Abstract
CD36 modulates platelet function via binding to oxidized LDL (oxLDL), cell-derived microparticles, and thrombospondin-1. We hypothesized that the level of platelet CD36 expression may be associated with inheritance of specific genetic polymorphisms and that this would determine platelet reactivity to oxLDL. Analysis of more than 500 subjects revealed that CD36 expression levels were consistent in individual donors over time but varied widely among donors (200-14 000 molecules per platelet). Platelet aggregometry and flow cytometry in a subset of subjects with various CD36 expression levels revealed a high level of correlation (r2 = 0.87) between platelet activation responses to oxLDL and level of CD36 expression. A genome-wide association study of 374 white subjects from the Cleveland Clinic ASCLOGEN study showed strong associations of single nucleotide polymorphisms in CD36 with platelet surface CD36 expression. Most of these findings were replicated in a smaller subset of 25 black subjects. An innovative gene-based genome-wide scan provided further evidence that single nucleotide polymorphisms in CD36 were strongly associated with CD36 expression. These studies show that CD36 expression on platelets varies widely, correlates with functional responses to oxLDL, and is associated with inheritance of specific CD36 genetic polymorphisms, and suggest that inheritance of specific CD36 polymorphisms could affect thrombotic risk.
Introduction
CD36 is an 88-kDa glycoprotein belonging to the Scavenger Receptor Type B family.1 It was identified initially as a protease-resistant platelet surface glycoprotein and named glycoprotein IV for its migration on sodium dodecyl sulfate-polyacrylamide gel electrophoresis.2 Since then, work in our laboratory and others has shown that CD36 is expressed on a broad array of other cell types,3,4 including microvascular endothelial cells, vascular smooth muscle cells, erythroid precursors, epithelia of breast, gut, and kidney, and cardiac and skeletal muscle. It is expressed on most phagocytic cells, including dendritic cells, microglia, monocytes/macrophages, and retinal pigment epithelium where it plays an important role in mediating recognition and uptake of oxidized phospholipids, apoptotic cells, and certain microbial cell wall components.5-7 Other well-characterized functions of CD36 include mediating endothelial cell antiangiogenic responses to thrombospondin-related proteins; fatty acid transport in gut epithelial cells, myocytes, and adipocytes3,4 ; and regulating oxidant stress.8
Although CD36 was recognized as a major platelet glycoprotein more than 3 decades ago, its role in platelet physiology has only recently been appreciated based on work by our group and others.9,10 In collaboration with Podrez et al,9 we identified platelet CD36 as a receptor for oxidized low-density lipoprotein (oxLDL) and showed that oxLDL induced platelet activation in a CD36-dependent manner.9 Using apoe null mice fed a high fat “Western” diet as a model of hyperlipidemia, we showed that cd36 gene deletion abrogated the associated prothrombotic state and platelet hyper-reactivity. This work defined CD36 as providing a mechanistic link between oxidant stress, hyperlipidemia, and thrombosis. Subsequently, we showed that platelet CD36 also functions as a receptor for cell-derived microparticles and thereby contributes to thrombus formation in settings of vascular injury and inflammation where microparticles are generated.11 Mechanistically, platelet CD36 engagement by oxLDL triggers a specific signal transduction pathway involving the src family kinases fyn and lyn and the MAP kinase jnk.12 CD36 also participates with CD47 in TSP-1 mediated down-regulation of inhibitory platelet signaling pathways involving adenyl and guanyl cyclases.13,14 Based on these findings, we hypothesized that the level of CD36 expression on platelets would modulate platelet reactivity; therefore, we set out to define the variability of expression in human subjects, relate it to platelet activation responses to oxLDL, and determine whether it is associated with inheritance of specific single nucleotide polymorphisms (SNPs) in the CD36 gene or in genes known to be involved in atherothrombotic risk.
The human CD36 gene is very large, extending up to 309.7 kb on band q11.2 of chromosome 7 and consisting of 15 or more exons of which 12 are coding.15,16 All but one of the 20 putative CD36 transcripts span a much smaller distance of 77.13 kb. The CD36 gene has been most extensively studied in Asian populations, including in Japan, Korea, Indonesia, Thailand, and China, where 3% to 8% of persons lack platelet CD36.16 This deficiency was initially defined in Japan as the Naka negative (Naka−) phenotype.17 Approximately 90% of Naka− subjects lack CD36 expression only in platelets; the remaining 10% are presumably CD36 null (type I deficiency) and to not express CD36 in any cells or tissues. Five mutations have been reported to be associated with type I deficiency in Asians. The type I Naka− phenotype is also common in African populations, although the associated null mutations are different from those reported in Asia.18
CD36 is highly polymorphic; in addition to the null mutations mentioned in the preceding paragraph, data from Ensembl Variation Build 60 (which is based on dbSNP Release 131), describe 2935 common genetic variants in or within 5 kb of the gene, all but one of which are SNPs. Some involve putative transcription factor binding sites17 or sites in the 5′-untranslated region, which are of potential significance because translational efficiency of the CD36 mRNA and thereby CD36 protein expression levels have been shown to be regulated by the 5′-untranslated region.19
Although the functional impact of CD36 deficiency has been well studied in different mouse and rat models, the impact of CD36 null mutations and polymorphisms in human biology are not well characterized. The cd36 null mouse strain generated in our laboratory shows dyslipidemia,20 defective fatty acid uptake in heart and muscle, alterations in insulin responsiveness,21 and protection from diet-induced atherosclerosis,22 thrombophilia,9 insulin resistance,23 and adipose inflammation.23 A small number of studies from Japan have implicated type I CD36 deficiency with increased risk for developing insulin resistance24 or cardiomyopathy,25 but these have not been consistently replicated. More than a dozen gene association studies have been reported linking CD36 SNPs to various phenotypes, including circulating high-density lipoprotein (HDL), LDL, and triglyceride levels,26-29 risks of metabolic syndrome and obesity,30,31 and risk of acute myocardial infarction (AMI) or stroke.32-34 Unfortunately, only a small number of these studies related CD36 SNPs or haplotypes to CD36 expression level. Love-Gregory et al have analyzed a cohort of black subjects and found associations of 5′ SNPs with metabolic syndrome and with HDL level.26 In a small subset of these subjects, they reported association of CD36 SNPs with peripheral blood monocyte CD36 levels.27 Interestingly, they found a negative correlation between monocyte CD36 levels and circulating HDL levels and a positive correlation with very low-density lipoprotein and apoB.
In the current study, we show that the level of platelet CD36 surface expression is highly variable among persons from a heterogeneous population. We also demonstrate that this variability affects the functional response of platelets to oxLDL and that the variability is associated with inheritance of specific genotypic polymorphisms at the CD36 locus in both whites and blacks.
Methods
Study populations
The initial study population included 32 healthy human volunteers who were students or employees at the Cleveland Clinic. We then recruited 500 patients through the Cleveland Clinic cardiac catheterization laboratory for validation studies of platelet CD36 expression measurement and 440 patients from the Cleveland Clinic ASCLOGEN (ASpirin and CLOpidogrel: GENotype vs Platelet Function Phenotype in Clinical Response) Trial for a gene association study. ASCLOGEN is a prospective clinical observational study of genotype-phenotype associations in patients undergoing percutaneous coronary intervention, who are on dual antiplatelet therapy. The study was designed to determine the influence of genotype on patient response to aspirin and clopidogrel therapy and on in-hospital and long-term outcomes. The study population included men and nonpregnant women at least 18 years old who had undergone percutaneous coronary intervention at the Cleveland Clinic. Subjects were excluded if they had taken ticlopidine, dipyridamole, steroidal drugs, or COX-2 inhibitors during the 2 weeks before enrollment or glycoprotein IIb/IIIa inhibitor within 7 days before enrollment or during percutaneous coronary intervention. Patients with known history of platelet disorders, with platelet counts <150 000/μL, or with hemoglobin < 10 g/100 mL or hematocrit < 30% were also excluded, as were those with a known allergy to aspirin or clopidogrel or who had a major surgical procedure in the week before enrollment. Clinical and demographic data on the 2 patient cohorts are shown in Table 1. All studies were approved by the Cleveland Clinic Institutional Review Board with informed consent from each subject in accordance with the Declaration of Helsinki. For platelet studies, blood samples were collected in 3.9% citrate buffer. Platelet-rich plasma (PRP) and washed platelets were isolated by sequential centrifugation. DNA was isolated from whole blood using the Puregene DNA purification kit (Gentra Systems).
Demographic and clinical phenotypes of study populations used for assessment of platelet CD36 expression levels and genetic analyses
| Variable . | Cardiac cath patients, n (%) . | ASCLOGEN white patients, n (%) . | ASCLOGEN black patients, n (%) . |
|---|---|---|---|
| Age, y (mean ± SD) | 62 ± 9 | 63 ± 10 | 58 ± 9 |
| Sex | |||
| Male | 313 (62.7) | 304 (81.3) | 18 (72) |
| Female | 186 (37.3) | 70 (18.7) | 7 (28) |
| Tobacco use | |||
| Present | 62 (12.9) | 59 (15.8) | 9 (36) |
| Past | 338 (70.3) | 243 (65.0) | 20 (80) |
| History | |||
| Atherosclerosis | 271 (54.6) | 374 (100) | 25 (100) |
| MI | 249 (51.2) | 168 (44.9) | 13 (52) |
| CABG/PCI | 221 (44.3) | 235 (62.8) | 15 (60) |
| Stroke | 37 (9.8) | 18 (4.8) | 1 (4) |
| Hypertension BP > 120/90 mmHg | 412 (85.3) | 311 (83.2) | 23 (92) |
| Diabetes | 140 (29.4) | 111 (29.7) | 11 (44) |
| Serum cholesterol ≥ 200 mg/dL | 104 (23.4) | 57 (20.1) | 6 (31.6) |
| LDL ≥ 120 mg/dL | 86 (19.9) | 68 (23.9) | 6 (33.3) |
| HDL ≤ 50(M) or ≤ 60(F) mg/dL | 279 (63) | 221 (81.0) | 15 (88.2) |
| Triglyceride ≥ 200 mg/dL | 80 (18) | 65 (23.0) | 3 (15.8) |
| Glucose ≥ 120 mg/dL | 108 (23.6) | 85 (23.4) | 7 (29.2) |
Platelet count ≤ 150 ×103/ L L | 37 (8.1) | 27 (7.3) | 1 (4.0) |
| BMI > 30 | 228 (44.3) | NA | NA |
| Variable . | Cardiac cath patients, n (%) . | ASCLOGEN white patients, n (%) . | ASCLOGEN black patients, n (%) . |
|---|---|---|---|
| Age, y (mean ± SD) | 62 ± 9 | 63 ± 10 | 58 ± 9 |
| Sex | |||
| Male | 313 (62.7) | 304 (81.3) | 18 (72) |
| Female | 186 (37.3) | 70 (18.7) | 7 (28) |
| Tobacco use | |||
| Present | 62 (12.9) | 59 (15.8) | 9 (36) |
| Past | 338 (70.3) | 243 (65.0) | 20 (80) |
| History | |||
| Atherosclerosis | 271 (54.6) | 374 (100) | 25 (100) |
| MI | 249 (51.2) | 168 (44.9) | 13 (52) |
| CABG/PCI | 221 (44.3) | 235 (62.8) | 15 (60) |
| Stroke | 37 (9.8) | 18 (4.8) | 1 (4) |
| Hypertension BP > 120/90 mmHg | 412 (85.3) | 311 (83.2) | 23 (92) |
| Diabetes | 140 (29.4) | 111 (29.7) | 11 (44) |
| Serum cholesterol ≥ 200 mg/dL | 104 (23.4) | 57 (20.1) | 6 (31.6) |
| LDL ≥ 120 mg/dL | 86 (19.9) | 68 (23.9) | 6 (33.3) |
| HDL ≤ 50(M) or ≤ 60(F) mg/dL | 279 (63) | 221 (81.0) | 15 (88.2) |
| Triglyceride ≥ 200 mg/dL | 80 (18) | 65 (23.0) | 3 (15.8) |
| Glucose ≥ 120 mg/dL | 108 (23.6) | 85 (23.4) | 7 (29.2) |
Platelet count ≤ 150 ×103/ L L | 37 (8.1) | 27 (7.3) | 1 (4.0) |
| BMI > 30 | 228 (44.3) | NA | NA |
MI indicates myocardial infarction; CABG/PCI, coronary artery bypass graft/percutaneous coronary intervention; BP, blood pressure; BMI, body mass index; and NA, not applicable.
Quantitative assay of platelet CD36 expression
PRP was incubated with phycoerythrin (PE)-conjugated anti-CD36 monoclonal antibody (clone 185-1G2; Santa Cruz Biotechnology) or isotype-matched control IgG, and mean fluorescence intensity (MFI) was then quantified by flow cytometry. A standard curve relating PE fluorescence intensity to PE surface density was generated using a mixture of PE-Quantibrite Beads (BD Biosciences) tagged with 4 defined amounts of PE ranging from 200 to 70 000 molecules per bead. The number of bound antibody molecules on each platelet was calculated from the standard curve using the platelet MFI and the molar ratio of PE to anti-CD36 IgG (supplied by the manufacturer). The stoichiometry of anti-CD36 binding to CD36 was assumed to be 1:1 for these studies.
Platelet aggregation and activation studies
oxLDL was prepared by incubation of LDL with 5μM copper sulfate at 37°C for 6 hours as previously described.20 PRP was incubated with 50 μg/mL oxLDL or native LDL as control for 30 minutes, and then a low concentration of adenosine diphosphate (ADP; 1 or 2μM) was added. Platelet aggregation was assessed turbidometrically using a dual-channel aggregometer (Chronolog). Platelet activation was also analyzed by flow cytometry with PE-conjugated anti–P-selectin IgG (BD Biosciences) to detect α-granule secretion.
Genotyping
Genotyping was performed using the Illumina Human Cardiovascular Disease (CVD) array, a gene-centric 50 000 SNP array designed to assess potentially relevant loci across a range of cardiovascular, metabolic, and inflammatory diseases.35 Twenty additional SNPs in the CD36, CYP2C19, and PR2Y12 genes, which are not on the array but previously reported in the literature as potentially relevant to the ASCLOGEN study goals, were genotyped by the 5′-allelic discrimination (TaqMan) assay as previously described.36
Genotype calling, annotation, and filtering of Illumina human CVD arrays
Genotypes were called using the Genotyping Module (Version 3.3.7) from Illumina's BeadStudio program (Version 3.1.3). Called genotypes were exported to text files and imported into R (Version 2.11.1) using the GenABEL R package (Version 1.6-5)37 for quality filtering and statistical analysis. Unique National Center for Biotechnology Information (NCBI) Build 37/hg19 chromosome and base pair locations were obtained from Ensembl Variation Build 59 (which is based on dbSNP Release 131) using the biomaRt R package (Version 2.6) for 48 090 of 49 111 measured genetic markers, 48 960 of which had rs identifiers and unique positions in NCBI Build 36/hg18. Gene associations and SNP types for each SNP were also obtained from Ensembl. Quality filtering by SNP was performed separately for the white and black subsets. For the white subset, the following criteria for SNP inclusion were used: call rate ≤ 0.97, minor allele frequency ≥ 0.01, and false discovery rate value > 0.01 from test of Hardy-Weinberg equilibrium. Because of the smaller sample size of the black subset, we used a higher minor allele frequency cutoff of 0.10 and lower SNP call rate threshold of 0.90.
Subject filtering
Subjects were included using the following criteria: per-subject SNP call rate ≥ 0.95, no sex mismatch, autosomal heterozygosity false discovery rate value > 0.01, inbreeding coefficient < 0.2, and Identical by state (IBS) score < 0.95. In addition, outliers defined as more than 6 SDs for whites or more than 4 SDs for blacks along the genetic principal components used to adjust for population stratification/substructure were dropped (n = 51).
Single SNP statistical analysis
For each SNP that passed our filtering step within each subset of samples, we fit an additive linear regression model with untransformed CD36 expression levels (MFI) as the response variable and predictor variables sex, first genetic principal component score,38 and SNP. SNP was coded as the number of copies of the coded allele. The first genetic principal component scores were calculated separately in the white and black subsets to help account for racial-specific population stratification. CD36 MFI values were restricted to those observed between 0 and 10, eliminating a few extreme values in the upper tail of the MFI distribution. After truncation, the CD36 MFI distribution was approximately symmetric and unimodal in both subsets. Fitted regression coefficients, P values for the test of the SNP coefficient equal to 0, and genomic control39 -adjusted SNP P values were obtained using the mlreg function in the GenABEL package. False discovery rate q values were calculated from the genomic control-adjusted P values with the R q value package (Version 1.22). In the CD36 cis analyses, we used the respective estimated genomic control λ-value from the genome-wide analyses of the white and black subsets. The R2 between SNPs was estimated from the observed genotypes for each subset using the GenABEL function r2fast.
We also calculated minimum permutation (min-p) P values40 for each subset analysis using 1000 and 10 000 random permutations of CD36 MFI values to assess genome-wide significance and significance of CD36 SNPs, respectively. These P values are empirical and do not rely on large-sample theory, which is questionable for the small black subset. For each permutation of CD36 MFI values, we ran the per-SNP analysis described in the preceding paragraph and recorded the minimum P value across all tested SNPs (either all filtered SNPs or all CD36 filtered SNPs). This was repeated 1000 or 10 000 times, generating 1000 of 10 000 minimum P values. Then we calculated the min-p P value for each SNP as the fraction of times the minimum permutation P value was greater than the observed P value.
In addition to performing the genome-wide association study (GWAS) analysis, we also performed a more focused “cis” analysis looking only at the 72 available SNPs in or near the CD36 locus (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Of these, 55 were common to the white and black sets, 14 were unique to the white set, and 3 were unique to the black set. These SNPs span 85.74 kb ranging from position 80 220 608 to 80 306 350 and fully cover 19 of the 20 known CD36 transcripts. The partially covered transcript had not yet been identified when our study was designed and initiated.
Multivariate SNP analyses
To jointly test the genetic association of all SNPs uniquely mapped to a given gene with CD36 MFI, we fit a least-squares kernel machine (LSKM) model41 using a weighted IBS kernel, with untransformed CD36 MFI as the response variable and predictor variables sex, first genetic principal component score, and an unspecified function of the joint vector of SNP genotypes mapped to a given gene as predictors. SNPs were coded in the kernel as the number of copies of the coded allele. The kernel weights were proportional to the inverse of the square root of the minor allele frequency of the SNP. Association of the set of SNPs from a gene with CD36 MFI was tested using the score test of Kwee et al.41 Only genes with at least 3 uniquely linked filtered SNPs and at least one uniquely linked filtered SNP in the gene were tested. SNPs that did not uniquely map to a gene were discarded. Because the LSKM methodology required complete genotype data, we imputed missing genotypes for typed SNPs in the white subset using IMPUTE, Version 2.1.242 with the pgs option.
Results
Platelet CD36 surface expression varies widely among persons
Among 32 normal healthy volunteers in the initial set, platelet CD36 surface expression levels ranged from 0 to 14 000 molecules per platelet (Figure 1A-B) with a mean of 6005, a median of 5766, and an SD of 1484. From this group of 32, a representative subgroup of 4 individual donors were studied repetitively over time periods as long as 3 years and showed consistent levels of CD36 expression on at least 5 analyses with coefficients of variance in all donors < 9% (Figure 1C). Analyses of the same subjects at different times in the day were also consistent (not shown). These results strongly suggest that a genetic component determines expression levels. As reported by others,43 we found that when platelets were activated CD36 levels increased by approximately 20% (not shown). This group of volunteers, however, were healthy nonsmokers with no history of cardiovascular disease and thus highly unlikely to have increased numbers of circulating activated platelets. In addition, we found no significant difference in the percentage increase in CD36 expression after ex vivo platelet activation among donors regardless of resting levels of expression, suggesting that the wide variability in platelet CD36 expression cannot be accounted for by differences in degrees of in vivo or ex vivo platelet activation among donors.
Variation of platelet CD36 surface expression in a normal population. PRP from human subjects was incubated with either PE-conjugated anti-CD36 IgG or its isotype control and then analyzed by flow cytometry. (A) Left panel: representative histogram from a subject expressing high CD36 levels. Middle panel: Representative histogram from a subject expressing low CD36 levels. Right panel: histogram from a CD36 null subject. (B) CD36 expression levels on platelets from 32 normal volunteer donors were quantified as described in the “Quantitative assay of platelet CD36 expression.” Each triangle represents a single donor at one point in time. (C) Four donors were analyzed at least 5 times over time periods of 3 to 6 months. Bar graph represents the mean and coefficient of variance of platelet CD36 expression.
Variation of platelet CD36 surface expression in a normal population. PRP from human subjects was incubated with either PE-conjugated anti-CD36 IgG or its isotype control and then analyzed by flow cytometry. (A) Left panel: representative histogram from a subject expressing high CD36 levels. Middle panel: Representative histogram from a subject expressing low CD36 levels. Right panel: histogram from a CD36 null subject. (B) CD36 expression levels on platelets from 32 normal volunteer donors were quantified as described in the “Quantitative assay of platelet CD36 expression.” Each triangle represents a single donor at one point in time. (C) Four donors were analyzed at least 5 times over time periods of 3 to 6 months. Bar graph represents the mean and coefficient of variance of platelet CD36 expression.
To replicate these findings, we studied platelets from a larger sample of 500 successive subjects recruited through the Cleveland Clinic Cardiac Catheterization Laboratory. Demographic and clinical data on this population are shown in Table 1. As with the normal volunteers, this group also showed a wide variability in platelet CD36 expression (Figure 2A) with a mean of 7876 ± 1924 molecules per platelet and a median of 7611 molecules per platelet. There was no significant difference in CD36 expression levels between male and female subjects (Figure 2B; P = .31). The narrower range of expression among this cohort compared with our original 32 subjects results from original group containing several persons of Asian ancestry who were platelet CD36 null (Naka−).
Variation of platelet CD36 surface expression in a larger population. Platelet CD36 expression was quantified by flow cytometry as in Figure 1 in a population of 500 patients presenting to the cardiac catheterization laboratory of Cleveland Clinic Foundation. (A) Each diamond represents a single patient. (B) CD36 expression levels in male and female patients. No significant difference was seen (P = .31).
Variation of platelet CD36 surface expression in a larger population. Platelet CD36 expression was quantified by flow cytometry as in Figure 1 in a population of 500 patients presenting to the cardiac catheterization laboratory of Cleveland Clinic Foundation. (A) Each diamond represents a single patient. (B) CD36 expression levels in male and female patients. No significant difference was seen (P = .31).
CD36 expression levels correlate with platelet reactivity to oxLDL
To assess the functional consequences of the variability in CD36 expression, we studied the effect of oxLDL on platelet aggregation and α-granule release in response to low-dose ADP (1-4μM) from donors with a wide range of CD36 expression levels. oxLDL induced a significant increase in the rate and extent of platelet aggregation (Figure 3A top left panel) in PRP from high CD36-expressing subjects but not from CD36 null donors (Figure 3A right lower panel). The maximal extent of aggregation to 2μM ADP increased from 21% to 44% when oxLDL was added (P < .05). Donors with intermediate (lower left) and low (upper right) CD36 expression levels showed correspondingly lower levels of response to oxLDL, increasing the maximum extent of aggregation by 5% to 15%. Similar results were seen using 4μM ADP or low doses of collagen (50 μg/mL) as an agonist. At maximum agonist concentrations (eg, 20μM ADP), oxLDL did not induce further increases in the extent of aggregation (not shown). We then assessed platelet P-selectin expression by quantitative flow cytometry in a series of donors and showed that platelet activation in response to low-dose ADP plus oxLDL correlated well (r2 = 0.87) with the level of CD36 surface expression (Figure 3B).
Platelet CD36 expression levels correlate with activation response to oxLDL. (A) PRP from 4 different healthy subjects expressing high, medium, low, or no platelet CD36 were incubated with oxLDL 50 μg/mL for 30 minutes and then stimulated with low-dose ADP (2μM). Aggregation was assessed turbidometrically with a dual-chamber aggregometer. Tracings show representative aggregometry curves from triplicate assays performed on platelets from 2 CD36 null donors and from 5 or more donors from each of the 3 other groups. oxLDL at this concentration by itself did not effect platelet aggregation. (B) Washed platelets from 7 normal healthy subjects with levels of CD36 expression ranging from 0 to more than 12 000 molecules/platelet were incubated with oxLDL 50 μg/mL for 30 minutes, and platelet α-granule release was quantified by flow cytometry using anti–P-selectin IgG. The graph plots mean fluorescence intensity of anti–P-selectin binding against surface expression of CD36 and shows a correlation coefficient (r2) of 0.87.
Platelet CD36 expression levels correlate with activation response to oxLDL. (A) PRP from 4 different healthy subjects expressing high, medium, low, or no platelet CD36 were incubated with oxLDL 50 μg/mL for 30 minutes and then stimulated with low-dose ADP (2μM). Aggregation was assessed turbidometrically with a dual-chamber aggregometer. Tracings show representative aggregometry curves from triplicate assays performed on platelets from 2 CD36 null donors and from 5 or more donors from each of the 3 other groups. oxLDL at this concentration by itself did not effect platelet aggregation. (B) Washed platelets from 7 normal healthy subjects with levels of CD36 expression ranging from 0 to more than 12 000 molecules/platelet were incubated with oxLDL 50 μg/mL for 30 minutes, and platelet α-granule release was quantified by flow cytometry using anti–P-selectin IgG. The graph plots mean fluorescence intensity of anti–P-selectin binding against surface expression of CD36 and shows a correlation coefficient (r2) of 0.87.
CD36 expression level is associated with inheritance of specific CD36 SNPs
Our targeted genome-wide scan across 34 088 filtered SNPs for association with platelet CD36 expression level in whites (n = 374) identified 24 SNPs with q values (false discovery rate) < 0.05 (Table 2). rs2058703, located in the 3′-untranslated region of BCL11A, was the only non-CD36 SNP to meet this statistical criterion. Six of these 24 SNPs, all within the CD36 locus, met the Bonferroni genome-wide significance threshold of 0.10 divided by 34 088 = 2.93 × 10−6. The genome-wide min-p P value for the strongest SNP rs3211870 was 0.057. Figure 4 shows a Manhattan plot for the data with the Bonferroni threshold shown as a horizontal black line. The estimated genomic control λ-inflation factor for the scan was 1.021.
SNPs showing strongest association with platelet CD36 expression in GWAS of whites
| SNP . | Chromosome . | Position . | A1/2 . | Gene . | CAF . | β . | P* . | q† . |
|---|---|---|---|---|---|---|---|---|
| rs3211870 | 7 | 80 287 209 | G/A | CD36 | 0.42 | 0.55 | .00000127 | 0.012 |
| rs17154155 | 7 | 80 234 243 | C/A | CD36 | 0.42 | 0.53 | .00000163 | 0.012 |
| rs3211849 | 7 | 80 283 323 | A/G | CD36 | 0.45 | 0.54 | .00000172 | 0.012 |
| rs1761662 | 7 | 80 238 463 | G/A | CD36 | 0.46 | 0.52 | .00000176 | 0.012 |
| rs7807607 | 7 | 80 226 885 | G/A | CD36 | 0.45 | 0.51 | .00000211 | 0.012 |
| rs3211842 | 7 | 80 282 636 | G/A | CD36 | 0.43 | 0.53 | .00000218 | 0.012 |
| rs1358337 | 7 | 80 288 385 | G/A | CD36 | 0.43 | 0.52 | .00000394 | 0.015 |
| rs1761667 | 7 | 80 244 939 | A/G | CD36 | 0.47 | 0.51 | .00000417 | 0.015 |
| rs2058703 | 2 | 60 679 226 | A/G | BCL11A | 0.45 | 0.49 | .00000423 | 0.015 |
| rs12706912 | 7 | 80 252 295 | A/C | CD36 | 0.44 | 0.51 | .00000515 | 0.015 |
| rs1049654 | 7 | 80 275 455 | C/A | CD36 | 0.46 | 0.50 | .00000560 | 0.015 |
| rs6960369 | 7 | 80 249 319 | G/A | CD36 | 0.07 | 1.01 | .00000577 | 0.015 |
| rs3211821 | 7 | 80 278 563 | G/A | CD36 | 0.46 | 0.50 | .00000584 | 0.015 |
| rs4545029 | 7 | 80 252 518 | C/A | CD36 | 0.47 | 0.50 | .00000689 | 0.015 |
| rs10480808 | 7 | 80 250 122 | A/T | CD36 | 0.44 | 0.51 | .00000807 | 0.015 |
| rs10499859 | 7 | 80 258 810 | G/A | CD36 | 0.47 | 0.50 | .00000921 | 0.015 |
| rs9784998 | 7 | 80 263 001 | G/A | CD36 | 0.07 | 0.98 | .00000951 | 0.015 |
| rs3211817 | 7 | 80 278 107 | A/C | CD36 | 0.07 | 0.98 | .00000981 | 0.015 |
| rs3211830 | 7 | 80 279 718 | G/A | CD36 | 0.07 | 0.98 | .00000951 | 0.015 |
| rs3211867 | 7 | 80 286 940 | C/A | CD36 | 0.07 | 0.98 | .00000951 | 0.015 |
| rs1924 | 7 | 80 291 398 | G/A | CD36 | 0.07 | 0.98 | .00000951 | 0.015 |
| rs11971740 | 7 | 80 245 921 | A/G | CD36 | 0.07 | 0.97 | .00000108 | 0.017 |
| rs4731642 | 7 | 80 248 001 | A/G | CD36 | 0.44 | 0.48 | .00000224 | 0.033 |
| rs1534314 | 7 | 80 271 108 | G/A | CD36 | 0.07 | 0.94 | .00000231 | 0.033 |
| SNP . | Chromosome . | Position . | A1/2 . | Gene . | CAF . | β . | P* . | q† . |
|---|---|---|---|---|---|---|---|---|
| rs3211870 | 7 | 80 287 209 | G/A | CD36 | 0.42 | 0.55 | .00000127 | 0.012 |
| rs17154155 | 7 | 80 234 243 | C/A | CD36 | 0.42 | 0.53 | .00000163 | 0.012 |
| rs3211849 | 7 | 80 283 323 | A/G | CD36 | 0.45 | 0.54 | .00000172 | 0.012 |
| rs1761662 | 7 | 80 238 463 | G/A | CD36 | 0.46 | 0.52 | .00000176 | 0.012 |
| rs7807607 | 7 | 80 226 885 | G/A | CD36 | 0.45 | 0.51 | .00000211 | 0.012 |
| rs3211842 | 7 | 80 282 636 | G/A | CD36 | 0.43 | 0.53 | .00000218 | 0.012 |
| rs1358337 | 7 | 80 288 385 | G/A | CD36 | 0.43 | 0.52 | .00000394 | 0.015 |
| rs1761667 | 7 | 80 244 939 | A/G | CD36 | 0.47 | 0.51 | .00000417 | 0.015 |
| rs2058703 | 2 | 60 679 226 | A/G | BCL11A | 0.45 | 0.49 | .00000423 | 0.015 |
| rs12706912 | 7 | 80 252 295 | A/C | CD36 | 0.44 | 0.51 | .00000515 | 0.015 |
| rs1049654 | 7 | 80 275 455 | C/A | CD36 | 0.46 | 0.50 | .00000560 | 0.015 |
| rs6960369 | 7 | 80 249 319 | G/A | CD36 | 0.07 | 1.01 | .00000577 | 0.015 |
| rs3211821 | 7 | 80 278 563 | G/A | CD36 | 0.46 | 0.50 | .00000584 | 0.015 |
| rs4545029 | 7 | 80 252 518 | C/A | CD36 | 0.47 | 0.50 | .00000689 | 0.015 |
| rs10480808 | 7 | 80 250 122 | A/T | CD36 | 0.44 | 0.51 | .00000807 | 0.015 |
| rs10499859 | 7 | 80 258 810 | G/A | CD36 | 0.47 | 0.50 | .00000921 | 0.015 |
| rs9784998 | 7 | 80 263 001 | G/A | CD36 | 0.07 | 0.98 | .00000951 | 0.015 |
| rs3211817 | 7 | 80 278 107 | A/C | CD36 | 0.07 | 0.98 | .00000981 | 0.015 |
| rs3211830 | 7 | 80 279 718 | G/A | CD36 | 0.07 | 0.98 | .00000951 | 0.015 |
| rs3211867 | 7 | 80 286 940 | C/A | CD36 | 0.07 | 0.98 | .00000951 | 0.015 |
| rs1924 | 7 | 80 291 398 | G/A | CD36 | 0.07 | 0.98 | .00000951 | 0.015 |
| rs11971740 | 7 | 80 245 921 | A/G | CD36 | 0.07 | 0.97 | .00000108 | 0.017 |
| rs4731642 | 7 | 80 248 001 | A/G | CD36 | 0.44 | 0.48 | .00000224 | 0.033 |
| rs1534314 | 7 | 80 271 108 | G/A | CD36 | 0.07 | 0.94 | .00000231 | 0.033 |
All SNPs with q value ≤ 0.05 using the white subset ordered by P value.
Position indicates base pair location using NCBI Build 37.p2, dbSNP Build 131; A1/2, allele 1/allele 2; CAF, coded allele, allele 2, frequency; and β, SNP estimated regression coefficient from the linear regression of CD36 MFI on sex, first genetic principal component, and SNP, where SNP was coded as number of copies of allele 2.
Genomic control-adjusted P value from F test of β = 0.
FDR q value from P value.
Manhattan plot of SNP-CD36 MFI associations from the white cohort. Chromosome and position information from NCBI Build 37/hg19. Black horizontal line indicates the 0.10 Bonferroni threshold.
Manhattan plot of SNP-CD36 MFI associations from the white cohort. Chromosome and position information from NCBI Build 37/hg19. Black horizontal line indicates the 0.10 Bonferroni threshold.
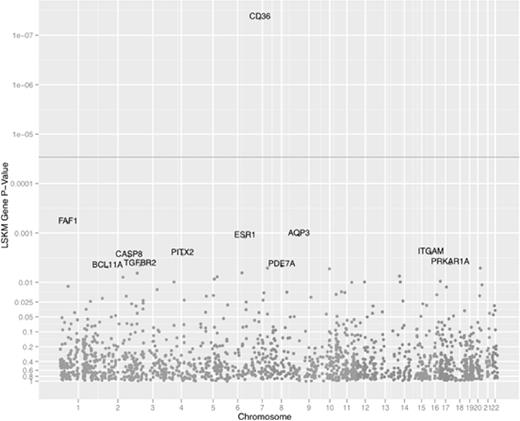
We also conducted a gene-based genome-wide scan on the white set using a one-degree of freedom LSKM score test. The LSKM test is a multi-SNP semiparametric test that jointly tests the association of all SNPs in a gene with the phenotype while accounting for the linkage disequilibrium among SNPs. It is a powerful intermediate approach between single SNP tests and multiple degrees of freedom haplotype tests. Figure 5 is a Manhattan plot of the gene-based scan across 1706 genes with at least 3 filtered SNPs. CD36 had the lowest gene-based P value, 4.55 × 10−8, for genetic association with platelet CD36 expression. This P value was considerably smaller than the smallest CD36 SNP P value. The FAF1 gene on chromosome 1 had the next smallest gene-based P value (6.19 × 10−4), but only CD36 met the Bonferroni threshold of .05 divided by 1706 = 2.93 × 10−5 or the q value threshold of 0.05.
Manhattan plot of gene-CD36 MFI associations from the LSKM genome-wide scan from the white cohort. Chromosome and gene position information from hg19. Dark gray horizontal line indicates the 0.05 Bonferroni threshold.
Manhattan plot of gene-CD36 MFI associations from the LSKM genome-wide scan from the white cohort. Chromosome and gene position information from hg19. Dark gray horizontal line indicates the 0.05 Bonferroni threshold.
A focused “cis” analysis considering only the SNPs located in or near the CD36 locus (listed in supplemental Table 1) identified 31 SNPs with min-p P values < .05 for genetic association with platelet CD36 expression levels (Table 3). Seven of the top 10 CD36 SNPs identified by this analysis have also been associated with various CD36-relevant phenotypes in other published studies,26-33 strengthening the validity of our findings. Lastly, we estimated linkage disequilibrium (LD), measured by R2, of each tested SNP with the strongest SNP (rs3211870). Most of the top SNPs were in strong LD with rs3211870 as indicated by the dark red color of their plotting symbols in Figure 6, although the observed CD36 LD structure was not very block-like. The statistical associations with CD36 expression levels quickly weakened just upstream of rs3211870, where the LD with rs3211870 also diminished.
CD36 SNPs showing strongest association with platelet CD36 expression level
| SNP . | Position . | A1/2 . | Whites . | Blacks . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CAF . | β . | P* . | PP† . | CAF . | β . | P* . | PP† . | |||
| rs3211870 | 21.384 | G/A | 0.42 | 0.55 | 0.000001.27 | < 0.0001 | 0.44 | 1.30 | 0.005 | 0.217 |
| rs17154155 | 74.350 | C/A | 0.42 | 0.53 | 0.000001.63 | < 0.0001 | 0.40 | 1.07 | 0.0256 | 0.517 |
| rs3211849 | 25.270 | A/G | 0.45 | 0.54 | 0.00000172 | < 0.0001 | 0.52 | 1.47 | 0.000544 | 0.058 |
| rs1761662 | 70.130 | G/A | 0.46 | 0.52 | 0.00000176 | < 0.0001 | 0.70 | 0.92 | 0.078 | 0.819 |
| rs7807607 | 81.708 | G/A | 0.45 | 0.51 | 0.00000211 | < 0.0001 | 0.62 | 0.77 | 0.208 | 0.985 |
| rs3211842 | 25.957 | G/A | 0.43 | 0.53 | 0.00000218 | < 0.0001 | 0.38 | 1.62 | 0.000287 | 0.041 |
| rs1358337 | 20.208 | G/A | 0.43 | 0.52 | 0.00000344 | < 0.0001 | 0.50 | 1.54 | 0.0000456 | 0.014 |
| rs1761667 | 63.654 | A/G | 0.47 | 0.51 | 0.00000417 | < 0.0001 | 0.54 | 1.83 | 0.0000276 | 0.011 |
| rs12706912 | 56.298 | A/C | 0.44 | 0.51 | 0.00000515 | 0.0001 | 0.48 | 1.39 | 0.0182 | 0.436 |
| rs1049654 | 33.138 | C/A | 0.46 | 0.50 | 0.00000560 | 0.0001 | 0.62 | 1.27 | 0.0131 | 0.371 |
| rs6960369 | 59.274 | G/A | 0.07 | 1.01 | 0.00000577 | 0.0001 | 0.27 | 0.24 | 0.74 | 1.000 |
| rs3211821 | 30.030 | G/A | 0.46 | 0.50 | 0.00000584 | 0.0001 | 0.70 | 1.01 | 0.0464 | 0.674 |
| rs4545029 | 56.075 | C/A | 0.47 | 0.50 | 0.00000659 | 0.0001 | 0.60 | 1.69 | 0.000147 | 0.028 |
| rs10480808 | 58.471 | A/T | 0.44 | 0.51 | 0.00000807 | 0.0003 | 0.44 | 1.65 | 0.000031 | 0.012 |
| rs10499859 | 49.783 | G/A | 0.47 | 0.50 | 0.00000921 | 0.0003 | 0.66 | 1.31 | 0.0269 | 0.530 |
| rs9784998 | 45.592 | G/A | 0.07 | 0.98 | 0.00000951 | 0.0003 | 0.24 | 1.43 | 0.0309 | 0.565 |
| rs3211817 | 30.486 | A/C | 0.07 | 0.98 | 0.00000951 | 0.0003 | NA | NA | NA | NA |
| rs3211830 | 28.875 | G/A | 0.07 | 0.98 | 0.00000951 | 0.0003 | 0.10 | 0.09 | 0.95 | 1.000 |
| rs3211867 | 21.653 | C/A | 0.07 | 0.98 | 0.00000951 | 0.0003 | 0.24 | 1.43 | 0.0309 | 0.565 |
| rs1924 | 17.195 | G/A | 0.07 | 0.98 | 0.00000951 | 0.0003 | 0.28 | 0.99 | 0.151 | 0.954 |
| rs11971740 | 62.672 | A/G | 0.07 | 0.97 | 0.0000108 | 0.0004 | 0.26 | 0.93 | 0.19 | 0.978 |
| rs4731642 | 60.592 | A/G | 0.44 | 0.48 | 0.0000224 | 0.0005 | 0.12 | 0.62 | 0.38 | 1.000 |
| rs1534314 | 37.485 | G/A | 0.07 | 0.94 | 0.0000231 | 0.0005 | 0.24 | 1.43 | 0.0309 | 0.565 |
| rs9918586 | 41.857 | A/G | 0.07 | 0.90 | 0.0000429 | 0.0010 | 0.30 | 0.96 | 0.098 | 0.875 |
| rs3211869 | 21.441 | T/A | 0.06 | 0.96 | 0.000113 | 0.0031 | 0.16 | 1.97 | 0.00486 | 0.214 |
| rs1537593 | 54.682 | G/A | 0.09 | 0.73 | 0.000146 | 0.0035 | 0.36 | 1.50 | 0.00688 | 0.259 |
| rs3211883 | 19.550 | T/A | 0.10 | 0.69 | 0.000298 | 0.0064 | 0.54 | 0.20 | 0.684 | 1.000 |
| rs10499858 | 72.747 | A/G | 0.06 | 0.81 | 0.000478 | 0.0100 | 0.14 | 1.66 | 0.026 | 0.521 |
| rs11771839 | 79.372 | C/A | 0.06 | 0.80 | 0.000512 | 0.0104 | 0.14 | 1.66 | 0.026 | 0.521 |
| rs2065668 | 87.985 | G/A | 0.35 | 0.40 | 0.000765 | 0.0147 | 0.26 | 0.38 | 0.472 | 1.000 |
| rs3211908 | 14.677 | G/A | 0.05 | 0.78 | 0.00212 | 0.0379 | NA | NA | NA | NA |
| SNP . | Position . | A1/2 . | Whites . | Blacks . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CAF . | β . | P* . | PP† . | CAF . | β . | P* . | PP† . | |||
| rs3211870 | 21.384 | G/A | 0.42 | 0.55 | 0.000001.27 | < 0.0001 | 0.44 | 1.30 | 0.005 | 0.217 |
| rs17154155 | 74.350 | C/A | 0.42 | 0.53 | 0.000001.63 | < 0.0001 | 0.40 | 1.07 | 0.0256 | 0.517 |
| rs3211849 | 25.270 | A/G | 0.45 | 0.54 | 0.00000172 | < 0.0001 | 0.52 | 1.47 | 0.000544 | 0.058 |
| rs1761662 | 70.130 | G/A | 0.46 | 0.52 | 0.00000176 | < 0.0001 | 0.70 | 0.92 | 0.078 | 0.819 |
| rs7807607 | 81.708 | G/A | 0.45 | 0.51 | 0.00000211 | < 0.0001 | 0.62 | 0.77 | 0.208 | 0.985 |
| rs3211842 | 25.957 | G/A | 0.43 | 0.53 | 0.00000218 | < 0.0001 | 0.38 | 1.62 | 0.000287 | 0.041 |
| rs1358337 | 20.208 | G/A | 0.43 | 0.52 | 0.00000344 | < 0.0001 | 0.50 | 1.54 | 0.0000456 | 0.014 |
| rs1761667 | 63.654 | A/G | 0.47 | 0.51 | 0.00000417 | < 0.0001 | 0.54 | 1.83 | 0.0000276 | 0.011 |
| rs12706912 | 56.298 | A/C | 0.44 | 0.51 | 0.00000515 | 0.0001 | 0.48 | 1.39 | 0.0182 | 0.436 |
| rs1049654 | 33.138 | C/A | 0.46 | 0.50 | 0.00000560 | 0.0001 | 0.62 | 1.27 | 0.0131 | 0.371 |
| rs6960369 | 59.274 | G/A | 0.07 | 1.01 | 0.00000577 | 0.0001 | 0.27 | 0.24 | 0.74 | 1.000 |
| rs3211821 | 30.030 | G/A | 0.46 | 0.50 | 0.00000584 | 0.0001 | 0.70 | 1.01 | 0.0464 | 0.674 |
| rs4545029 | 56.075 | C/A | 0.47 | 0.50 | 0.00000659 | 0.0001 | 0.60 | 1.69 | 0.000147 | 0.028 |
| rs10480808 | 58.471 | A/T | 0.44 | 0.51 | 0.00000807 | 0.0003 | 0.44 | 1.65 | 0.000031 | 0.012 |
| rs10499859 | 49.783 | G/A | 0.47 | 0.50 | 0.00000921 | 0.0003 | 0.66 | 1.31 | 0.0269 | 0.530 |
| rs9784998 | 45.592 | G/A | 0.07 | 0.98 | 0.00000951 | 0.0003 | 0.24 | 1.43 | 0.0309 | 0.565 |
| rs3211817 | 30.486 | A/C | 0.07 | 0.98 | 0.00000951 | 0.0003 | NA | NA | NA | NA |
| rs3211830 | 28.875 | G/A | 0.07 | 0.98 | 0.00000951 | 0.0003 | 0.10 | 0.09 | 0.95 | 1.000 |
| rs3211867 | 21.653 | C/A | 0.07 | 0.98 | 0.00000951 | 0.0003 | 0.24 | 1.43 | 0.0309 | 0.565 |
| rs1924 | 17.195 | G/A | 0.07 | 0.98 | 0.00000951 | 0.0003 | 0.28 | 0.99 | 0.151 | 0.954 |
| rs11971740 | 62.672 | A/G | 0.07 | 0.97 | 0.0000108 | 0.0004 | 0.26 | 0.93 | 0.19 | 0.978 |
| rs4731642 | 60.592 | A/G | 0.44 | 0.48 | 0.0000224 | 0.0005 | 0.12 | 0.62 | 0.38 | 1.000 |
| rs1534314 | 37.485 | G/A | 0.07 | 0.94 | 0.0000231 | 0.0005 | 0.24 | 1.43 | 0.0309 | 0.565 |
| rs9918586 | 41.857 | A/G | 0.07 | 0.90 | 0.0000429 | 0.0010 | 0.30 | 0.96 | 0.098 | 0.875 |
| rs3211869 | 21.441 | T/A | 0.06 | 0.96 | 0.000113 | 0.0031 | 0.16 | 1.97 | 0.00486 | 0.214 |
| rs1537593 | 54.682 | G/A | 0.09 | 0.73 | 0.000146 | 0.0035 | 0.36 | 1.50 | 0.00688 | 0.259 |
| rs3211883 | 19.550 | T/A | 0.10 | 0.69 | 0.000298 | 0.0064 | 0.54 | 0.20 | 0.684 | 1.000 |
| rs10499858 | 72.747 | A/G | 0.06 | 0.81 | 0.000478 | 0.0100 | 0.14 | 1.66 | 0.026 | 0.521 |
| rs11771839 | 79.372 | C/A | 0.06 | 0.80 | 0.000512 | 0.0104 | 0.14 | 1.66 | 0.026 | 0.521 |
| rs2065668 | 87.985 | G/A | 0.35 | 0.40 | 0.000765 | 0.0147 | 0.26 | 0.38 | 0.472 | 1.000 |
| rs3211908 | 14.677 | G/A | 0.05 | 0.78 | 0.00212 | 0.0379 | NA | NA | NA | NA |
All CD36 SNPs with PP values ≤ 0.05 using the white subset ordered by P value.
Position indicates base pair location using NCBI Build 37.p2, dbSNP Build 131; A1/2, allele 1/allele 2; CAF, coded allele, allele 2, frequency; β, SNP estimated regression coefficient from the linear regression of CD36 MFI on sex, first genetic principal component, and SNP, where SNP was coded as number of copies of allele 2; and NA, not applicable (SNP was dropped during QC filtering).
Genomic control-adjusted P value from F test of β = 0.
min-p permutation P value of β = 0 based on 10 000 permutations restricted to CD36 SNPs. Regressions fit separately for white and black subsets.
CD36 SNP P values for association with CD36 MFI.P values are listed in supplemental Table 1. CAF indicates coded allele frequency; and R2, estimated r2 of SNP with top SNP rs3211870, estimated separately for each subset (white and black). Dark gray horizontal line represents the Bonferroni 0.05 threshold for the CD36 cis analysis. SNPs with dark labels have been associated with lipid, metabolic, or CVD phenotypes in the literature (supplemental Table 2).
CD36 SNP P values for association with CD36 MFI.P values are listed in supplemental Table 1. CAF indicates coded allele frequency; and R2, estimated r2 of SNP with top SNP rs3211870, estimated separately for each subset (white and black). Dark gray horizontal line represents the Bonferroni 0.05 threshold for the CD36 cis analysis. SNPs with dark labels have been associated with lipid, metabolic, or CVD phenotypes in the literature (supplemental Table 2).
We also analyzed genetic associations with platelet CD36 expression levels in the smaller black cohort (25 subjects; 58 SNPs). As shown in Table 3, of the top 31 SNPs identified in the cis analysis of the white subset, 19 also showed evidence of genetic association at P < .05 in the black cohort. Importantly, directions of the estimated effects were consistent between analyses for all of the SNPs, and 6 of the 10 most strongly associated CD36 SNPs in the black cohort have also been associated with various CD36-relevant phenotypes in other published studies.26,27
Although the size of the black cohort was very small, we were able to perform a genome-wide scan (28 446 SNPs) of the cohort and identified rs7803660, an intronic SNP in the EGFR gene, as the sole SNP to meet the Bonferroni significance threshold; the genomic control-corrected P value was 7.54 × 10−7, and the genome-wide min-p P value was .691 based on 1000 permutations, indicating that our smallest observed P value was not unusual. Although not reaching the Bonferroni threshold, 3 of the top 15 SNPs by P value were in the CD36 locus; among these, rs1761667 had the smallest P value (2.76 × 10−5). The estimated genomic control λ-inflation factor for the scan was 1.087.
Discussion
This study represents the first systematic examination of platelet CD36 expression in normal human subjects and reveals a surprisingly wide range. Among those persons who are not Naka− (ie, platelet CD36 null), expression on resting platelets ranged from approximately 2000 to 14 000 molecules/platelet. Importantly, this wide range of expression was also observed in a larger population of subjects of mixed age, sex, and ethnicity who presented to the Cleveland Clinic Cardiac Catheterization Laboratory. CD36 surface expression has been reported to increase in response to platelet activation.43 Because CD36 is expressed on the membranes of α-granules and the open canalicular system,44 this is probably the result of increased accessibility of this pool of CD36 to antibodies after platelets are activated. Indeed, we also observed an approximately 20% increment in anti-CD36 antibody binding after platelet activation. This effect was similar among donors regardless of their basal level of CD36 expression (data not shown); thus, variations in platelet activation in vivo or during blood processing ex vivo cannot account for the large degree of individual CD36 expression variance observed. Platelet CD36 expression levels were consistent in individual donors over time periods as long as 3 years and did not vary significantly with the time of day of phlebotomy.
Although these data cannot define the degree of heritability of CD36 expression levels, they strongly suggest that a significant component of the population variability is genetically determined. CD36 is a large and highly polymorphic gene, yet systematic studies relating its genetic polymorphisms to protein expression are extremely limited. In one of the few such reports, Love-Gregory et al analyzed a cohort of 74 black subjects and found associations of 5′-SNPs with total level of CD36 protein in peripheral blood monocytes.27 We now, for the first time, report strong associations of SNPs in the CD36 gene with platelet surface CD36 expression levels. We used multiple statistical methods to analyze data obtained from 440 subjects from the Cleveland Clinic ASCLOGEN study. Both SNP-based and gene-based GWAS in the white cohort (Figures 4, 5) identified the CD36 locus as highly associated with platelet CD36 expression levels with significant P values, even after rigorous Bonferroni correction. In addition, a more highly powered parallel analysis looking only at the available SNPs in or near CD36 also showed that multiple CD36 SNPs were strongly associated with CD36 expression in both white and black cohorts (Table 3). Although we did not measure monocyte CD36 expression levels, our data are concordant with the smaller study of Love-Gregory et al27 (Table 4) in that minor alleles in 4 CD36 SNPs that they reported to be associated with monocyte CD36 expression in blacks were associated with the same direction of platelet CD36 expression in our student in both the white and black populations. Their study also included a small subset of 57 black subjects in whom total platelet CD36 was assessed by semiquantitative Western blot. Mostly weak associations with CD36 SNPs were found in that analysis, including some identified in both our white and black cohorts.
CD36 SNPs associated with published phenotypes and showing evidence of association with platelet CD36 expression
| SNP . | Rank* . | P† . | Lit P‡ . | Direction§ . | Phenotype . | Reference . |
|---|---|---|---|---|---|---|
| rs3211870 | 1 | 0.00000127 | 0.0079 | −+ | HDL | 26 |
| rs3211870 | 1 | 0.00000127 | 0.005 | ++ | TMP | 27 |
| rs3211849 | 3 | 0.00000172 | 0.029 | −+ | HDL | 26 |
| rs7807607 | 5 | 0.00000211 | 0.07 | ++ | MS | 30 |
| rs3211842 | 6 | 0.00000218 | 0.00074 | +− | HDL | 26 |
| rs1358337 | 7 | 0.00000344 | 0.00066 | −+ | HDL | 26 |
| rs1761667 | 8 | 0.00000417 | 0.00035 | ++ | TMP | 27 |
| rs1761667 | 8 | 0.00000417 | 0.028 | +− | FFA | 29 |
| rs1049654 | 10 | 0.00000560 | 0.0028 | −+ | HDL | 26 |
| rs10499859 | 15 | 0.00000921 | 0.003 | − | TMP | 27 |
| rs10499859 | 15 | 0.00000921 | 0.00028 | −+ | HDL | 26 |
| rs3211867 | 19 | 0.00000951 | 0.003 | ++ | Obesity | 31 |
| rs9784998 | 20 | 0.00000951 | 0.026 | ++ | TMP | 27 |
| rs9784998 | 20 | 0.00000951 | 0.015 | +− | HDL | 26 |
| rs3211883 | 27 | 0.000298 | 0.007 | ++ | Obesity | 31 |
| rs3211908 | 31 | 0.00212 | 0.0005 | ++ | Obesity | 31 |
| rs1984112 | 35 | 0.00788 | 0.008 | +− | FFA | 29 |
| rs2151916 | 43 | 0.0221 | 0.02 | +− | LDL | 28 |
| SNP . | Rank* . | P† . | Lit P‡ . | Direction§ . | Phenotype . | Reference . |
|---|---|---|---|---|---|---|
| rs3211870 | 1 | 0.00000127 | 0.0079 | −+ | HDL | 26 |
| rs3211870 | 1 | 0.00000127 | 0.005 | ++ | TMP | 27 |
| rs3211849 | 3 | 0.00000172 | 0.029 | −+ | HDL | 26 |
| rs7807607 | 5 | 0.00000211 | 0.07 | ++ | MS | 30 |
| rs3211842 | 6 | 0.00000218 | 0.00074 | +− | HDL | 26 |
| rs1358337 | 7 | 0.00000344 | 0.00066 | −+ | HDL | 26 |
| rs1761667 | 8 | 0.00000417 | 0.00035 | ++ | TMP | 27 |
| rs1761667 | 8 | 0.00000417 | 0.028 | +− | FFA | 29 |
| rs1049654 | 10 | 0.00000560 | 0.0028 | −+ | HDL | 26 |
| rs10499859 | 15 | 0.00000921 | 0.003 | − | TMP | 27 |
| rs10499859 | 15 | 0.00000921 | 0.00028 | −+ | HDL | 26 |
| rs3211867 | 19 | 0.00000951 | 0.003 | ++ | Obesity | 31 |
| rs9784998 | 20 | 0.00000951 | 0.026 | ++ | TMP | 27 |
| rs9784998 | 20 | 0.00000951 | 0.015 | +− | HDL | 26 |
| rs3211883 | 27 | 0.000298 | 0.007 | ++ | Obesity | 31 |
| rs3211908 | 31 | 0.00212 | 0.0005 | ++ | Obesity | 31 |
| rs1984112 | 35 | 0.00788 | 0.008 | +− | FFA | 29 |
| rs2151916 | 43 | 0.0221 | 0.02 | +− | LDL | 28 |
All SNPs from supplemental Table 1 with P < .05 from the white subset analysis ordered by P value.
TMP indicates total monocyte CD36 protein; MS, metabolic syndrome; and FFA, free fatty acids.
Rank indicates rank of SNP by P value in supplemental Table 1.
P value from supplemental Table 1.
Reported P value in study for association with phenotype.
Direction of SNP effect on CD36 expression, direction of SNP effect reported in study for association with phenotype.
Although few data have been published linking CD36 SNPs to expression levels, there are multiple reports associating CD36 SNPs with phenotypes relevant to the known functions of CD36. These include serum levels of HDL, LDL, triglycerides, and fatty acids as well as risk of atherothrombotic events, obesity, and metabolic syndrome.26-34 The directions of our CD36 expression associations were consistent with those studies: SNPs associated with higher CD36 are those that have been linked to obesity, metabolic syndrome, and dyslipidemia, whereas SNPs associated with low levels are those linked to low circulating fatty acids and high HDL. Table 4 lists 14 CD36 SNPs with P values < .05 that have been associated with other phenotypes in the literature and shows the direction of effect we found on platelet CD36 expression and the direction of other phenotype effects reported in the literature.
The mechanisms by which CD36 genetic polymorphisms influence platelet CD36 expression remain to be determined. The finding that many of the SNPs associated with platelet CD36 levels have also been reported to be associated with lipid phenotypes suggests that the mechanisms may not be entirely megakaryocyte specific. It is possible that the identified SNPs could be located in genomic areas important to CD36 gene transcription or mRNA translation. Our data showed that the SNP with the strongest association with platelet CD36 expression was in strong LD with the other associated SNPs. Thus, it is also quite possible that these SNPs are in LD with other rarer polymorphisms in the coding region of the gene that effect transcription, translation, mRNA stability, or protein stability.
Although our studies showed the highest level of significance for SNPs in the CD36 gene, it is probable that a component of platelet CD36 expression variability relates to polymorphisms/mutations in genes outside the CD36 locus acting in a “trans” manner. Little is known if and how megakaryocyte CD36 gene expression is regulated, but in other cells, such as monocytes, adipocytes, hepatocytes, and myocytes, CD36 gene expression can be regulated by a number of signaling pathways and transcription factors, including peroxisome proliferator-activated receptor-γ, LXR, OCT-2, estrogen, growth hormone, protein kinase C activators, interleukin-4, transforming growth factor-β, and macrophage colony-stimulating factor.3,45-48 It is thus interesting that a SNP outside of the CD36 locus in the BCL11A gene was identified in our GWAS with a P value < 10−5. BCL11a has also been identified in GWAS of fetal hemoglobin levels in patients with inherited hemoglobinopathies.49 The protein product is a transcriptional regulator known to be active in hematopoietic cells; thus, it is interesting to speculate that it could also be involved in regulating platelet CD36, especially because CD36 is known to function as a receptor for malaria-infected erythrocytes1 and CD36 deficiency tracks geographically with sickle cell disease and other hemoglobinopathies.18
The complex CD36 regulatory pathways also make it probable that nongenetic factors contribute to the variability of platelet CD36 expression. CD36 expression in monocytes can be effected by certain drugs (statins, vitamin E, peroxisome proliferator-activated receptor-γ agonists, and HIV protease inhibitors), high-fat diet, oxidant stress, and hyperglycemia.3,4,48 Some studies have reported that platelet CD36 expression was increased by cardiopulmonary bypass,50 or strenuous exercise,43 but in these studies the increases were small and were associated with evidence of platelet activation.
The biologic significance of our findings relates to the potential role of CD36 in promoting thrombus formation. We thus demonstrated, for the first time, that levels of CD36 in healthy subjects correlated closely (r2 = 0.87) with degree of platelet reactivity to oxLDL, a model CD36 ligand, and hypothesize that SNPs or haplotypes that associate with high or low CD36 expression levels might be predictive for thrombotic risk. These data are consistent with our studies of cd36 null mice8,9,11 that showed partial protection from prothrombotic states induced by hyperlipidemia, circulating microparticles, and/or oxidant injury to the vessel wall. We propose that CD36-mediated platelet signals generated in the setting of chronic diseases associated with oxidant stress, hyperlipidemia, or microparticle generation may lower the threshold for platelet activation by other agonists and hence predispose to thrombosis. Our data are also consistent with those of Knowles et al who studied a cohort of subjects presenting with their initial cardiovascular event and found a significant association between a CD36 SNP and those presenting with AMI versus stable angina.32 Similarly, Pellikka et al found an association between a CD36 SNP and risk of fatal prehospitalization AMI.33 A whole genome scan of approximately 9600 subjects enrolled in the Family Blood Pressure Program identified a region of chromosome 7 containing the CD36 locus associated with stroke or AMI.34 These studies are consistent with our hypothesis that CD36 levels could impact thrombotic risk associated with underlying atherosclerosis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ying Hu for technical assistance.
This work was supported by the National Heart, Lung, and Blood Institute Specialized Center of Clinically Oriented Research on Thrombosis (P50 HL81011; R.L.S.).
National Institutes of Health
Authorship
Contribution: A.G. designed, performed, and analyzed experiments and wrote the paper; J.B. and L.Z. performed statistical genetics analysis and contributed to writing the paper; K.C. performed platelet activation experiments; G.M. and Q.W. helped design, perform, and/or interpret genotyping assays; M.F. and K.M. provided input to experimental design and interpretation; R.M.A. coordinated subject accrual and sample procurement; and R.L.S. contributed to overall project design and analyses of experiments and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A.G. was a PhD candidate at Cleveland State University, and this work was submitted in partial fulfillment of the thesis requirement.
The current affiliation of A.G. is Howard Hughes Medical Institute, University of Michigan Life Sciences Institute, Ann Arbor, MI.
Correspondence: Roy L. Silverstein, Department of Cell Biology, Lerner Research Institute NC10, Cleveland Clinic Foundation, 9500 Euclid Ave, Cleveland, OH 44195; e-mail: silverr2@ccf.org.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal