4% of U937 cells, compared with 8% apoptosis in OCI-AML3 cells.
Depletion of NPM1 down-regulates the expression of leukemogenic markers induces G0/G1 accumulation and markedly reduces clonogenic survival of AML cells. (A) Top panel, HL-60 and OCI-AML3 cells were transfected with control siRNA or NPM1 siRNA for 24 hours. After 24 hours, total RNA was extracted and semi-quantitative RT-PCR was performed for WT-NPM1, Mt-NPM1, HOXA9, Meis1, and FLT3. The levels of β-actin mRNA served as the loading control. Bottom panel, quantitative PCR reactions were also performed with SYBR Green to assess NPM1 depletion in the siRNA transfected cells. Relative expression of NPM-1 was normalized to GAPDH. (B) HL-60 and OCI-AML3 cells were transfected with control (−) or NPM1 (+) siRNA and incubated for 48 hours. At the end of incubation, nuclear and cytosolic fractions were isolated and immunoblot analyses were performed for NPM1, HOXA9, Meis1, and FLT3. Expression levels of β-actin and EZH2 served as the loading and fraction controls for the cytosolic and nuclear extracts, respectively. (C) HL-60 and OCI-AML3 cells were transfected with control or NPM1-siRNA and incubated for 96 hours. Then, cells were fixed and stained with propidium iodide and cell cycle status was determined by flow cytometry. Values represent the mean of 3 independent experiments + SEM. (+) indicates G0/G1 values significantly different (P < .05) compared with control siRNA transfected cells. (D) HL-60, OCI-AML3, and U937 cells were transfected with control or NPM1 siRNA and incubated for 48 hours. Then, colony growth in semisolid media was assessed after 8 days. Columns represent the mean of 3 independent experiments; bars represent the SEM.
Depletion of NPM1 down-regulates the expression of leukemogenic markers induces G0/G1 accumulation and markedly reduces clonogenic survival of AML cells. (A) Top panel, HL-60 and OCI-AML3 cells were transfected with control siRNA or NPM1 siRNA for 24 hours. After 24 hours, total RNA was extracted and semi-quantitative RT-PCR was performed for WT-NPM1, Mt-NPM1, HOXA9, Meis1, and FLT3. The levels of β-actin mRNA served as the loading control. Bottom panel, quantitative PCR reactions were also performed with SYBR Green to assess NPM1 depletion in the siRNA transfected cells. Relative expression of NPM-1 was normalized to GAPDH. (B) HL-60 and OCI-AML3 cells were transfected with control (−) or NPM1 (+) siRNA and incubated for 48 hours. At the end of incubation, nuclear and cytosolic fractions were isolated and immunoblot analyses were performed for NPM1, HOXA9, Meis1, and FLT3. Expression levels of β-actin and EZH2 served as the loading and fraction controls for the cytosolic and nuclear extracts, respectively. (C) HL-60 and OCI-AML3 cells were transfected with control or NPM1-siRNA and incubated for 96 hours. Then, cells were fixed and stained with propidium iodide and cell cycle status was determined by flow cytometry. Values represent the mean of 3 independent experiments + SEM. (+) indicates G0/G1 values significantly different (P < .05) compared with control siRNA transfected cells. (D) HL-60, OCI-AML3, and U937 cells were transfected with control or NPM1 siRNA and incubated for 48 hours. Then, colony growth in semisolid media was assessed after 8 days. Columns represent the mean of 3 independent experiments; bars represent the SEM.
Knockdown of NPM1 induces differentiation and sensitizes AML cells expressing Mt NPM1 to ATRA-induced differentiation
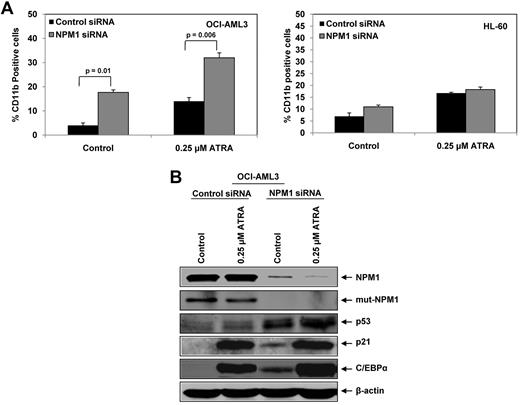
Next we determined the effects of NPM1 depletion on the induction of differentiation in HL-60 and OCI-AML3 cells. NPM1 knockdown in OCI-AML3 cells by siRNA was associated with induction of p53 and the differentiation markers, p21 and C/EBPα (Figure 2A). This was not observed in HL-60, suggesting that WT NPM1 expression does not block differentiation of AML cells. In addition, we determined the effects of NPM1 knockdown in OCI-AML2 cells, which are known to express both WT p53 and WT NPM1. As compared with control shRNA, while NPM1 shRNA caused significant depletion of NPM1 levels, it did not affect the levels of CEBP-α, p53, or p21 in OCI-AML2 cells (Figure 2A). Knockdown of NPM1 also induced the morphologic features of granulocytic/monocytic differentiation in 50% of OCI-AML3 cells (P < .004). In contrast, NPM1 knockdown induced minimal morphologic differentiation in HL-60 cells (∼ 12%; Figures 2B-D). As a further confirmation, 96 hours post NPM1 siRNA transfection and knockdown of NPM1, 2-color flow cytometry for NPM1 and CD11b showed a significant increase in the number of CD11b-positive OCI-AML3 cells that also showed a decrease in NPM1 staining (P < .0002). Taken together, these findings demonstrate that knockdown of NPM1 can induce the features of granulocytic/monocytic differentiation in AML cells with Mt NPM1. However, this was not observed in similarly treated HL-60 cells. The inhibitory effect of NPM1 siRNA on the colony growth of AML cells with WT NPM1 (eg, HL-60), observed after 10 days of culture in semisolid medium, may however be because of reduction in FLT-3 levels and slight increase in granulocytic/monocytic differentiation represented by increase in CD11b expression. NPM1 knockdown did not induce CD11b or morphologic differentiation in OCI-AML2 cells (data not shown).
Depletion of NPM1 induces differentiation of mutant NPM1 expressing AML cells and abolishes OCI-AML3 cell-induced AML in NOD/SCID mice. (A) HL-60, OCI-AML3, and OCI-AML2 cells were transfected with control or NPM1 siRNA and incubated for 72 hours. Then, immunoblot analyses were performed for NPM1, C/EBPα, p53, and p21 on the total cell lysates. The expression of β-actin in the lysates served as the loading control. (B) Top panel, representative light microscopic images of HL-60 and OCI-AML3 cells transfected with control or NPM1 siRNA for 96 hours. Bottom panel, 96 hours post transfection, control and NPM1 siRNA transfected HL-60 and OCI-AML3 cells were cytospun onto glass slides, Wright-Giemsa stained and the percentage of differentiated cells was determined by light microscopy. Values represent the mean of 3 independent experiments ± SEM. (C) OCI-AML3 cells were transfected with control or NPM1 siRNA for 96 hours. At the end of treatment, cells were fixed, permeabilized and stained with DyLight 647-conjugated anti-NPM1 antibody followed by flow cytometry. For determination of CD11b expression, cells were washed with 1X PBS and stained with anti-CD11b antibody. Representative flow histograms of NPM1 and CD11b expression levels are presented. (D) OCI-AML3 cells were transfected with control or NPM1 siRNA for 96 hours. Then, cells were washed and stained with CD11b antibody and the percentages of CD11b positive cells were determined by flow cytometry. Values represent the mean percentage of CD11b positive cells from 3 independent experiments ± SEM. (E) Female NOD/SCID mice were irradiated and then injected in the lateral tail vein with OCI-AML3 sh-NT or OCI-AML3 sh-NPM1 (n = 5 mice per group). The mice were monitored daily for symptoms of leukemia. Survival of the mice in OCI-AML3 sh-NT and OCI-AML3 sh-NPM1 is represented by Kaplan-Meier plot. Median survival of the OCI-AML3 sh-NT leukemia bearing mice was 16 days. (F) When the mice were humanely killed, the spleens were collected and measured. Top panel, the box plot displays the range of spleen size from OCI-AML3 sh-NT and sh-NPM1 leukemia bearing mice. The median spleen size for OCI-AML3 sh-NT mice was 1.6 cm compared with 1.2 cm in the OCI-AML3 sh-NPM1 mice. Bottom panel, representative images of the spleens from 2 OCI-AML3 sh-NT and sh-NPM1 leukemia bearing mice are shown.
Depletion of NPM1 induces differentiation of mutant NPM1 expressing AML cells and abolishes OCI-AML3 cell-induced AML in NOD/SCID mice. (A) HL-60, OCI-AML3, and OCI-AML2 cells were transfected with control or NPM1 siRNA and incubated for 72 hours. Then, immunoblot analyses were performed for NPM1, C/EBPα, p53, and p21 on the total cell lysates. The expression of β-actin in the lysates served as the loading control. (B) Top panel, representative light microscopic images of HL-60 and OCI-AML3 cells transfected with control or NPM1 siRNA for 96 hours. Bottom panel, 96 hours post transfection, control and NPM1 siRNA transfected HL-60 and OCI-AML3 cells were cytospun onto glass slides, Wright-Giemsa stained and the percentage of differentiated cells was determined by light microscopy. Values represent the mean of 3 independent experiments ± SEM. (C) OCI-AML3 cells were transfected with control or NPM1 siRNA for 96 hours. At the end of treatment, cells were fixed, permeabilized and stained with DyLight 647-conjugated anti-NPM1 antibody followed by flow cytometry. For determination of CD11b expression, cells were washed with 1X PBS and stained with anti-CD11b antibody. Representative flow histograms of NPM1 and CD11b expression levels are presented. (D) OCI-AML3 cells were transfected with control or NPM1 siRNA for 96 hours. Then, cells were washed and stained with CD11b antibody and the percentages of CD11b positive cells were determined by flow cytometry. Values represent the mean percentage of CD11b positive cells from 3 independent experiments ± SEM. (E) Female NOD/SCID mice were irradiated and then injected in the lateral tail vein with OCI-AML3 sh-NT or OCI-AML3 sh-NPM1 (n = 5 mice per group). The mice were monitored daily for symptoms of leukemia. Survival of the mice in OCI-AML3 sh-NT and OCI-AML3 sh-NPM1 is represented by Kaplan-Meier plot. Median survival of the OCI-AML3 sh-NT leukemia bearing mice was 16 days. (F) When the mice were humanely killed, the spleens were collected and measured. Top panel, the box plot displays the range of spleen size from OCI-AML3 sh-NT and sh-NPM1 leukemia bearing mice. The median spleen size for OCI-AML3 sh-NT mice was 1.6 cm compared with 1.2 cm in the OCI-AML3 sh-NPM1 mice. Bottom panel, representative images of the spleens from 2 OCI-AML3 sh-NT and sh-NPM1 leukemia bearing mice are shown.
NPM1 knockdown abolishes OCI-AML3 cell-induced AML in NOD/SCID mice
We next determined the effects of NPM1 knockdown on the in vivo efficacy of OCI-AML3 cells to induce lethal AML, after injection of the cells into the tail vein of the recipient NOD/SCID mice. This was compared with the in vivo effects of OCI-AML3 cells transduced with a nontargeted (NT) shRNA (control mice). Notably, Kaplan Meier plot in Figure 2E demonstrates that mice bearing OCI-AML3 cells transduced with NPM1 shRNA, showing knockdown of NPM1 (Figure 2E inset), demonstrated dramatically improved survival, compared with the control mice (P < .01). Mice injected with OCI-AML3 cells transduced with NPM1 shRNA were humanely killed on day 25, after all the control mice had developed lethal AML. As shown in the box plot in Figure 2F, mice injected with OCI-AML3 cells transduced with NPM1 shRNA demonstrated significantly less spleen enlargement (median spleen size 1.2 cm) compared with the control mice (median spleen size 1.6 cm; P = .008).
Knockdown of NPM1 sensitizes AML cells expressing Mt NPM1 to ATRA-induced differentiation
We next determined the effects of NPM1 depletion on ATRA-induced differentiation of AML cells. As shown in Figure 3A (left panel), treatment with ATRA alone induced differentiation of OCI-AML3 cells, as indicated by an increase in percentage of CD11b positive cells. Importantly, knockdown of NPM1 significantly enhanced ATRA–induced differentiation of OCI-AML3 cells, with a further increase in the percentage of CD11b-positive cells (NPM1 siRNA-treated ATRA-induced CD11b-positive cells: 33%; vs the control siRNA-treated ATRA-induced CD11b-positive cells: 17%). We also determined the effect of NPM1 knock down on ATRA-induced CD11b expression in HL-60 cells. In contrast to the effects observed in OCI-AML3 cells, NPM1 knockdown did not significantly increase CD11b expression in ATRA treated HL-60 cells (Figure 3A right panel). We next examined the effects of NPM1 knockdown on ATRA-mediated induction in the levels of p53, p21, and C/EBPα. As shown in Figure 3B, treatment with 0.25μM of ATRA alone for 72 hours appreciably induced p53, p21, and C/EBPα in OCI-AML3 cells. ATRA treatment also caused a slight decline in the levels of Mt NPM1 in OCI-AML3 cells (Figure 3B). In addition, siRNA mediated NPM1 knockdown further enhanced ATRA-induced p53, p21, and C/EBPα in OCI-AML3 cells (Figure 3B). Here, ATRA also reduced the levels of total NPM1 in OCI-AML3 cells (Figure 3B). NPM1 knockdown in HL-60 cells did not induce sensitivity to ATRA (supplemental Figure 2).
Knockdown of NPM1 enhances ATRA- induced differentiation of AML cells. (A) OCI-AML3 and HL-60 cells were transfected with control or NPM1-siRNA for 24 hours. After this, cells were treated with 0.25μM ATRA and incubated for an additional 72 hours. Differentiation was evaluated by staining the cells with Alexa 488-conjugated–anti-CD11b antibody followed by flow cytometry. Columns represent the mean of 3 independent experiments. Bars represent the SEM. (B) OCI-AML3 cells were transfected with control or NPM1 siRNA for 24 hours. Then, cells were treated with 0.25μM ATRA for an additional 72 hours. At the end of treatment, immunoblot analyses were performed for NPM1, Mt-NPM1, p53, p21, and C/EBPα on the total cell lysates. The expression levels of β-actin in the lysates served as the loading control.
Knockdown of NPM1 enhances ATRA- induced differentiation of AML cells. (A) OCI-AML3 and HL-60 cells were transfected with control or NPM1-siRNA for 24 hours. After this, cells were treated with 0.25μM ATRA and incubated for an additional 72 hours. Differentiation was evaluated by staining the cells with Alexa 488-conjugated–anti-CD11b antibody followed by flow cytometry. Columns represent the mean of 3 independent experiments. Bars represent the SEM. (B) OCI-AML3 cells were transfected with control or NPM1 siRNA for 24 hours. Then, cells were treated with 0.25μM ATRA for an additional 72 hours. At the end of treatment, immunoblot analyses were performed for NPM1, Mt-NPM1, p53, p21, and C/EBPα on the total cell lysates. The expression levels of β-actin in the lysates served as the loading control.
NPM1 depletion enhances ATRA- and Ara-C–induced apoptosis of AML cells with Mt NPM1
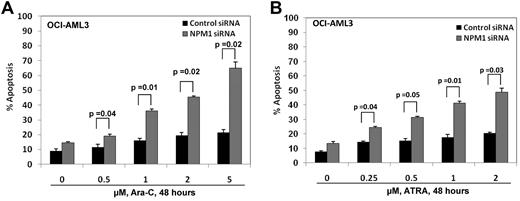
We next determined the effects of NPM1 knockdown on ATRA and Ara-C–induced apoptosis of Mt NPM1c+ expressing cells. Control or NPM1 siRNA-treated OCI-AML3 cells were exposed to 0.25-2.0μM of ATRA or 0.5-5.0μM Ara-C for 48 hours (Figure 4A-B). After this, the percentage of apoptotic cells was determined by annexin V staining followed by flow cytometry. Treatment with ATRA dose-dependently induced apoptosis to a significantly greater extent in OCI-AML3 cells treated with NPM1 siRNA versus the control siRNA-treated cells (P < .05; Figure 4B). NPM1 siRNA did not sensitize HL-60 cells to ATRA-induced apoptosis (data not shown). In addition, treatment with Ara-C also dose-dependently induced significantly more apoptosis of OCI-AML3 cells treated with NPM1 siRNA versus control siRNA treated cells (P ≤ .05; Figure 4A). These findings demonstrate that knockdown of NPM1 sensitizes OCI-AML3 cells to the lethal effects of ATRA and Ara-C.
Depletion of NPM1 by siRNA significantly enhances sensitivity of Mt NPM1 expressing AML cells to Ara-C and ATRA. (A,B) OCI-AML3 cells were transfected with control or NPM1 siRNA for 24 hours, followed by treatment with 0.5-5μM Ara-C or 0.25-2μM of ATRA for an additional 48 hours. Cells were stained with annexin V and propidium iodide and the percentages of apoptotic cells were determined by flow cytometry. Columns represent the mean of 3 independent experiments. Bars represent the SEM.
Depletion of NPM1 by siRNA significantly enhances sensitivity of Mt NPM1 expressing AML cells to Ara-C and ATRA. (A,B) OCI-AML3 cells were transfected with control or NPM1 siRNA for 24 hours, followed by treatment with 0.5-5μM Ara-C or 0.25-2μM of ATRA for an additional 48 hours. Cells were stained with annexin V and propidium iodide and the percentages of apoptotic cells were determined by flow cytometry. Columns represent the mean of 3 independent experiments. Bars represent the SEM.
NSC348884 disrupts NPM1 oligomerization and differentially induces apoptosis in Mt NPM1-expressing cells
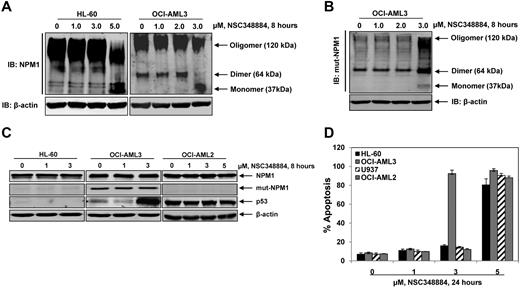
NPM1 forms dimers and higher order oligomers through its N-terminal oligomerization domain (amino acids 1-110).15 NSC348884 was identified as a small molecule inhibitor that disrupts NPM1 oligomerization.16 To examine the ability of NSC348884 to disrupt NPM1 oligomerization in AML cells, HL-60 and OCI-AML3 cells were treated with 1.0-5.0μM of NSC348884 for 8 hours and the disruption of NPM1 oligomers was analyzed by native PAGE. While treatment with 2μM of NSC348884 for 8 hours only slightly increased the monomeric form (37 kDa) of NPM1, treatment with 3.0μM of NSC348884 significantly disrupted the dimer (∼ 64 kDa) formation, and increased the monomeric form in OCI-AML3 but not in HL-60 cells (Figure 5A). However, exposure to higher concentrations of NSC348884 (5.0μM) also disrupted NPM1 oligomers in HL-60 cells (Figure 5A). In OCI-AML3 cells, exposure to 3.0μM NSC348884 also disrupted the oligomers of Mt NPM1 (Figure 5B). NSC348884 treatment had no effect on the levels of the Mt or WT NPM1, but increased the levels of p53 in OCI-AML3 cells (Figure 5C). In contrast, NSC348884 treatment did not alter the levels of WT NPM1 or p53 in OCI-AML2 cells (Figure 5C). We next determined the effects of NSC348884 treatment on the viability of HL-60, U937, OCI-AML3 and OCI-AML2 cells. As demonstrated in Figure 5D, treatment with 1.0 or 3.0μM of NSC348884 did not induce apoptosis of HL-60, U937 or OCI-AML2 cells, but induced marked apoptosis of OCI-AML3 cells. Higher concentrations of NSC348884 (≥ 5.0μM) induced apoptosis in all cell types (Figure 5D). Exposure of OCI-AML3 cells to 3.0μM of NSC348884 induced PARP cleavage and internucleosomal DNA fragmentation associated with apoptosis (Figure 6A top and bottom panels, respectively). During the 12 hours after starting treatment with 3.0μM NSC348884, marked apoptosis as well as loss of viability were evident in OCI-AML3 but not in HL-60 cells (Figure 6B). Induction of apoptosis was further assessed by morphologic examination of NSC348884-treated cells. As compared with HL-60 cells, which showed no response, NSC348884-treated OCI-AML3 cells exhibited cell shrinkage, nuclear condensation, membrane blebbing, and DNA fragmentation (Figure 6C). The necrotic cell death marker HMGB1 is released into the supernatant of cultured cells during necrosis.38 HMGB1 was not detected in NSC348884-treated cells (data not shown), suggesting that NSC348884 does not induce necrotic cell death. We also determined the effect of NSC348884 in OCI-AML3 cells in which NPM1 had been knocked down by NPM1 siRNA treatment. As shown in supplemental Figure 3, there was no difference in the extent of apoptosis induced by treatment with 3.0 or 5.0μM NSC348884 in the control OCI-AML3 cells versus those cells in which NPM1 was knocked down.
Small molecule inhibitor NSC348884 disrupts NPM1 oligomerization and induces apoptosis in OCI-AML3 cells. (A) HL-60 and OCI-AML3 cells were treated with the indicated concentrations of NSC348884 for 8 hours. After this, total cell lysates were prepared and native gel electrophoresis and immunoblot analysis of NPM1 was performed to assess the disruption of NPM1 oligomerization. The expression levels of β-actin in the lysates served as the loading control. (B) OCI-AML3 cells were treated with the indicated concentrations of NSC348884 for 8 hours. After this, total cell lysates were prepared and native gel electrophoresis and immunoblot analysis of mutant NPM1 was performed to assess the disruption of mutant NPM1 oligomerization. The expression levels of β-actin in the lysates served as the loading control. (C) HL-60, OCI-AML3, and OCI-AML2 cells were treated with indicated concentrations of NSC348884 for 8 hours. After this, total cell lysates were prepared and immunoblot analyses were performed for NPM1, Mt-NPM1 and p53. The expression levels of β-actin in the lysates served as the loading control. (D) HL-60, OCI-AML3, U937, and OCI-AML2 cells were treated with the indicated concentrations of NSC348884 for 24 hours. Then, cells were stained with annexin V and propidium iodide and the percentages of apoptotic cells were determined by flow cytometry. Columns represent the mean of 3 independent experiments; bars represent the SEM.
Small molecule inhibitor NSC348884 disrupts NPM1 oligomerization and induces apoptosis in OCI-AML3 cells. (A) HL-60 and OCI-AML3 cells were treated with the indicated concentrations of NSC348884 for 8 hours. After this, total cell lysates were prepared and native gel electrophoresis and immunoblot analysis of NPM1 was performed to assess the disruption of NPM1 oligomerization. The expression levels of β-actin in the lysates served as the loading control. (B) OCI-AML3 cells were treated with the indicated concentrations of NSC348884 for 8 hours. After this, total cell lysates were prepared and native gel electrophoresis and immunoblot analysis of mutant NPM1 was performed to assess the disruption of mutant NPM1 oligomerization. The expression levels of β-actin in the lysates served as the loading control. (C) HL-60, OCI-AML3, and OCI-AML2 cells were treated with indicated concentrations of NSC348884 for 8 hours. After this, total cell lysates were prepared and immunoblot analyses were performed for NPM1, Mt-NPM1 and p53. The expression levels of β-actin in the lysates served as the loading control. (D) HL-60, OCI-AML3, U937, and OCI-AML2 cells were treated with the indicated concentrations of NSC348884 for 24 hours. Then, cells were stained with annexin V and propidium iodide and the percentages of apoptotic cells were determined by flow cytometry. Columns represent the mean of 3 independent experiments; bars represent the SEM.
Treatment with NSC348884 induces apoptosis in mutant but not wild type NPM1 expressing AML cells. (A) HL-60 and OCI-AML3 cells were treated with the indicated concentrations of NSC348884 for 8 hours. At the end of treatment, total cell lysates were prepared and immunoblot analyses were performed for total PARP and cleaved PARP. The expression levels of β-actin in the lysates served as the loading control. Alternatively, evidence of apoptosis was demonstrated by DNA laddering assay. (B) HL-60 and OCI-AML3 cells were treated with 3μM NSC348884 for the indicated times. Then, cells were stained with annexin V and propidium iodide and the percentages of apoptotic cells were measured by flow cytometry. Alternatively, the percentages of nonviable cells were determined by trypan blue dye uptake in a hemocytometer. (C) HL-60 and OCI-AML3 cells were treated with the indicated concentrations of NSC348884 for 8 hours. Then, cells were cytospun onto glass slides and morphologic features of apoptosis were observed by light microscopy at 40× magnification. Representative images of untreated and NSC348884 treated HL-60 and OCI-AML3 cells are displayed.
Treatment with NSC348884 induces apoptosis in mutant but not wild type NPM1 expressing AML cells. (A) HL-60 and OCI-AML3 cells were treated with the indicated concentrations of NSC348884 for 8 hours. At the end of treatment, total cell lysates were prepared and immunoblot analyses were performed for total PARP and cleaved PARP. The expression levels of β-actin in the lysates served as the loading control. Alternatively, evidence of apoptosis was demonstrated by DNA laddering assay. (B) HL-60 and OCI-AML3 cells were treated with 3μM NSC348884 for the indicated times. Then, cells were stained with annexin V and propidium iodide and the percentages of apoptotic cells were measured by flow cytometry. Alternatively, the percentages of nonviable cells were determined by trypan blue dye uptake in a hemocytometer. (C) HL-60 and OCI-AML3 cells were treated with the indicated concentrations of NSC348884 for 8 hours. Then, cells were cytospun onto glass slides and morphologic features of apoptosis were observed by light microscopy at 40× magnification. Representative images of untreated and NSC348884 treated HL-60 and OCI-AML3 cells are displayed.
Expression of Mt NPM1 enhances sensitivity of cultured or primary AML cells to NSC348884 and/or ATRA, which is reduced by coexpression of FLT3-ITD
Because it has been previously described that coexpression of FLT3-ITD adversely impacts the therapeutic outcome in AML with Mt NPM1,21,22 we determined the effect of transfection with FLT3-ITD on the sensitivity of OCI-AML3 cells to treatment with NSC348884. Figure 7A (inset) demonstrates that OCI-AML3 transiently transfected with FLT3-ITD lentivirus (OCI-AML3/FLT3-ITD cells) exhibit several fold higher expression of FLT3-ITD, without causing any change in the levels of NPM1. As compared with the vector control cells, OCI-AML3/FLT3-ITD cells were significantly less sensitive to treatment with NSC348884 (P < .01; Figure 7A). In addition, OCI-AML3/FLT3-ITD cells were also significantly less sensitive to apoptosis induced by cotreatment with NSC348884 and ATRA (Figure 7B). However, combination of NSC348884 and ATRA induced significantly more apoptosis than either agent alone in both OCI-AML3/FLT3-ITD and the control cells (Figure 7B). Based on the differential lethal activity of NSC348884 against cultured OCI-AML3 cells with NPM1c+, we next determined whether treatment with NSC348884 would be relatively more lethal against primary AML cells with Mt NPM1c+. We screened 20 primary AML samples and discovered 9 of these to be positive for the presence of Mt NPM1, as determined by qPCR analysis described in Methods (supplemental Figure 4), and by immunofluorescence analysis (data not shown). Six of the 9 samples expressing Mt NPM1 were also later detected to be positive for FLT3-ITD, as determined by a semi-quantitative RT-PCR assay using a set of specific primers flanking FLT3 (supplemental Figure 4), as has been previously described.39 Overall, primary AML cells were relatively less sensitive to NSC348884 than OCI-AML3 cells (Figure 7C vs 7A). Approximately 80% of OCI-AML3 cells showed apoptosis, after exposure to 3.0μM NSC348884, while an exposure to 5.0μM was required to induce similar level of apoptosis in the primary AML cells with Mt NPM1. Lower levels of NSC348884 (1.0 or 3.0μM) induced significantly less apoptosis in primary AML cells with or without Mt NPM1 (Figure 7C). Exposure to 5.0μM NSC348884 induced significantly more apoptosis of primary AML cells with Mt NPM1 expression compared with normal CD34+ cells and primary AML cells expressing WT NPM1 (P < .001). Importantly, primary AML cells with coexpression of Mt NPM1 and FLT3-ITD were significantly less sensitive to NSC348884 than primary AML cells with only Mt NPM1 (Figure 7C). We further determined the effects of treatment with ATRA or Ara-C alone and in combination with NSC348884 against primary AML and normal CD34+ cells. The combination of 3.0μM NSC348884 and ATRA was more active in AML cells than either agent alone (P < .05). In addition, cotreatment with 0.25 or 0.5μM ATRA and 3.0μM NSC348884 induced significantly more apoptosis of AML cells with Mt NPM1 than treatment with ATRA or NSC348884 alone (P < .01; Figure 7D). Although cotreatment with NSC348884 enhanced Ara-C–induced apoptosis of AML cells, the combination was not significantly more cytotoxic in Mt NPM1 expressing AML cells compared with those expressing WT NPM1 (supplemental Figure 5). Importantly, cotreatment with ATRA and NSC348884 was significantly less lethal against AML cells coexpressing Mt NPM1 and FLT3-ITD (P < .01; Figure 7D). Collectively, these data indicate that, while the presence of NPM1 mutation sensitizes AML cells to NSC348884 alone or in combination with ATRA, coexpression of FLT3- ITD attenuates this effect.
Expression of mutant NPM1 enhances sensitivity of primary AML cells to NSC348884 and/or ATRA. (A) OCI-AML3 vector and FLT3-ITD overexpressing cells were treated with the indicated concentrations of NSC348884 for 24 hours. After treatment, cells were stained with annexin V and propidium iodide and the percentages of apoptotic cells were determined by flow cytometry. Columns represent the mean of 3 independent experiments; bars represent the SEM. Inset shows the expression of FLT3-ITD and NPM1 levels in OCI-AML3 vector and FLT3-ITD transfected cells. The expression levels of β-actin in the lysates served as the loading control. (B) OCI-AML3 vector and FLT3-ITD overexpressing cells were treated with the indicated concentrations of NSC348884 and/or ATRA for 48 hours. After treatment, cells were stained with annexin V and propidium iodide and the percentages of apoptotic cells were determined by flow cytometry. Columns represent the mean of 3 independent experiments. Bars represent the SEM. * indicates values significantly greater (P < .05) in the combination compared with treatment with either agent alone. † indicates values significantly less (P < .01) in OCI-AML3 cells with FLT3-ITD overexpression. (C,D) Primary patient-derived AML cells expressing wild-type (n = 4) or Mt NPM1 (n = 3), Mt-NPM1/FLT3-ITD (n = 2), and normal CD34+ progenitor cells (n = 3) were treated with the indicated concentrations of NSC348884 for 48 hours (C), or treated with the indicated concentrations of NSC348884 and/or ATRA for 48 hours (D). After treatment, cells were stained with annexin V and propidium iodide and the percentages of apoptotic cells were determined by flow cytometry. Columns represent the mean of 3 independent experiments; bars represent the SEM. In panel C, * indicates values significantly greater (P < .01) in the primary AML cells with Mt NPM1 compared with primary AML cells with WT NPM1 and primary CD34+ progenitor cells. † indicates values significantly less (P < .01) in primary AML cells with Mt NPM1/FLT3-ITD compared with AML cells with Mt NPM1 alone. In panel D, * indicates % values significantly greater (P < .05) in the combination treatments compared with treatment with either agent alone. † indicates values significantly less (P < .01) in Mt NPM1/FLT3-ITD AML cells compared with AML cells with Mt NPM1 alone in the combination treatments.
Expression of mutant NPM1 enhances sensitivity of primary AML cells to NSC348884 and/or ATRA. (A) OCI-AML3 vector and FLT3-ITD overexpressing cells were treated with the indicated concentrations of NSC348884 for 24 hours. After treatment, cells were stained with annexin V and propidium iodide and the percentages of apoptotic cells were determined by flow cytometry. Columns represent the mean of 3 independent experiments; bars represent the SEM. Inset shows the expression of FLT3-ITD and NPM1 levels in OCI-AML3 vector and FLT3-ITD transfected cells. The expression levels of β-actin in the lysates served as the loading control. (B) OCI-AML3 vector and FLT3-ITD overexpressing cells were treated with the indicated concentrations of NSC348884 and/or ATRA for 48 hours. After treatment, cells were stained with annexin V and propidium iodide and the percentages of apoptotic cells were determined by flow cytometry. Columns represent the mean of 3 independent experiments. Bars represent the SEM. * indicates values significantly greater (P < .05) in the combination compared with treatment with either agent alone. † indicates values significantly less (P < .01) in OCI-AML3 cells with FLT3-ITD overexpression. (C,D) Primary patient-derived AML cells expressing wild-type (n = 4) or Mt NPM1 (n = 3), Mt-NPM1/FLT3-ITD (n = 2), and normal CD34+ progenitor cells (n = 3) were treated with the indicated concentrations of NSC348884 for 48 hours (C), or treated with the indicated concentrations of NSC348884 and/or ATRA for 48 hours (D). After treatment, cells were stained with annexin V and propidium iodide and the percentages of apoptotic cells were determined by flow cytometry. Columns represent the mean of 3 independent experiments; bars represent the SEM. In panel C, * indicates values significantly greater (P < .01) in the primary AML cells with Mt NPM1 compared with primary AML cells with WT NPM1 and primary CD34+ progenitor cells. † indicates values significantly less (P < .01) in primary AML cells with Mt NPM1/FLT3-ITD compared with AML cells with Mt NPM1 alone. In panel D, * indicates % values significantly greater (P < .05) in the combination treatments compared with treatment with either agent alone. † indicates values significantly less (P < .01) in Mt NPM1/FLT3-ITD AML cells compared with AML cells with Mt NPM1 alone in the combination treatments.
Discussion
Findings presented here demonstrate that knockdown of NPM1 induces p53, p21 and C/EBPα, lowers HOXA9 and Meis1 levels, and inhibits cell cycle progression and colony growth of AML cells expressing Mt NPM1. Knockdown of NPM1 also induced differentiation and sensitized AML cells expressing Mt NPM1 to ATRA and Ara-C–induced apoptosis. In addition, disruption of NPM1 oligomerization by treatment with NSC348884 induced more apoptosis of AML cells expressing Mt NPM1, compared with AML cells expressing WT NPM1. NSC348884 also sensitized Mt but not WT NPM1 expressing AML cells to ATRA-induced apoptosis. Several studies have highlighted that the leukemogenic potential of the coexpression of HOXA9 and Meis1 is associated with increased proliferation and suppressed differentiation.28,29,40 In addition, activation of HOX genes has been documented in AML cells expressing Mt NPM1.41 HOX genes are also over-expressed in hematopoietic stem cells, and their expression is reduced with differentiation.37,40,41 The precise mechanism by which the altered status of NPM1 regulates HOXA9 was not explored in our studies. However, the physiologic role of NPM1 in the ribosomal mRNA biogenesis is well documented and the attenuation of the nucleolar WT NPM1 may affect the expression of specific mRNA, including HOXA9, thereby reducing HOXA9 protein levels.1,5,6,14
Loss of C/EBPα levels, or its functional loss through mutations, has been shown to increase proliferation and suppress differentiation of myeloid progenitor cells, thereby promoting leukemogenesis.42,43 Consistent with this, our findings demonstrate that knockdown of NPM1, which resulted in up-regulation of p21 and C/EBPα, is associated with cell cycle arrest, growth inhibition and differentiation of OCI-AML3 cells. Furthermore, the flow cytometric analysis presented here clearly show that, as a marker of differentiation, CD11b expression was induced in those cells in which NPM1 levels were also down regulated. Based on the preclinical studies that have shown ATRA to be an active differentiating agent for AML cells,44 ATRA has been tested in combination with induction chemotherapy for AML.45 In a randomized clinical trial of patients with AML, addition of ATRA to intensive chemotherapy yielded higher relapse-free and overall survival, especially in patients with Mt NPM1.45 However, in a different report, addition of ATRA was shown to have no overall effect on the treatment outcome in AML, regardless of the status of NPM1.46 Preclinical studies have also shown that overexpression of NPM1 confers resistance to ATRA, and levels of NPM1 are down regulated during differentiation and apoptosis.2,47 Consistent with these reports, our findings show that OCI-AML3 cells expressing Mt NPM1, which also have reduced levels of the nucleolar WT NPM1, are relatively more sensitive to ATRA. In addition, further down modulation of NPM1 sensitized OCI-AML3 cells to ATRA-mediated up-regulation of p53, p21 and C/EBPα. This was associated with sensitization of OCI-AML3 cells to ATRA- and Ara-C–induced apoptosis. Unlike OCI-AML3 cells, knockdown of NPM1 did not sensitize HL-60 cells with WT NPM1 to ATRA. These findings suggest that targeted depletion of NPM1 may selectively sensitize AML cells expressing Mt NPM1 to ATRA and Ara-C. Knockdown of NPM1 could potentially be sensitizing AML cells expressing Mt NPM1 to ATRA and Ara-C–induced apoptosis through multiple contributing mechanisms. Future studies will need to focus on elucidating the underlying mechanism(s) of this observation. It is also noteworthy that shRNA mediated knockdown of NPM1 in OCI-AML3 cells abolished their ability to induce lethal AML in NOD/SCID mice. This may be because of differentiation and apoptosis induced by NPM1 knockdown in OCI-AML3 cells. However, further studies will have to focus on whether inducible knockdown of NPM1 in the in vivo setting in established AML would lead to eradication of AML in the mice.
Recently, a small peptide inhibitor, CIGB-300, that inhibits casein kinase (CK) 2–mediated phosphorylation of NPM1, was shown to induce massive apoptosis of lung cancer cells.48 Separately, NSC348884, which disrupts the oligomer formation by NPM1, was also shown to be lethal for several different cancer cell-types.16 In the present studies, we demonstrate for the first time that AML cells expressing Mt NPM1 are more sensitive to the disruptive effects of NSC348884 on NPM1 oligomerization than AML cells expressing WT NPM1. In addition, treatment with NSC348884 is shown here for the first time to induce more apoptosis of AML cells expressing Mt NPM1 versus AML or normal CD34+ progenitor cells expressing WT NPM1. This suggests the possibility that AML cells expressing Mt NPM1 are more “addicted” to the biologic consequences of the combined presence of Mt NPM1c+ and the reduced levels of the nucleolar WT NPM1, because the knockdown by siRNA or inhibition of oligomerization by NSC348884 of both the Mt and WT NPM1 was also differentially more cytotoxic against AML cells with Mt NPM1. The dependence on this altered status of NPM1 in AML cells expressing Mt NPM1 is also highlighted by the observation that these cells exhibit a characteristic and different gene expression signature notable for up-regulation of genes that are involved in the stem cell maintenance, regardless of the presence of additional chromosomal alterations.21,41 It seems clear that the normal progenitor cells, or even the AML cells with the WT NPM1, lack this dependence such that these cells are less susceptible to the agents that cause either targeted knockdown of the levels or inhibit oligomerization and function of the Mt and WT NPM1. Treatment with NSC348884 also sensitized AML cells with Mt NPM1 to ATRA, pointing to a potential targeted differentiation therapy for AML with Mt NPM1. Coexpression of several other mutations has been detected in AML cells that also express Mt NPM1 and are cytogenetically normal. These include FLT3-ITD, and more recently, IDH1 and IDH2.49 This has been correlated with poorer relapse free and overall survival of AML patients, after standard therapy.49 Consistent with this, our findings also demonstrated that OCI-AML3 cells with ectopic expression of FLT3-ITD were less sensitive than control OCI-AML3 cells to treatment with NSC348884 alone or NSC348884 and ATRA. Similar results were obtained in primary AML cells that coexpressed Mt NPM1 and FLT3-ITD, compared with primary AML cells expressing Mt NPM1 alone. Collectively, these observations suggest that novel combination therapies should be considered for AML that express Mt NPM1 with or without coexpression of FLT3-ITD. These combinations could include anti-NPM1 agents and other agents, such as ATRA, or other novel agents that target FLT3. Other agents that target the metabolic consequences of the mutant IDH1 and IDH2 may also have to be considered.50 It is also clear that further in vitro and in vivo studies are warranted to pre-clinically develop and test combinations of anti-NPM1 agents with other novel agents that would be lethal for AML cells with precisely characterized genetic mutations in conjunction with the presence of Mt NPM1.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
These studies were partially supported by a grant from Georgia Cancer Coalition to H.J.K.
Authorship
Contribution: R.B., W.F., R.R., D.G.C., S.N., U.M., H.M., L.C., S.V., and K.H. performed the in vitro studies with the cultured and primary AML cells; S.A., C.W., J.M., and C.U. procured and assisted in performing the studies on primary AML cells; H.J.K. provided primary FLT3-ITD expressing AML cells for the in vitro studies; and K.N.B. planned and supervised the in vitro studies and prepared the report.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kapil N. Bhalla, MD, KUMC Cancer Center, The University of Kansas Medical Center, 3901 Rainbow Blvd, 4030 Robinson, MS 1027, Kansas City, KS; e-mail: kbhalla@kumc.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal