Abstract
Inhibitor of DNA binding protein 4 (ID4) is a member of the dominant-negative basic helix-loop-helix transcription factor family that lacks DNA binding activity and has tumor suppressor function. ID4 promoter methylation has been reported in acute myeloid leukemia and chronic lymphocytic leukemia (CLL), although the expression, function, and clinical relevance of this gene have not been characterized in either disease. We demonstrate that the promoter of ID4 is consistently methylated to various degrees in CLL cells, and increased promoter methylation in a univariable analysis correlates with shortened patient survival. However, ID4 mRNA and protein expression is uniformly silenced in CLL cells irrespective of the degree of promoter methylation. The crossing of ID4+/− mice with Eμ-TCL1 mice triggers a more aggressive murine CLL as measured by lymphocyte count and inferior survival. Hemizygous loss of ID4 in nontransformed TCL1-positive B cells enhances cell proliferation triggered by CpG oligonucleotides and decreases sensitivity to dexamethasone-mediated apoptosis. Collectively, this study confirms the importance of the silencing of ID4 in murine and human CLL pathogenesis.
Introduction
Chronic lymphocytic leukemia (CLL) is the most prevalent type of adult leukemia and has an extremely heterogeneous natural history. Approximately 90% of patients are older than 50 years, with the median age of 72 years at diagnosis.1 CLL is characterized by clonal overgrowth of CD5-, CD19-, and CD23-positive B cells.2,3 Prognostic factors, including IgVH gene mutational status, ZAP70 expression, cytogenetic abnormalities, and a variety of other biomarkers, have been applied to predict survival of patients with CLL. However, our understanding of environmental or molecular initiating events associated with CLL progression is limited, in part because of our inability to serially study the process of leukemia transformation and the importance of genes found to be silenced in tumor cells versus normal B cells. Developing novel strategies to address these obstacles will contribute enormously to our knowledge of disease initiation and progression.
The recent introduction of several mouse models of CLL (reviewed in Pekarsky et al4 ) provides important tools that could be used to determine the importance of loss or gain of function of genes in human CLL. The TCL1 oncogene is expressed in approximately 90% of human CLL cells. Transgenic mice with Eμ-driven, B cell–specific expression of TCL15 initially are healthy but gradually develop a B-cell leukemia with features of human CLL. These include unmutated IgVH status, increased expression of Bcl-2, epigenetic silencing by methylation, and aberrantly expressed microRNA genes mmu-mir-15a and mmu-mir-16-1, as has been described in human CLL.5-9 In addition, the disease phenotype includes expansion of nonclonal B1 lymphocytes at 3-5 months with eventual transformation to a mature B-cell leukemia very similar to human CLL. With disease progression, enlarged lymph nodes, spleen, liver, and elevated blood lymphocyte counts are noted, ultimately resulting in death at a median of 11 months.5,8 Data recently published by our group demonstrate that the Eμ-TCL1 transgenic mouse also has a pattern of epigenetic silencing similar to human CLL.9,10 Collectively, these characteristics indicate that the Eμ-TCL1 mouse model of CLL is a useful tool for defining the relevance of novel genes found to be uniformly silenced in CLL compared with normal B cells.
Inhibitor of DNA binding protein 4 (ID4) is a member of the dominant-negative basic helix-loop-helix (bHLH) transcription factor family (ID1-4).11 Members of this family lack a DNA binding domain but retain the ability to bind and thus inhibit the function of other bHLH proteins. Such binding predominately results in a tumor suppressor role of ID4 in colorectal,12 prostate,13,14 and gastric15 cancers, whereas in breast16 and bladder17 cancer, it has oncogenic features.16-18 In a study that used an interleukin-15 transgenic mouse model of natural killer (NK) cell leukemia, the authors demonstrated that ID4 was silenced by methylation in transformed lymphocytes.19 Studies with YAC-1 lymphocytes transfected with ID4 demonstrated both increased apoptosis and decreased proliferation in vitro and in vivo relative to the vector control, thereby suggesting a tumor-suppressor role. ID4 was also shown to be methylated in tumor cells from 87% of acute myeloid leukemia patients and 100% of CLL patients.19 This high degree of promoter methylation has been previously reported in CLL with DAPK1,20 the only gene to date firmly identified to be associated with familial predisposition to CLL. Thus, a strong rationale can be made for the importance of the ID4 gene in the development of CLL. Herein, we use the Eμ-TCL1 transgenic model of CLL to demonstrate the importance of ID4 in CLL pathogenesis and provide justification for future detailed study of this gene's function in leukemogenesis.
Methods
Mice, human samples, and cell lines
ID4+/− mice on a CD1 background were provided by Dr Fred Sablitzky, University of Nottingham.21 ID4+/− mice were crossed with homozygous TCL1-tg mice on a C3H/B6 background. The first generation of ID4+/−TCL1-tg and ID4+/+TCL1-tg mice obtained from these crosses were used for the studies described herein. Mice were kept in a pathogen-free barrier facility, and all animal experiments were performed under protocols approved by The Ohio State University Institutional Animal Care and Use Committee. B cells were isolated from mouse spleens by Ficoll density gradient centrifugation and magnetic-activated cell sorting (Miltenyi Biotec). Murine B cells were at least 80% CD19-positive by flow cytometry. B cells were also isolated by the use of Rosette-Sep (Stem Cell Technologies) from the peripheral blood of healthy donors or patients with CLL as defined by National Cancer Institute criteria22 seen at The Ohio State University (OSU). In these samples, cells were routinely at least 90% CD19-positive. A second set of samples was obtained before treatment from CLL patients enrolled on CALGB 9712, a randomized phase 2 study of concurrent versus sequential rituximab and fludarabine. The demographics of the patients and treatment outcome of this study have been published.23,24 Sampling was performed according to institutional review board–approved protocols after receipt of written informed consent according to the Declaration of Helsinki.
DNA and RNA isolation, immunoblot analysis, and real-time reverse-transcription polymerase chain reaction
Genomic DNA was isolated with the use of published protocols.25 Plasmid DNA was obtained by QIAprep Spin Miniprep kit (QIAGEN). RNA isolation by Trizol (Invitrogen) was used for SYBR-Green (Bio-Rad) and TaqMan (Applied Biosystems) reverse-transcription polymerase chain reaction (RT-PCR) following the manufacturers' recommendations and protocols. Anti-ID4, tubulin, and actin antibodies for immunoblots were obtained from Santa Cruz Biotechnologies. The specificity of this antibody for ID4 was validated by immunoblotting lysates from HEK 293 T cells transfected with an empty vector or a vector containing human ID4 cDNA. A total of 60 μg of protein was loaded per lane for CLL and B cells, whereas 5 μg of protein was loaded from the positive and negative control HEK 293 cells. Protein expression was detected by a chemiluminescent detection system (Amersham Pharmacia Biotech); the intensity was calculated relative to the tubulin or actin loading control in each lane. Chemiluminescent images were digitally quantified on an AlphaInnotec instrument (Cell Biosciences).
MassARRAY analysis
Functional analysis of cell proliferation
ID4+/−TCL1-tg and ID4+/+TCL1-tg mice were injected intraperitoneally with 2.5 μg/g CpG oligonucleotide every 4 days for 7 total injections. At 4 days after the last injection, 5-bromodeoxyuridine (BrdU) in phosphate-buffered saline at a dose of 50 pg/g body weight was injected intraperitoneally daily for 4 days, at which point CD19-positive B cells were isolated. Cells were fixed, permeabilized, treated with DNase, and stained with a fluorescein isothiocyanate-labeled anti-BrdU monoclonal antibody according to the manufacturer's instructions (FITC BrdU Flow kit; BD Biosciences). Samples were analyzed on a FACScan cytometer (BD Biosciences).
Histopathology, blood smear preparation, and flow cytometry
Mouse peripheral blood cells were collected from the cavernous sinus, and smears were immediately prepared and stained by May-Grunwald-Giemsa. For peripheral blood immunophenotyping, cells were treated with 0.165M ammonium chloride to eliminate red cells, and flow cytometric analysis was performed by the use of antibodies specific for murine CD19 and immunoglobulin M (BD Biosciences). A CLL phenotype in these mice was based on the observation of elevated (≥ 20 000/μL) circulating lymphocytes by blood smear. For tissue immunophenotyping, tissues were fixed in 10% buffered formalin and embedded in paraffin. Sections were stained with hematoxylin and eosin by the use of standard protocols and analyzed by pathologists at OSU.
Affymetrix microarray analysis
Total RNA was prepared from CD19-positive splenic B cells isolated from 1-month-old ID4+/−TCL1-tg and ID4+/+TCL1-tg mice by use of the RNEasy column purification Kit (QIAGEN). To obtain enough material for both microarray and validation PCR, splenic B cells from 3 mice were combined as 1 sample; a total of 3 samples from each group were used for analysis. Isolated RNA was hybridized to Affymetrix GeneChip mouse Genome 430 2.0 array (Affymetrix), which contains 45 101 probe-sets. Scanned image files were analyzed by GENECHIP 3.2 (Affymetrix). Background correction and normalization was performed and gene expression level was summarized over probes by use of the RMA method.28 A filtering method based on the percentage of samples with expression value above noise level was applied to filter out probe-sets with little or no expression, resulting in 13 987 probe-sets. Generalized linear models were used to detect differentially expressed genes between ID4+/−TCL1-tg and ID4+/+TCL1-tg mouse groups. To improve the estimates of variability and statistical tests for differential expression, a variance shrinkage method was used.29 The significance level of P = 7.15 × 10−5 was used by controlling the average number of false positives at 1.30 Fold changes of at least 1.5 were used to further reduce the list of significant probe-sets after controlling the number of false positives. The significant up- or down-regulated genes in the microarray data were then analyzed by the gene ontology annotation tool EASE software.31 All microarray data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE25100 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE25100).
Statistical analysis
Average ID4 methylation across amplicons and ID4 mRNA expression were treated as continuous variables in all analyses. Data from animal models and cell-line experiments were compared by the use of 2-sample t tests. P values were adjusted by Bonferroni or the Holm method to control the type I error rate at .05. Kaplan-Meier survival curves were obtained for ID4+/−TCL1-tg and ID4+/+TCL1-tg mice groups, and the difference between curves was tested by use of the log-rank test.
Nonparametric Wilcoxon rank sum tests were used to compare differences in methylation between groups of human samples. Associations between ID4 methylation and baseline clinical, demographic, and molecular features of patients registered to CALGB 9712 were analyzed with the use of t tests from generalized linear models. The proportional hazards model was used to generate predicted survival curves at certain levels (40%, 60%, and 80%) of methylation, and the association between ID4 methylation and overall survival, with and without adjusting for other variables, was tested with the Wald test. The proportional hazards assumption was checked for each variable and hazard ratios are presented with 95% confidence intervals. In the more exploratory analyses that used clinical data, P values were not adjusted for multiple testing. These analyses were performed by the CALGB Statistical Center.
Results
ID4 promoter is differentially methylated in human CLL cells and demonstrates increasing promoter methylation on the basis of disease status
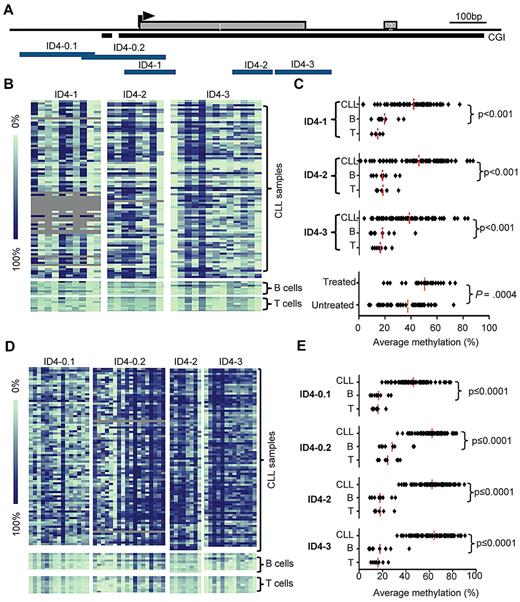
We examined the quantitative methylation of ID4 in samples from 84 CLL patients seen at OSU. Five amplicons, ID4-0.1, ID4-0.2, ID4-1, ID4-2, and ID4-3, located within a CpG island across the first exon (Figure 1A) of the ID4 gene, were investigated for their DNA methylation status by the use of MassARRAY (Figure 1B). Three amplicons, ID4-1, 2, and 3, showed very similar levels of DNA methylation across the CLL patient samples with averages of 41.8%, 45.8%, and 38.5%, respectively. Within each amplicon, these methylation levels were significantly different compared with the average methylation of normal B cells (average = 19.4%, 17.9%, and 17.9%, respectively; P < .001 for all; Figure 1C). Samples from OSU patients used in this study were further divided into 2 groups: those obtained when patients had asymptomatic disease and were without treatment (less advanced), and those obtained from previously treated patients (advanced disease). Samples from patients with more advanced disease (average methylation, 50.4%; SD, 13.9%) had significantly greater levels of ID4 promoter methylation compared with those with less advanced disease (average methylation, 37.6%; SD, 15.4; P = .0004, Figure 1C), suggesting that ID4 methylation may be a later event in CLL.
Methylation of ID4 promoter-associated CpG island in CLL patients. (A) Schematic representation of the ID4 gene showing the location of the CpG island (CGI; black bar) and the amplicons used for MassARRAY based methylation analysis (ID4-0.1, 0.2, 1, 2, and 3; blue bars). The arrowhead indicates the predicted transcription start site. (B) Graphical display of quantitative DNA methylation data on amplicons ID4-1, 2, and 3. Each square represents a CpG unit, and each row represents a sample. Gray indicates unavailable data. Color coding for DNA methylation levels is explained by the bar on the left. Samples included 85 CLL PBMC samples obtained from patients seen at OSU; normal control patients were 9 sorted CD19+ B-cell samples and 8 CD3+ T-cell samples from healthy donors. (C) Plot of average percentage methylation over amplicons ID4-1, 2, and 3, in OSU CLL sample set and controls (normal B and T cells), and further separated by treatment status. P values were based on Mann-Whitney U tests. (D) Graphical display of DNA methylation in amplicons ID4-0.1, 0.2, 2, and 3. Included are 82 PBMC pretreatment samples from CLL patients enrolled on CALGB 9712, and same controls used for Figure 1B. (E) Plot of average percentage methylation over amplicons ID4-0.1, 0.2, 2, and 3, in CLL and controls (normal B and T cells) samples. P values were determined on the basis of Mann-Whitney U tests.
Methylation of ID4 promoter-associated CpG island in CLL patients. (A) Schematic representation of the ID4 gene showing the location of the CpG island (CGI; black bar) and the amplicons used for MassARRAY based methylation analysis (ID4-0.1, 0.2, 1, 2, and 3; blue bars). The arrowhead indicates the predicted transcription start site. (B) Graphical display of quantitative DNA methylation data on amplicons ID4-1, 2, and 3. Each square represents a CpG unit, and each row represents a sample. Gray indicates unavailable data. Color coding for DNA methylation levels is explained by the bar on the left. Samples included 85 CLL PBMC samples obtained from patients seen at OSU; normal control patients were 9 sorted CD19+ B-cell samples and 8 CD3+ T-cell samples from healthy donors. (C) Plot of average percentage methylation over amplicons ID4-1, 2, and 3, in OSU CLL sample set and controls (normal B and T cells), and further separated by treatment status. P values were based on Mann-Whitney U tests. (D) Graphical display of DNA methylation in amplicons ID4-0.1, 0.2, 2, and 3. Included are 82 PBMC pretreatment samples from CLL patients enrolled on CALGB 9712, and same controls used for Figure 1B. (E) Plot of average percentage methylation over amplicons ID4-0.1, 0.2, 2, and 3, in CLL and controls (normal B and T cells) samples. P values were determined on the basis of Mann-Whitney U tests.
Given the heterogeneous nature of this first set of patients, we studied a second set of 82 peripheral blood mononuclear cell (PBMC) samples obtained from patients with symptomatic CLL who participated in a previously reported, prospective chemoimmunotherapy trial of fludarabine and rituximab (CALGB 9712).22,28 These patients had an average follow-up of more than 9 years from initiating treatment and were characterized for genomic abnormalities (interphase cytogenetics, IgVH mutational status) known to be important in predicting CLL outcome. Interestingly, the levels of ID4 methylation in this set resembles more the levels seen in the CLL samples from patients with advanced disease in set 1 (supplemental Figure 1, available on the Blood web site; see the Supplemental Materials link at the top of the online article). ID4-1 was then additionally substituted by 5′ extended amplicons ID4-0.1 and ID4-0.2. These amplicons are located partially outside the CpG island but cover the ID4 promoter region as determined in the previous study. The average ID4 methylation within each of the amplicons studied when this patient set was used (ID4-0.1, 0.2, 2, and 3) was 46.4%, 62.4%, 64.2%, and 61.2%, respectively. Compared with normal B cells (with average methylation levels of 16.8%, 28.2%, 18.4%, and 30.4%, respectively) methylation in CLL patient cells was significantly greater (P < .0001 for all; Figure 1D,E).
Similar results were observed comparing methylation of CLL patient cells with normal T cells. The average methylation for CLL samples over all 4 amplicons was 58.5% (SD, 10.2%), and these values across the amplicons were used in the subsequent analyses. With respect to clinical features (supplemental Table 1), increased age at time of registration and elevated white blood cell counts (WBC) were significantly associated with increased methylation (P = .007 and P = .01, respectively) whereas no difference of ID4 methylation was found in sorted B cells from healthy patients ranging in age from 29 to 64 years (supplemental Figure 2). In contrast, common prognostic factors such as Rai stage (P = .26), IgVH mutational status (P = .60), and high-risk genomic features del(11q22.3)/ del(17p13.1) (P = .69) were not associated with ID4 methylation. As a sole variable, the increasing level of ID4 promoter methylation was associated with decreasing overall survival (P = .03), although some of this relationship may be explained by age and WBC, 2 variables shown to be of prognostic significance in other treatment trials (supplemental Table 2). In a multivariable model including ID4 methylation, age, and lymphocyte count, ID4 methylation was not isolated as a statistically significant independent predictor, although this may be attributable to sample number (supplemental Figure 3). Larger studies of CLL patients receiving chemoimmunotherapy with extended follow-up will be required to fully address this question.
ID4 expression is reduced early in CLL patients irrespective of promoter methylation
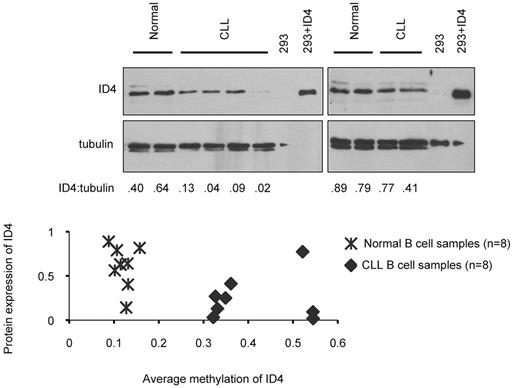
Despite variable levels of ID4 methylation, mRNA expression of ID4 in all CLL patient samples investigated was effectively undetectable (38-40 PCR cycles required to reach detection threshold). Analysis of ID4 protein expression in CLL cells and normal B cells (Figure 2) revealed that ID4 protein levels in CLL cells were diminished approximately two-thirds compared with B cells from healthy volunteers (CLL cell: normal B-cell ID4 protein ratio = 0.35; P = .0002). The uniform reduction of ID4 mRNA expression to below detectable levels suggests ID4 transcriptional silencing is an early event in human CLL, and that methylation of the ID4 promoter is observed with progression of the disease.
ID4 methylation and protein expression in CLL cells. Top, Immunoblot analysis of ID4 expression in normal B cells or CLL cells. Lysate from HEK 293 T cells transfected with vector alone or ID4 cDNA were included as controls. Tubulin expression served as a loading control. Blots shown are representative of a total of 8 samples for each normal and CLL CD19+ B cells, all run in duplicate. Raw intensities of ID4 and tubulin were background-corrected and ratios of ID4 over tubulin were log2-transformed for each sample. Analysis of variance model was used and each immunoblot was treated as a block. Differences of ID4 levels between CLL and normal were evaluated by 2-sided t test. ID4 expression in CLL cells is 0.35-fold the level in normal B cells (P = .0002). Bottom, Correlation between protein expression and methylation level of ID4 was calculated by Pearson correlation coefficients. On the basis of the results obtained from 8 healthy and 8 CLL B-cell samples, the correlation coefficient is −0.53 (P = .034) between average methylation across all amplicons and ID4 protein expression.
ID4 methylation and protein expression in CLL cells. Top, Immunoblot analysis of ID4 expression in normal B cells or CLL cells. Lysate from HEK 293 T cells transfected with vector alone or ID4 cDNA were included as controls. Tubulin expression served as a loading control. Blots shown are representative of a total of 8 samples for each normal and CLL CD19+ B cells, all run in duplicate. Raw intensities of ID4 and tubulin were background-corrected and ratios of ID4 over tubulin were log2-transformed for each sample. Analysis of variance model was used and each immunoblot was treated as a block. Differences of ID4 levels between CLL and normal were evaluated by 2-sided t test. ID4 expression in CLL cells is 0.35-fold the level in normal B cells (P = .0002). Bottom, Correlation between protein expression and methylation level of ID4 was calculated by Pearson correlation coefficients. On the basis of the results obtained from 8 healthy and 8 CLL B-cell samples, the correlation coefficient is −0.53 (P = .034) between average methylation across all amplicons and ID4 protein expression.
ID4 is transcriptionally silenced before transformation in the TCL1 transgenic mouse model of CLL
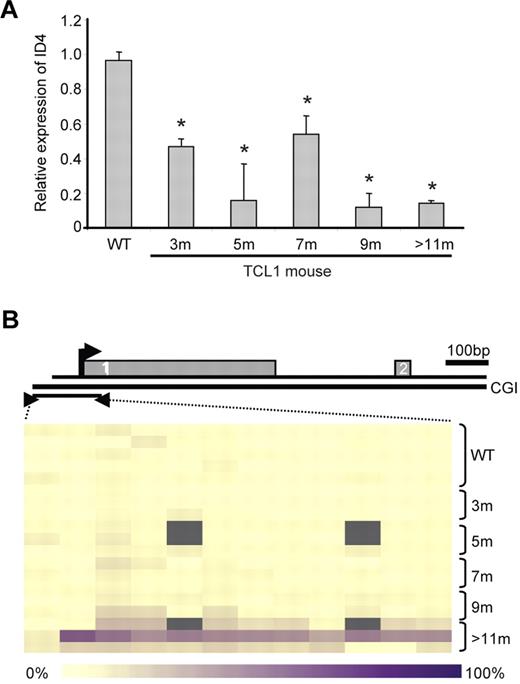
Given the uniform transcriptional silencing of ID4 in all CLL patients examined, we next used the Eμ-TCL1 transgenic mouse model of CLL to determine when ID4 silencing occurs during the development of disease. This model of CLL shows a long period of latency, with early oligoclonal B cells appearing at 3 months.5 Progression of CLL occurs gradually with increased peripheral WBC count and enlarged spleen and lymph nodes, and death typically occurs at 9-12 months of age. In CD19-selected splenocytes derived from TCL1 transgenic mice, we found that ID4 mRNA expression was already decreased at 3 months compared with that in splenocytes of age-matched control mice (Figure 3A). Corresponding to diminished mRNA expression, there was progressive loss of ID4 protein level beginning at 3 months in the isolated B cells (supplemental Figure 4). As with other genes regulated by methylation in this mouse model and also in human CLL, ID4 promoter methylation was not observed at the 3-month time point, but rather at 9 months of age or later (Figure 3B). The transcriptional silencing and subsequent promoter methylation of ID4 was therefore similar to that observed in human CLL. These data collectively justify use of the Eμ-TCL1 transgenic model to determine the contribution of ID4 loss to disease progression.
ID4 expression in B cells from Eμ-TCL1 transgenic mice. (A) Quantitative ID4 expression in wild-type (WT) and Eμ-TCL1 transgenic mouse spleen cells by real-time RT-PCR. The expression in Eμ-TCL1-tg mouse samples is shown relative to the expression in WT cells (defined as 1). Error bars indicate SD. *Indicates statistically significant differences based on 2-sample t tests. (B) ID4 methylation in WT and Eμ-TCL1-tg mouse spleen cells at different time points as measured by quantitative MassARRAY based methylation analysis. Gray indicates unavailable data. The diagram above it depicts the relative location of the amplicon with respect to exon 1 and exon 2 of ID4. The bar at the bottom indicates the color coding for the DNA methylation levels (yellow, 0% methylation; dark purple, 100% methylation).
ID4 expression in B cells from Eμ-TCL1 transgenic mice. (A) Quantitative ID4 expression in wild-type (WT) and Eμ-TCL1 transgenic mouse spleen cells by real-time RT-PCR. The expression in Eμ-TCL1-tg mouse samples is shown relative to the expression in WT cells (defined as 1). Error bars indicate SD. *Indicates statistically significant differences based on 2-sample t tests. (B) ID4 methylation in WT and Eμ-TCL1-tg mouse spleen cells at different time points as measured by quantitative MassARRAY based methylation analysis. Gray indicates unavailable data. The diagram above it depicts the relative location of the amplicon with respect to exon 1 and exon 2 of ID4. The bar at the bottom indicates the color coding for the DNA methylation levels (yellow, 0% methylation; dark purple, 100% methylation).
Early haploid loss of ID4 accelerates TCL1-induced CLL
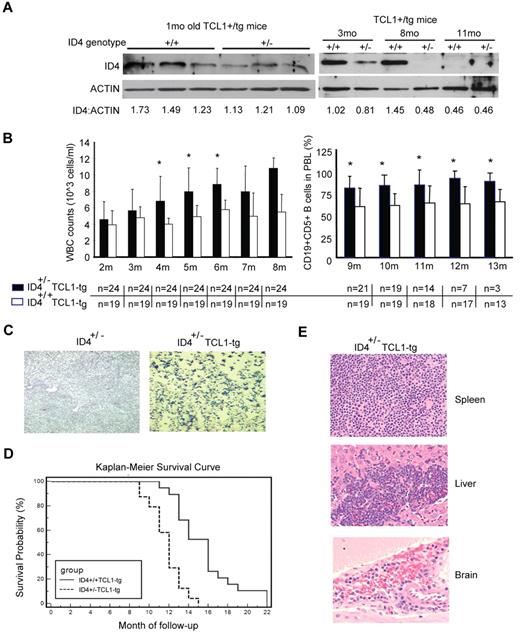
The early transcriptional silencing of ID4 in human CLL cells prevents determining its relevance to CLL progression. Because ID4 is transcriptionally silenced in a progressive manner before the development of overt leukemia in the TCL1 mouse model of CLL, we used this system to examine the importance of ID4 in early CLL pathogenesis. We hypothesized that earlier reduction of ID4 would accelerate CLL progression and shorten survival. We therefore knocked down ID4 expression in TCL1 transgenic mice through genetic crossing with an ID4 hemizygous (ID4+/−) strain of mice that were previously described.21 A total of 43 F1 littermates with only a single allele of TCL1 transgene, including 24 ID4+/−TCL1-tg and 19 ID4+/+TCL1-tg mice, were monitored from birth until death with serial assessment of peripheral blood lymphocyte counts. Decreased expression of ID4 protein in the ID4+/−TCL1-tg mice was confirmed by immunoblot analysis at the ages of 1, 3, and 8 months compared with ID4+/+TCL1-tg mice (Figure 4A). Increased B-cell expansion in ID4+/−TCL1-tg mice, starting at the age of 4 months, but not in ID4+/+TCL1-tg mice or control ID4 heterozygous knockout mice (Figure 4B,C, P = .0008), suggests that haploid loss of ID4 results in accelerated CLL development in TCL1 transgenic mice. More importantly, the survival curves between ID4+/− and ID4+/+ mice showed a significant difference (P < .0001), where the median survival of the ID4+/− mice was 12 months (range, 9-15 months), and the median survival was 16 months (range, 11-22 months) for the ID4+/+ TCL1 mice (Figure 4D).
Haploid loss of ID4 in Eμ-TCL1-tg mice leads to accelerated CLL disease progression. (A) ID4 protein expression in CD19+-selected B cells from Eμ-TCL1-tg mice without (+/+) or with (+/−) haploid loss of ID4. Numbers at the bottom give the relative amounts of ID4 protein. (B) Left: WBC counts from blood smears obtained from mice aged from 2 to 8 months. Right: 100 μL of peripheral blood lymphocytes (PBL) were collected each month and measured by flow cytometry for the percentage of CD19+CD5+ cells in mice aged from 9 to 13 months. Bottom: Number of live animals from each group that was available and analyzed. *Indicates statistically significant differences based on 2-sample t tests. (C) Representative blood smear stained with Wright-Giemsa showing an increased number of circulating lymphocytes in Eμ-TCL1-tg mice with haploid loss of ID4 (age, 14 months), but not in age-matched control heterozygous ID4 mice. (D) Kaplan-Meier survival curves from F1 littermates of Eμ-TCL1-tg mice with or without the haploid loss of ID4. (E) Histologic immunophenotyping of spleen, liver, and brain from Eμ-TCL1 mice with haploid loss of ID4. Top: Obliterative lymphoma is present in the periarteriolar lymphoid sheaths (white pulp) of spleen. Middle: Evidence of lymphocyte infiltration is suggested in approximately 25% of the liver sections by intrasinusoidal neoplastic cells. Bottom: Marked hippocampal neuronal necrosis was observed in the section from brain, as well as moderate multifocal cerebral, cerebellar, and meningeal hemorrhage, indicating the infiltration of malignant lymphocytes in the brain.
Haploid loss of ID4 in Eμ-TCL1-tg mice leads to accelerated CLL disease progression. (A) ID4 protein expression in CD19+-selected B cells from Eμ-TCL1-tg mice without (+/+) or with (+/−) haploid loss of ID4. Numbers at the bottom give the relative amounts of ID4 protein. (B) Left: WBC counts from blood smears obtained from mice aged from 2 to 8 months. Right: 100 μL of peripheral blood lymphocytes (PBL) were collected each month and measured by flow cytometry for the percentage of CD19+CD5+ cells in mice aged from 9 to 13 months. Bottom: Number of live animals from each group that was available and analyzed. *Indicates statistically significant differences based on 2-sample t tests. (C) Representative blood smear stained with Wright-Giemsa showing an increased number of circulating lymphocytes in Eμ-TCL1-tg mice with haploid loss of ID4 (age, 14 months), but not in age-matched control heterozygous ID4 mice. (D) Kaplan-Meier survival curves from F1 littermates of Eμ-TCL1-tg mice with or without the haploid loss of ID4. (E) Histologic immunophenotyping of spleen, liver, and brain from Eμ-TCL1 mice with haploid loss of ID4. Top: Obliterative lymphoma is present in the periarteriolar lymphoid sheaths (white pulp) of spleen. Middle: Evidence of lymphocyte infiltration is suggested in approximately 25% of the liver sections by intrasinusoidal neoplastic cells. Bottom: Marked hippocampal neuronal necrosis was observed in the section from brain, as well as moderate multifocal cerebral, cerebellar, and meningeal hemorrhage, indicating the infiltration of malignant lymphocytes in the brain.
All mice had evidence of active leukemia at the time of death. One possibility of delayed onset of CLL in this experiment compared with the reported onset of CLL in other groups (11-14 months) is that our mice in these tests have significant parts of the CD1 genome, which was contributed by the ID4 knockout strain. Autopsies in mice with leukemia revealed enlarged spleens with significant hyperplasia of the white pulp and lymphocyte infiltration in livers from both genotypes. Histopathology analysis on 9 ID4+/−TCL1-tg mice showed that all had lymphoma in nonlymphoid tissues such as the intestine, brain, pancreas, or kidney compared with only 1 of 5 ID4+/+TCL1-tg mice (Figure 4E). Collectively, these data support the role of ID4 in TCL1-driven CLL progression.
Disrupted apoptosis and enhanced proliferation capabilities of ID4+/−TCL1+-tg lymphocytes
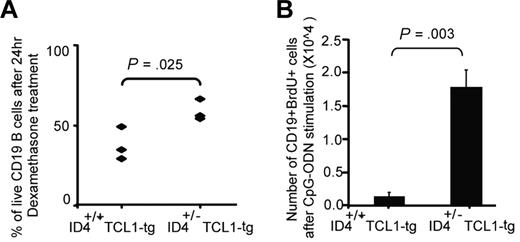
How haploid loss of ID4 in nontransformed TCL1-tg B cells influences progression and aggressiveness of the leukemia is unclear. Previous work suggests that introduction of ID4 into transformed B cells enhances spontaneous apoptosis and diminishes engraftment.19 To ascertain the influence of haploid loss of ID4 on nontransformed B cells, we investigated whether B cells from ID4+/−TCL1-tg mice show changes in apoptosis and/or proliferation relative to ID4+/+TCL1-tg nontransformed B cells. These studies were conducted using isolated CD19-positive spleen cells obtained from 1-month-old mice 24 hours after treatment either with vehicle control or 1μM dexamethasone. Cells from both groups showed similar percentage of spontaneous apoptosis after the treatment of vehicle control (35%-40%). However, haploid loss of ID4 in TCL1+-tg lymphocytes significantly decreased sensitivity to dexamethasone-mediated apoptosis (P = .025, Figure 5A). We next determined whether proliferation of B lymphocytes differed in the ID4+/− TCL1 mice compared with ID4+/+ TCL1 mice. One-month-old mice of each genotype were injected every 4 days with immune-stimulatory CpG oligonucleotide, followed by BrdU. Incorporation of BrdU was then assessed in CD19-positive cells by flow cytometry. In these experiments, cells from ID4+/− TCL1 mice showed significant increase in proliferation compared with the ID4+/+ control mice (P = .003; Figure 5B). Together, these results indicate that partial loss of ID4 in B cells before the development of leukemia results in diminished dexamethasone-induced apoptosis as well as increased proliferation, behaviors that may contribute to leukemic development and progression.
Haploid loss of ID4 in TCL1-tg B cells inhibits dexamethasone-mediated apoptosis in vitro and promotes TLR9 agonist-stimulated cell proliferation in vivo. (A) CD19+ cells were isolated from the spleens of 1-month-old Eμ-TCL1-tg mice with or without haploid loss of ID4. Isolated cells were cultured in hybridoma medium supplemented with 1% β-mercaptoethanol. After 24 hours of treatment with 1μM dexamethasone, cells were analyzed by annexin/PI flow cytometry. P value is determined on the basis of a 2-sample t test. (B) Three mice from both groups were injected with CpG oligonucleotide every 4 days for a total of 7 injections. Four days after the last injection, BrdU was injected daily for another 4 days, and CD19+ cells were isolated. The total number of CD19+BrdU+ cells was then determined by flow cytometry. P value is based on a 2-sample t test.
Haploid loss of ID4 in TCL1-tg B cells inhibits dexamethasone-mediated apoptosis in vitro and promotes TLR9 agonist-stimulated cell proliferation in vivo. (A) CD19+ cells were isolated from the spleens of 1-month-old Eμ-TCL1-tg mice with or without haploid loss of ID4. Isolated cells were cultured in hybridoma medium supplemented with 1% β-mercaptoethanol. After 24 hours of treatment with 1μM dexamethasone, cells were analyzed by annexin/PI flow cytometry. P value is determined on the basis of a 2-sample t test. (B) Three mice from both groups were injected with CpG oligonucleotide every 4 days for a total of 7 injections. Four days after the last injection, BrdU was injected daily for another 4 days, and CD19+ cells were isolated. The total number of CD19+BrdU+ cells was then determined by flow cytometry. P value is based on a 2-sample t test.
Partial deletion of ID4 generates a defined gene signature in B cells
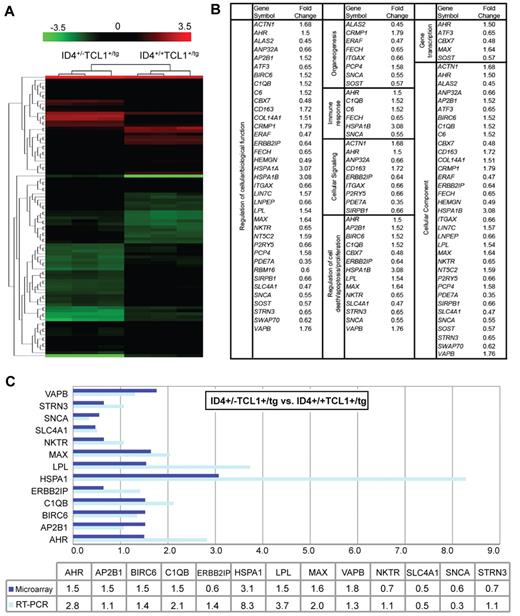
Given the phenotypic acceleration of TCL1-driven leukemia as well as disrupted apoptosis and increased proliferation in untransformed B-cells, we next sought to determine whether partial loss of ID4 with coexpression of human TCL1 generates a distinct gene signature in nontransformed B cells. Gene expression analysis was performed on CD19-selected B cells, comparing ID4+/−TCL1-tg mice with ID4+/+TCL1-tg mice (n = 3 for each group). As can be seen in Figure 6A, ID4+/− (column 1) and ID4+/+ (column 2) mice with a single allele of human TCL1 showed gene expression profiles with distinct patterns of gene down-regulation and up-regulation. Ninety-seven probe sets were differentially expressed (P ≤ 7.15 × 10−5) by at least 1.5-fold between the groups (Figure 6B). The signature included genes involved in the regulation of cell proliferation, differentiation, and signaling relating to the regulation of cellular function. Among these categories, 14 genes were identified as potential target genes that could be critical for disrupted apoptosis and proliferation noted with ID4 loss. Given the potential importance of these factors in facilitating ID4-accelerated CLL progression, 13 of these target genes involved in apoptosis were validated by real-time PCR (Figure 6C, bottom). This group showed similar fold changes as that observed using microarray data.
Microarray analysis of CD19+ B cells from 1-month-old ID4+/−TCL1-tg and ID4+/+TCL1-tg mice. (A) Heat map of the expression level of probe sets in Affymetrix array with at least a 1.5-fold increase or a 1.5-fold decrease in expression of ID4+/−TCL1-tg relative to ID4+/+TCL1-tg mouse B cells obtained from 1-month-old mice. Data are presented from 3 CD19+ B-cell samples in each group of 3 mice. Red and green colors indicate high and low expression, respectively. (B) Significantly up- or down-regulated genes in the microarray data were analyzed by the EASE gene ontology annotation tool. The enriched GO terms are listed. (C) Microarray (dark blue bars) and RT-PCR expression (light blue bars) of the 13 potential target genes involved in apoptosis. Fold changes in expression of ID4+/−TCL1-tg relative to ID4+/+TCL1-tg mouse B cells are shown graphically and in a table.
Microarray analysis of CD19+ B cells from 1-month-old ID4+/−TCL1-tg and ID4+/+TCL1-tg mice. (A) Heat map of the expression level of probe sets in Affymetrix array with at least a 1.5-fold increase or a 1.5-fold decrease in expression of ID4+/−TCL1-tg relative to ID4+/+TCL1-tg mouse B cells obtained from 1-month-old mice. Data are presented from 3 CD19+ B-cell samples in each group of 3 mice. Red and green colors indicate high and low expression, respectively. (B) Significantly up- or down-regulated genes in the microarray data were analyzed by the EASE gene ontology annotation tool. The enriched GO terms are listed. (C) Microarray (dark blue bars) and RT-PCR expression (light blue bars) of the 13 potential target genes involved in apoptosis. Fold changes in expression of ID4+/−TCL1-tg relative to ID4+/+TCL1-tg mouse B cells are shown graphically and in a table.
Discussion
In this report we confirm the high degree of ID4 promoter methylation in human CLL previously reported by our group9 and extended this observation to demonstrate that the level of promoter methylation as quantified by MassARRAY correlates with more advanced disease and inferior survival from the time of initial treatment with chemoimmunotherapy. Of interest, ID4 mRNA transcript and protein were absent or strongly reduced in CLL patient cells, whereas methylation levels were quite variable, thereby suggesting that transcriptional silencing mechanisms other than promoter methylation are initially responsible for the decreased expression of this gene. This could be accomplished through alterations in transcription factors or miRNAs targeting ID4. Alternatively, it is possible that ID4 methylation represents a marker for global epigenetic changes in the CLL genome and in this way represents many alterations occurring in parallel. Future work will be needed to address these possibilities. Given the similarities between human CLL and murine Eμ-TCL1 leukemia previously documented,9 we confirmed that ID4 was similarly silenced in mice leukemia cells through a transcriptional repression followed by promoter methylation. We then crossed this model with ID4 hemizygous mice and determined that partial loss of ID4 accelerates development of leukemia in these animals. These studies also demonstrated increased proliferation and disrupted apoptosis in B lymphocytes from the hemizygous ID4 TCL1 mice compared with ID4 homozygous controls. Furthermore, a specific gene expression profile, characterized by gain or loss of genes associated with apoptosis and also proliferation, differentiates ID4 hemizygous from ID4 homozygous TCL1 mice. Future efforts to generate a conditional knockout of ID4 in B cells might further elucidate the role of this protein. Irrespective of such experiments, these findings provide evidence for the role of ID4 silencing in the pathogenesis of CLL.
A continued challenge in biomedical research is determining the importance of single genes to the contribution of leukemic transformation. In CLL, this is difficult because the disease has a long natural history and often goes undetected because of a lack of symptoms early in the disease. Although a precursor to CLL called monoclonal B-cell lymphocytosis has been described,32-34 the study of these cells serially for molecular aberrations is extremely challenging. Our group has recently characterized the Eμ-TCL1 mouse model as an excellent preclinical model to study epigenetic changes over the course of the disease.9 Herein, we have used this model in a different manner, introducing haploinsufficiency of ID4 before transformation to determine whether this accelerates development of leukemia and/or produces more aggressive disease. A similar investigation by Enzler and colleagues35 was recently published using Eμ-TCL1 mice crossed with transgenic mice overexpressing the B-cell growth factor BAFF. In this study, gain of BAFF expression by non-B cells diminished apoptosis of the leukemia cells and led to more rapid progression of disease and compromised survival, as we observed here with genetic loss of ID4 expression. Both studies support the use of this Eμ-TCL1 transgenic mouse model of CLL to identify genes of potential pathogenic importance in human CLL. A limitation of the study remains the mixed backgrounds of 3 strains used in the experiment, which could possibly change the murine CLL disease phenotype. Efforts to back-cross to a homogenous background would diminish this variability, although our initial sample size calculations still provided sufficient sample size to demonstrate a highly significant difference between these 2 groups using an objective study end point (mouse death).
ID4 was identified as a potentially relevant contributor to CLL pathogenesis through an epigenetic screen of an interleukin-15 transgenic NK-cell leukemia.19 This study identified ID4 as one of the few genes to be consistently methylated at the time of development of acute leukemia in this model. As part of this evaluation, ID4 transfection into transformed murine B cells enhanced FAS-mediated apoptosis and also prolonged survival of cells engrafted into immunocompromised mice.19 Our studies in nontransformed ID4+/− hemizygous B cells support that partial loss of this gene promotes both increased proliferation of B cells in vivo after CpG stimulation and also disrupted apoptosis after dexamethasone treatment. Outside of the studies in the NK-cell leukemia model and those outlined here, the function of ID4 in neoplastic transformation has not been explored. In solid tumors, including breast,36,37 gastric,15 colon,12 and prostate13,14 cancers, as well as myelodysplasia,38 adverse outcomes have been associated with ID4 methylation and silencing, as we observe in CLL. Through its interaction with specific transcription factors, ID4 can mediate different effects depending upon the cellular context, similar to what is proposed for microRNA. Given the importance of ID4 silencing in CLL progression, it will be important to focus on identifying binding partners of ID4 in B cells.
The distinct gene profile of TCL1 mice with hemizygous versus homozygous ID4 provides several potential target genes. The 2 most prominent examples in this list include MAX and ERBB2IP. MAX is a transcription factor and member of the bHLH leucine zipper family that can form homodimers and heterodimers with other family members, including the oncogene MYC, which is involved in regulation of apoptotic signaling.39 ERBB2IP is a member of the leucine-rich repeat and PDZ domain family. It regulates ERBB2 function and localization through binding of the unphosphorylated form of ERBB2, and affects the RAS signaling pathway by disrupting RAS-RAF interaction.40 Three genes overexpressed in ID4+/− mice that are involved in apoptosis are also of interest. Prognostic factor lipoprotein lipase is reported as an antiapoptotic gene in CLL patients that is strongly associated with poor outcome and IgVH gene mutation status41,42 ; Hspa1b (HSP70 inducible) is a molecular chaperone that has been well characterized to disrupt apoptosis; and aryl hydrocarbon receptor is a ligand-activated transcription factor that influences cell proliferation and differentiation, and is overexpressed in adult T-cell leukemia.43 Our current efforts are focused on understanding how loss of ID4 influences expression of these genes and their contribution to the ID4 phenotype of disrupted apoptosis and proliferation in B cells.
In addition to the findings showing the importance of ID4 loss in CLL progression, this study demonstrates variability of promoter methylation of a specific gene despite uniform transcriptional silencing. Furthermore, ID4 promoter methylation was significantly greater in previously treated CLL patients compared with untreated patients. In a second study of samples derived from CLL patients at the time of initial treatment, we noted that increased ID4 promoter methylation was associated with increased age and WBC count but not other pretreatment characteristics, including Rai stage, IgVH gene mutational status, and interphase cytogenetic abnormalities. Increase in ID4 methylation was also modestly associated with shorter survival after chemoimmunotherapy. The strength of this association with survival was confounded by the association with age and WBC count and will require confirmation in a larger patient dataset. Regardless, these studies indicate that quantitative promoter methylation of ID4 may be a potential contributor to poor outcome in CLL.
In conclusion, we provide further evidence that ID4 protein plays a role in the pathogenesis of CLL. Despite the potential association of promoter methylation with clinical outcome, ID4 expression is silenced in virtually all CLL patients independent of promoter methylation. Reduction of ID4 expression in a mouse model of human CLL accelerated the disease, increased B-cell proliferation, and increased the apoptotic threshold. Together, our results demonstrate that further study of ID4 function and development of potential therapeutics to re-express this gene are warranted.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We acknowledge Drs Richard Larson and Kanti Rai from the Cancer and Leukemia Group B, who provided clinical input into the design of CALGB 9712 from which the patient samples are derived.
This project was supported by The National Cancer Institute (P01 CA101956, P50-CA140158, and P30 CA 16058), The Leukemia and Lymphoma Society, and the D. Warren Brown Foundation.
National Institutes of Health
Authorship
Contribution: S.-S.C. was involved in planning the experiments, performing the mouse and in vitro studies, writing the first draft of the manuscript and approving the final version; R.C. was involved in planning the experiments related to methylation, reviewing drafts of the paper, and approving the final version; D.M.L. was involved in performing a subset of the experiments, reviewing drafts of the paper, and approving the final version; L.Y. was involved in planning and analyzing the experiments, reviewing drafts of the manuscript, and approving the final version; J.Q. was involved in performing a subset of the experiments, reviewing drafts of the paper, and approving the final version; A.S.R. was involved in planning and analyzing the experiments, reviewing drafts of the manuscript, and approving the final version; D.A.W. was involved in performing a subset of the experiments, reviewing drafts of the paper, and approving the final version; K.E.W. was involved in performing a subset of the experiments, reviewing drafts of the paper, and approving the final version; A.J.J. was involved in planning and supervising the animal experiments, reviewing drafts of the paper, and approving the final version; F.S. initiated the collaboration, provided the ID4 knockout mice, reviewed drafts of the paper, and approved the final version of the manuscript; and C.P. and J.B. obtained funding, designed and supervised all of the experiments performed, assisted in drafting the manuscript, and approved the final version of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John C. Byrd, MD, B302 Starling-Loving Hall, 320 West 10th Avenue, Columbus, OH 43210; e-mail: john.byrd@osumc.edu; and Christoph Plass, PhD, German Cancer Research Center, Im Neuenheimer Feld 280, 69120 Heidelberg, Germany; e-mail: c.plass@dkfz-heidelberg.de.
References
Author notes
C.P. and J.C.B. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal