Abstract
Hepcidin, a hormone produced mainly by the liver, has been shown to inhibit both intestinal iron absorption and iron release from macrophages. Hemojuvelin, a glycophosphatidyl inositol–linked membrane protein, acts as a bone morphogenetic protein coreceptor to activate hepcidin expression through a SMAD signaling pathway in hepatocytes. In the present study, we show in mice that loss of hemojuvelin specifically in the liver leads to decreased liver hepcidin production and increased tissue and serum iron levels. Although it does not have any known function outside of the liver, hemojuvelin is expressed at very high levels in cardiac and skeletal muscle. To explore possible roles for hemojuvelin in skeletal muscle, we analyzed conditional knockout mice that lack muscle hemojuvelin. The mutant animals had no apparent phenotypic abnormalities. We found that systemic iron homeostasis and liver hepcidin expression were not affected by loss of hemojuvelin in skeletal muscle regardless of dietary iron content. We conclude that, in spite of its expression pattern, hemojuvelin is primarily important in the liver.
Introduction
Hepcidin is a peptide hormone secreted by the liver that plays a central role in iron homeostasis. Ferroportin, the only known mammalian cellular iron exporter, is expressed on the basolateral membrane of intestinal enterocytes and on the plasma membrane of macrophages. Hepcidin binding to ferroportin leads to internalization and degradation of ferroportin in lysosomes, which decreases iron absorption from the diet and iron release from macrophages that recycle iron from senescent erythrocytes. High levels of hepcidin result in iron deficiency and low levels result in iron overload. This homeostatic mechanism has been confirmed by studies in humans and in mice: loss-of-function mutations in the hepcidin gene (HAMP) cause juvenile hemochromatosis with iron overload,1 inactivation of the hepcidin gene in mice leads to iron overload,2 overexpression of hepcidin in patients with hepatic adenomas causes iron refractory anemia,3 and transgenic mice that overexpress hepcidin develop severe iron deficiency anemia before birth.4
As a central regulator of body iron metabolism, hepcidin is modulated by the same factors that alter iron homeostasis, including changes in body iron stores and in the rates of erythropoiesis, inflammation, anemia, and hypoxia. Hepcidin levels are increased in response to oral and parenteral iron loading and decreased under iron-deficient conditions.5 Inflammation stimulates hepcidin expression by increasing IL-6 levels. The IL-6 ligand-receptor interaction results in activation of the JAK/STAT signaling cascade and binding of the transcriptional activator STAT3 to the hepcidin promoter.6,7 Conversely, anemia and hypoxia are associated with decreased hepcidin levels.8 The von Hippel-Lindau/hypoxia-inducible transcription factor (VHL/HIF) signaling pathway may link hypoxia and hepcidin regulation in vivo. HIF proteins have been reported to bind to the hepcidin promoter and reduce hepcidin expression in the liver in response to hypoxia or iron deficiency,9,10 but a more recent study disputed these findings.11
Studies of patients with autosomal recessive hemochromatosis, an iron-overload disorder, have revealed that several hepatocyte proteins are required for appropriate hepcidin regulation, including HFE,12 TFR2,13 and the hemojuvelin gene HJV (HFE2).14 Mutations in either HAMP or HJV cause a more severe form of hemochromatosis that presents in the first or second decade of life. In juvenile hemochromatosis patients who carry HJV mutations, urinary hepcidin is undetectable, suggesting that HJV is a key regulator of hepcidin.14 To date, approximately 30 disease-linked mutations have been identified in the HJV gene, including missense and nonsense mutations. These mutations are predicted to lead to loss of function of HJV, thus reducing hepcidin expression in hepatocytes.15 Disruption of Hjv in the mouse results in decreased hepcidin expression and increased iron deposition in the liver, pancreas, and heart, but decreased iron levels in tissue macrophages.16,17
HJV encodes hemojuvelin, a member of the repulsive guidance molecule family that resides on the cell membrane as a glycosylphosphatidyl inositol-linked protein. It functions as a bone morphogenetic protein (BMP) coreceptor to activate hepcidin expression through a BMP/SMAD signaling pathway.18 Recently, BMP6 was shown to be the key ligand binding to hemojuvelin in a complex with at least one of the type I BMP receptors.19,20 This ligand-receptor complex initiates a signaling cascade leading to phosphorylation of Smad proteins that associate with Smad4, resulting in nuclear translocation and activation of hepcidin transcription. Accordingly, disease-associated mutations in HJV result in decreased BMP/SMAD signaling and decreased hepcidin expression.18 These findings are supported by the discovery that loss of Smad4 in hepatocytes leads to a failure of hepcidin expression and massive iron overload in the mouse.21
In addition to residing on the cell membrane, hemojuvelin can be cleaved and secreted out of cells in a soluble form. Soluble hemojuvelin can selectively bind to BMP ligands and inhibit endogenous and BMP-induced hepcidin expression.22,23 Administration of soluble hemojuvelin decreases hepcidin expression in vivo, leading to increased ferroportin expression and increased serum iron levels in mice.22 Soluble hemojuvelin is produced by cleavage by furin, a proprotein convertase, at position 332-335 (RNRR).24,25 Furin is induced by iron deficiency and hypoxia in association with stabilization of HIF-1α, suggesting that soluble hemojuvelin production may play an important role in iron deficiency and hypoxia-mediated hepcidin regulation.25 Subsequently, the serine protease TMPRSS6 was shown to be mutated in patients with iron-refractory iron deficiency anemia.26 TMPRSS6 cleaves hemojuvelin on the cell membrane and inhibits hepcidin expression.27 In contrast to the soluble hemojuvelin produced by furin cleavage, the products of TMPRSS6 cleavage do not inhibit BMP-induced hepcidin expression.28
Previous studies showed that HJV is expressed in the heart and muscle at levels exceeding its expression in the liver.14 However, hepcidin is absent in skeletal muscle and is expressed at very low levels in the heart.5 These observations suggest that HJV might have a distinct role in nonhepatic tissues. We hypothesized that soluble hemojuvelin is produced by skeletal muscle and circulates to the liver to regulate hepcidin expression. To test our hypothesis, we investigated hepcidin expression and iron homeostasis in mice lacking hemojuvelin in skeletal muscle.
Methods
Targeting of the murine Hjv locus
We isolated Hjv genomic clones from a strain 129/SvJ mouse library (Stratagene). To construct a targeting vector with exon 4 of Hjv flanked by LoxP sites (Figure 1A), we first retrieved a 10-kb fragment containing all 4 exons of Hjv. We then targeted a LoxP-Neo-LoxP to intron 3, followed by removal of the neomycin (Neo) cassette using Cre recombinase, leaving a single LoxP site in intron 3. A second FRT-Neo-FRT-LoxP cassette was targeted downstream of the 3′ end of exon 4. Linearized vector was electroporated into 129 J1 embryonic stem cells. Correct homologous recombination was confirmed by Southern blot analysis of flanking sequences from both ends of the targeted region. Embryonic stem cell clones with normal karyotypes were injected into C57BL/6 blastocysts. Transmission of the targeted alleles was confirmed by Southern blot analysis. The animals were maintained on an inbred 129S6/SvEvTac background. Subsequent genotyping was carried out by extracting DNA from snipped toe/tail samples and subjecting it to Southern blot and/or PCR analysis. Mouse lines carrying Cre recombinase transgenes under the control of tissue-specific promoters were obtained from The Jackson Laboratory. Conditional knockout mice were obtained by intercrossing male mice of genotype Hjv fl/+;Cre/+ with female mice of genotype Hjv fl/fl. Hjv fl/fl;Cre/+ mice were then compared with littermate control Hjv fl/fl mice.
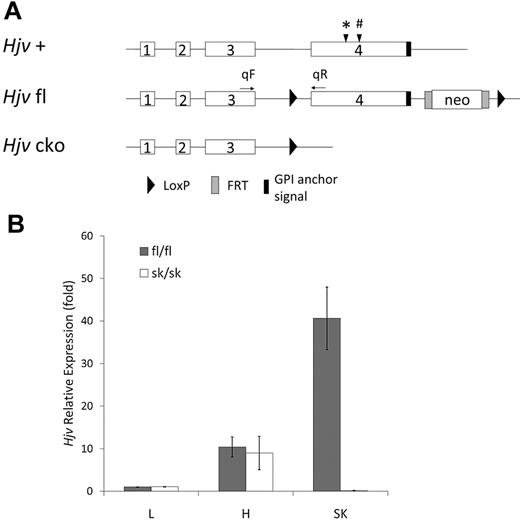
Targeted disruption of the Hjv gene. (A) Schematic representation of the mouse wild-type Hjv genomic sequence (+), LoxP-flanked allele with FRT-flanked Neo sequence (fl), and conditional knock out (cko) alleles. Positions of sequences coding for features of the protein are indicated as follows: (*) indicates the Tmprss6 cleavage site28 and (#) the Furin cleavage site.24,25 (B) Hjv mRNA relative expression in liver (L), heart (H), and skeletal muscle (SK) by quantitative RT-PCR. fl/fl indicates mice with the homozygous fl allele (n = 4); sk/sk, mice with the homozygous cko allele in skeletal muscle (n = 4). Quantitative PCR primers of Hjv were qF, the forward primer in exon 3, and qR, the reverse primer in exon 4. Rpl19 was used as an internal control. The Hjv expression level in liver from Hjv fl/fl mice was assigned as 1. Error bars represent SD.
Targeted disruption of the Hjv gene. (A) Schematic representation of the mouse wild-type Hjv genomic sequence (+), LoxP-flanked allele with FRT-flanked Neo sequence (fl), and conditional knock out (cko) alleles. Positions of sequences coding for features of the protein are indicated as follows: (*) indicates the Tmprss6 cleavage site28 and (#) the Furin cleavage site.24,25 (B) Hjv mRNA relative expression in liver (L), heart (H), and skeletal muscle (SK) by quantitative RT-PCR. fl/fl indicates mice with the homozygous fl allele (n = 4); sk/sk, mice with the homozygous cko allele in skeletal muscle (n = 4). Quantitative PCR primers of Hjv were qF, the forward primer in exon 3, and qR, the reverse primer in exon 4. Rpl19 was used as an internal control. The Hjv expression level in liver from Hjv fl/fl mice was assigned as 1. Error bars represent SD.
Animal care and analysis
All mice were born and housed in the barrier facility at Duke University according to protocols approved by the Duke University Institutional Animal Care and Use Committee. Animals were maintained on a standard diet containing 380 ppm iron (Isopro RMH 3000, 5P76; Prolab). We analyzed mice on 3 diets: standard diet, an iron-deficient diet, and an iron-rich diet. For nonstandard diet experiments, recently weaned, 3-week-old animals were maintained on a low-iron diet (2-5 ppm iron; Teklad TD 99397, Harlan Laboratories) with iron-free water or a high-iron diet (380 ppm iron plus 2% carbonyl iron; Teklad TD 10066, Harlan Laboratories) with regular water for 5 weeks. Food and water were provided ad libitum. All mice were analyzed at 8 weeks of age.
Iron analysis in blood and tissues
Under Avertin anesthesia (Sigma-Aldrich), whole blood was collected from the retro-orbital sinuses through capillary tubes (Fisher Scientific) into serum separator tubes (Becton Dickinson). Serum was prepared and serum iron levels were determined using a serum iron/unsaturated iron binding capacity kit (Thermo-DMA) according to the manufacturer's instructions. Nonheme iron concentrations of the liver, spleen, heart, and skeletal muscle (quadriceps femoris) were determined as described previously.29
RNA extraction, RT-PCR, and quantitative RT-PCR
Total liver RNA was isolated from flash-frozen tissue using the RNeasy mini kit (QIAGEN). Total skeletal and cardiac muscle RNA was isolated using the RNeasy Fibrous Tissue Mini Kit (QIAGEN,). Total RNA was reverse transcribed using the iScript cDNA Synthesis Kit (Bio-Rad) according to the manufacturer's protocol. Expression of wild-type Hjv transcript was sought by quantitative PCR using forward primer 5′-TTGGCCCTTGCTAGATAACG-3′, located within exon 3 and reverse primer 5′-TCAATGCATTCCTGCATGTT-3′, located within exon 4. Expression of a partial Hjv transcript was sought by quantitative PCR using forward primer 5′-TCTGACCTGAGTGAGACTGC-3′, located within exon 1, and reverse primer 5′-GATGATGAGCCTCCTACCTA-3′, located within exon 2. Quantitative PCR to assess Hamp1, Id1, and Bmp6 transcript abundance was performed as described previously.20
Statistical analysis
The unpaired 2-tailed Student t test was performed with Microsoft Excel (PC 2007 version) software. P < .05 was considered statistically significant.
Results
Generation of conditional hemojuvelin-knockout mice
The mouse genome was modified to generate a Cre recombinase–sensitive conditional knockout allele of hemojuvelin (Hjv fl) containing one LoxP site in intron 3, a Neo sequence flanked by FRT sites, and another LoxP site in an untranscribed sequence downstream of exon 4 (Figure 1A). In the absence of Cre recombinase activity, the Hjv fl gene retains an intact promoter sequence and encodes full-length wild-type murine hemojuvelin. The Neo sequence in the Hjv fl allele did not interfere with the expression or function of Hjv; Hjv fl/fl mice had similar iron levels in the liver and spleen compared with wild-type mice (data not shown). Cre-mediated recombination was expected to lead to a conditional knockout allele (Hjv cko) lacking exon 4, which encodes 208 amino acids and contains the glycophosphatidyl inositol anchor signal sequence and the furin and matriptase-2 cleavage sites.
We established germline-competent 129S6/SvEvTac mice carrying the Hjv fl allele. Male mice heterozygous for the Hjv fl allele (Hjv fl/+ mice) were intercrossed to C57BL/6 hemizygous HSA-Cre/+ transgenic female mice [B6.Cg-Tg(ACTA1-cre)79Jme/J strain; The Jackson Laboratory] that express Cre recombinase restricted to skeletal muscle to obtain male Hjv fl/+;HSA-Cre/+ mice. We next intercrossed male Hjv fl/+;HSA-Cre/+ mice with female Hjv fl/fl mice to obtain Hjv fl/fl;HSA-Cre/+ mice (hereafter referred to as Hjv sk/sk mice). Their littermates with the Hjv fl/fl genotype but no Cre transgene were used as controls in the analyses. All offspring had a mixed 129S6/SvEvTac;C57BL/6 genetic background.
Heterozygous Hjv sk/+ and homozygous Hjv sk/sk mice had no apparent abnormalities. The numbers of offspring of both genotypes followed a Mendelian inheritance pattern, suggesting normal viability during the gestational and neonatal periods. Wild-type Hjv mRNA was analyzed using RT-PCR primers from exon 3 and 4. Quantitative PCR results showed that wild-type Hjv mRNA expression was undetectable in skeletal muscle from Hjv sk/sk mice, but was normal in liver and heart compared with Hjv fl/fl mice (Figure 1B). The predicted, truncated Hjv transcript including the first 3 exons was assayed using RT-PCR primers from exons 1 and 2. Quantitative PCR analysis demonstrated that the presence of the truncated Hjv transcript in skeletal muscle of Hjv sk/sk mice was expressed at approximately 30% of wild-type levels. Because of the lack of a reliable antibody against murine hemojuvelin, we could not test whether a truncated hemojuvelin protein was produced.
Iron overload phenotype in hemojuvelin liver-specific knockout mice
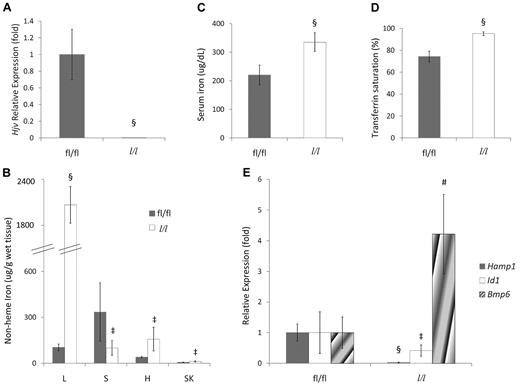
To confirm that deletion of Hjv exon 4 leads to loss of hemojuvelin function, we evaluated iron homeostasis in Hjv liver-specific conditional knockout mice by intercrossing male Hjv fl/+;Alb-Cre/+ transgenic mice with female Hjv fl/fl mice. The Alb-Cre transgene [B6.Cg-Tg(Alb-cre)21Mgn/J strain; The Jackson Laboratory] has been bred onto a 129S6/SvEvTac background, allowing us to carry out this experiment on a pure genetic background. Although the truncated Hjv mRNA was expressed at approximately 50% of wild-type levels (data not shown), Hjv full-length mRNA expression was not detected in the livers of Hjv fl/fl;Alb-Cre/+ mice (hereafter referred to as Hjv l/l mice; Figure 2A). Hjv l/l mice exhibited higher nonheme iron levels in the liver, heart, and muscle, but lower nonheme iron levels in the spleen (Figure 2B) compared with littermate Hjv fl/fl mice. Hjv l/l mice also had increased serum iron and transferrin saturation (Figure 2C-D). Hepatic hepcidin and Id1 mRNA expression was decreased in Hjv l/l mice, whereas Bmp6 mRNA expression was increased (Figure 2E). Whereas we cannot rule out the possibility that a truncated protein is produced from the deleted allele, the fact that all of these features recapitulate the phenotype found in Hjv-null mice17 strongly suggests that deletion of Hjv exon 4 leads to loss of function of Hjv.
Hjv liver-specific knockout mice exhibit iron-overload phenotypes. (A) Relative Hjv mRNA expression in liver determined by quantitative RT-PCR. fl/fl indicates mice with the homozygous fl allele; l/l, homozygous fl mice expressing the Cre transgene in liver. The Hjv expression level in the livers of fl/fl mice was assigned a value of 1. (B) Serum iron concentration. (C) Serum transferrin saturation. (D) Nonheme tissue iron concentrations (μg/g wet weight) of fl/fl and Hjv liver-specific knockout (l/l) mice. L indicates liver; S, spleen; H, heart; and SK, skeletal muscle. (E) Hepatic Hamp1, Id1, and Bmp6 relative to Rpl19 mRNA expression by quantitative RT-PCR. Results are reported as the -fold change from Hjv fl/fl mice. (A-E) Mean values from analysis of 8-week-old mice (fl/fl mice: 3 male and 2 female, l/l mice: 4 male and 2 female) are graphed. Error bars represent SD. ‡P < .05, #P < .005, and §P < .0005 compared with Hjv fl/fl mice.
Hjv liver-specific knockout mice exhibit iron-overload phenotypes. (A) Relative Hjv mRNA expression in liver determined by quantitative RT-PCR. fl/fl indicates mice with the homozygous fl allele; l/l, homozygous fl mice expressing the Cre transgene in liver. The Hjv expression level in the livers of fl/fl mice was assigned a value of 1. (B) Serum iron concentration. (C) Serum transferrin saturation. (D) Nonheme tissue iron concentrations (μg/g wet weight) of fl/fl and Hjv liver-specific knockout (l/l) mice. L indicates liver; S, spleen; H, heart; and SK, skeletal muscle. (E) Hepatic Hamp1, Id1, and Bmp6 relative to Rpl19 mRNA expression by quantitative RT-PCR. Results are reported as the -fold change from Hjv fl/fl mice. (A-E) Mean values from analysis of 8-week-old mice (fl/fl mice: 3 male and 2 female, l/l mice: 4 male and 2 female) are graphed. Error bars represent SD. ‡P < .05, #P < .005, and §P < .0005 compared with Hjv fl/fl mice.
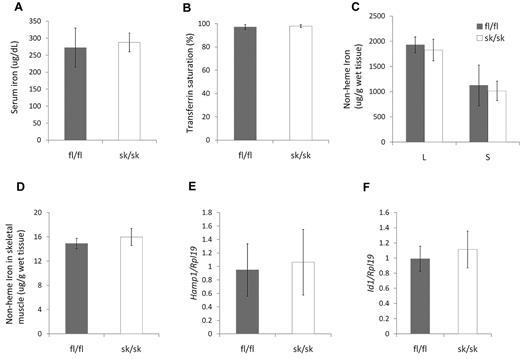
Iron homeostasis in hemojuvelin skeletal muscle–specific knockout mice
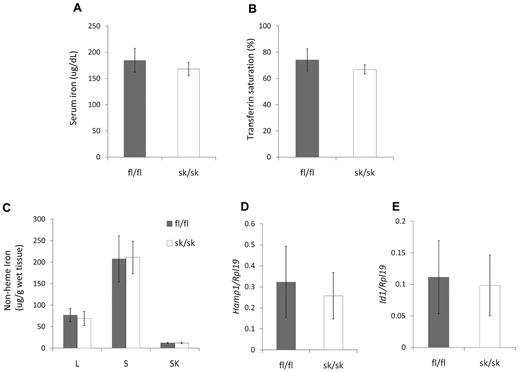
Soluble hemojuvelin has been shown to decrease hepcidin expression both in vitro and in vivo.22,23 Considering the high expression levels of Hjv mRNA and the lack of hepcidin in skeletal muscle, we hypothesized that Hjv in skeletal muscle serves as a source of soluble hemojuvelin, which circulates to the liver to regulate hepatic hepcidin expression and systemic iron homeostasis. We examined parameters of iron homeostasis in hemojuvelin skeletal muscle–specific knockout mice at 8 weeks of age. Hjv sk/sk mice did not show significant differences in serum iron (Figure 3A), transferrin saturation (Figure 3B), or nonheme iron concentration in liver, spleen, or skeletal muscle (Figure 3C) compared with Hjv fl/fl mice. Hepatic Hamp1 and Id1 mRNA expression in Hjv sk/sk mice was similar to that of Hjv fl/fl mice (Figure 3D-E). Younger Hjv sk/sk mice (age 4-6 weeks) also had tissue iron levels similar to control Hjv fl/fl mice (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Iron homeostasis is not altered in Hjv skeletal muscle–specific knockout mice. (A) Serum iron concentration. (B) Serum transferrin saturation. (C) Nonheme tissue iron concentrations (μg/g wet weight) of Hjv fl/fl and skeletal muscle–specific knockout (sk/sk) male mice. L indicates liver; S, spleen; and SK, skeletal muscle. (D) Hepatic Hamp1 relative to Rpl19 mRNA expression. (E) Hepatic Id1 relative to Rpl19 mRNA expression. Mean values from analysis of 8-week-old male mice (n = 8 per genotype) are graphed. Error bars represent SD. No statistically significant differences were observed.
Iron homeostasis is not altered in Hjv skeletal muscle–specific knockout mice. (A) Serum iron concentration. (B) Serum transferrin saturation. (C) Nonheme tissue iron concentrations (μg/g wet weight) of Hjv fl/fl and skeletal muscle–specific knockout (sk/sk) male mice. L indicates liver; S, spleen; and SK, skeletal muscle. (D) Hepatic Hamp1 relative to Rpl19 mRNA expression. (E) Hepatic Id1 relative to Rpl19 mRNA expression. Mean values from analysis of 8-week-old male mice (n = 8 per genotype) are graphed. Error bars represent SD. No statistically significant differences were observed.
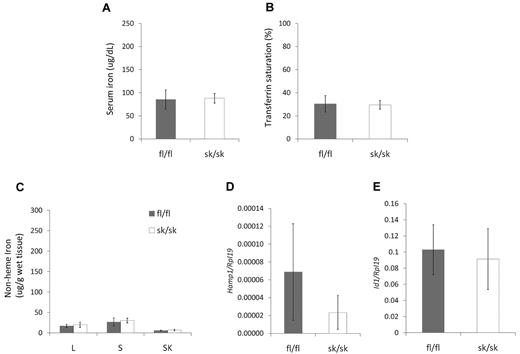
We next investigated whether Hjv fl/fl and Hjv sk/sk mice would respond differently to an iron-deficient diet. We weaned 3-week-old pups with Hjv fl/fl and Hjv sk/sk genotypes from an Hjv fl/+;HSA1-Cre/+ and Hjv fl/fl intercross, and fed them an iron-deficient diet for 5 weeks before serum and tissue analysis. Both Hjv fl/fl and Hjv sk/sk mice on the iron-deficient diet showed significant decreases in serum iron (Figure 4A), transferrin saturation (Figure 4B), and nonheme iron in the liver, spleen, and skeletal muscle (Figure 4C) compared with mice on a standard diet (Figure 3). Hepatic Hamp1 mRNA expression was much lower in all mice on the iron-deficient diet (Figure 4D). However, hepatic Id1 mRNA expression did not show a significant change from that observed in mice on a standard diet in either cohort (Figure 4E). In summary, no differences in the parameters of iron homeostasis were observed between Hjv fl/fl and Hjv sk/sk mice on an iron-deficient diet.
Iron homeostasis in Hjv skeletal muscle–specific knockout mice on iron-deficient diet. (A) Serum iron concentration. (B) Serum transferrin saturation. (C) Nonheme tissue iron concentrations (μg/g wet weight) of Hjv fl/fl and skeletal muscle-specific knockout (sk/sk) female mice. L indicates liver; S, spleen; and SK, skeletal muscle. (D) Hepatic Hamp1 relative to Rpl19 mRNA expression. Note that all values were very low compared with mice on a standard diet. (E) Hepatic Id1 relative to Rpl19 mRNA expression. Mean values from analysis of 8-week-old female mice (n = 8 per genotype) are graphed. Error bars represent SD. No statistically significant differences were observed.
Iron homeostasis in Hjv skeletal muscle–specific knockout mice on iron-deficient diet. (A) Serum iron concentration. (B) Serum transferrin saturation. (C) Nonheme tissue iron concentrations (μg/g wet weight) of Hjv fl/fl and skeletal muscle-specific knockout (sk/sk) female mice. L indicates liver; S, spleen; and SK, skeletal muscle. (D) Hepatic Hamp1 relative to Rpl19 mRNA expression. Note that all values were very low compared with mice on a standard diet. (E) Hepatic Id1 relative to Rpl19 mRNA expression. Mean values from analysis of 8-week-old female mice (n = 8 per genotype) are graphed. Error bars represent SD. No statistically significant differences were observed.
We also investigated whether an iron-rich diet differentially alters systemic iron homeostasis in Hjv fl/fl and Hjv sk/sk mice. We fed newly weaned, 3-week-old Hjv fl/fl and Hjv sk/sk mice an iron-rich diet for 5 weeks. We observed a significant increase in serum iron (Figure 5A), transferrin saturation (Figure 5B), and nonheme iron concentrations in the liver, spleen (Figure 5C), and skeletal muscle (Figure 5D) compared with mice on a standard diet (Figure 3). Hepatic Hamp1 and Id1 mRNA expression was also increased on an iron-rich diet compared with a standard diet (Figure 5E-F). However, there was no difference between Hjv fl/fl and Hjv sk/sk mice in any tested parameters of iron homeostasis.
Iron homeostasis in Hjv skeletal muscle–specific knockout mice on iron-rich diet. (A) Serum iron concentration. (B) Serum transferrin saturation. (C) Nonheme tissue iron concentrations (μg/g wet weight) of Hjv fl/fl and skeletal muscle–specific knockout (sk/sk) male mice. L indicates liver; and S, spleen. (D) Nonheme iron concentrations in skeletal muscle. (E) Hepatic Hamp1 relative to Rpl19 mRNA expression. (F) Hepatic Id1 relative to Rpl19 mRNA expression. Mean values from analysis of 8-week-old male mice (n = 4 for fl/fl and n = 6 for sk/sk) are graphed. Error bars represent SD. No statistically significant differences were observed.
Iron homeostasis in Hjv skeletal muscle–specific knockout mice on iron-rich diet. (A) Serum iron concentration. (B) Serum transferrin saturation. (C) Nonheme tissue iron concentrations (μg/g wet weight) of Hjv fl/fl and skeletal muscle–specific knockout (sk/sk) male mice. L indicates liver; and S, spleen. (D) Nonheme iron concentrations in skeletal muscle. (E) Hepatic Hamp1 relative to Rpl19 mRNA expression. (F) Hepatic Id1 relative to Rpl19 mRNA expression. Mean values from analysis of 8-week-old male mice (n = 4 for fl/fl and n = 6 for sk/sk) are graphed. Error bars represent SD. No statistically significant differences were observed.
Discussion
Hemojuvelin functions as a BMP coreceptor to activate hepcidin expression through a BMP/SMAD signaling pathway.18 In the present study, we demonstrated that loss of hemojuvelin specifically in the liver leads to an iron overload phenotype similar to that observed in Hjv-null mice. Hepatic hepcidin and Id1 mRNA expression was decreased in these mice, whereas Bmp6 mRNA expression was increased, which is consistent with iron overload. These data confirm both that hepatic hemojuvelin is critical in maintaining liver hepcidin expression and systemic iron homeostasis and that the conditional knockout allele does not encode a functional hemojuvelin protein.
Hemojuvelin is also thought to be cleaved and secreted out of cells in a soluble form. Soluble hemojuvelin can selectively bind to BMP ligands and inhibit endogenous, BMP-induced hepcidin expression in vitro.22,23 Considering that hemojuvelin and furin are highly expressed in skeletal muscle, we hypothesized that soluble hemojuvelin produced by skeletal muscle might regulate hepatic hepcidin expression.
We tested our hypothesis by generating a strain of mice in which the hemojuvelin gene was specifically inactivated in skeletal muscle. These Hjv conditional-knockout mice did not show any differences in iron homeostasis compared with control mice and had a similar response to both iron-deficient and iron-rich diets. These data indicate that hemojuvelin in skeletal muscle is dispensable for systemic iron homeostasis under the conditions tested.
There are several limitations to our study and the conclusions we can draw. All mice used in our experiments were of a mixed 129S6/SvEvTac;C57BL/6 genetic background, and we cannot rule out the possibility that the resulting increased variability masked a very subtle difference between Hjv sk/sk and Hjv fl/fl control mice. Although hemojuvelin mRNA is highly expressed in skeletal muscle, we cannot confirm that hemojuvelin protein is also expressed at high levels because of the lack of a reliable antibody against murine hemojuvelin. In addition, the production of soluble hemojuvelin has not been definitively proven in vivo in general or in muscle in particular. It may be that as-yet-unidentified stress conditions are necessary to induce cleavage of soluble hemojuvelin in skeletal muscle and thus alter systemic iron homeostasis. Finally, hemojuvelin is also expressed at high levels in cardiac muscle, and cardiac hemojuvelin may compensate for the loss of hemojuvelin in skeletal muscle. By generating a strain of mice with inactivated hemojuvelin in both cardiac and skeletal muscle, we could determine whether the loss of hemojuvelin in both cardiac and skeletal muscle affects systemic iron homeostasis.
The function of hemojuvelin in skeletal muscle remains unknown. In the present study, the loss of hemojuvelin in skeletal muscle did not lead to abnormalities in muscle histology (data not shown), and patients with juvenile hemochromatosis have not been reported to have muscle abnormalities. Nonheme iron in skeletal muscle from Hjv sk/sk mice was similar to that of control mice. Id1 mRNA expression was also similar, suggesting that hemojuvelin does not regulate an analogous BMP/SMAD signaling pathway in skeletal muscle.
In conclusion, we generated mice lacking hemojuvelin in the liver and replicated iron-overload phenotypes found in hemojuvelin-null mice. We also demonstrated that loss of hemojuvelin in skeletal muscle did not appreciably affect systemic iron homeostasis regardless of dietary iron content. However, the reason for high hemojuvelin expression in muscle remains obscure.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Paul Schmidt for assistance with technical aspects of this work and members of the Andrews laboratory for helpful discussions.
This work was supported by a Cooley's Anemia Foundation Research Fellowship (to W.C.), by a Roche Foundation for Anemia Research grant (to N.C.A.), and by National Institutes of Health grant R01 DK066373 (to N.C.A.).
National Institutes of Health
Authorship
Contribution: W.C. designed and performed experiments, analyzed results, and wrote the manuscript; F.W.H. designed experiments and made the Hjv conditional-knockout vector; T.B.d.R. performed some of the Hjv liver-specific knockout mice experiments; and N.C.A. guided experiments and contributed to writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nancy C. Andrews, MD, PhD, Duke University School of Medicine, DUMC 2927, Durham, NC 27710; e-mail: nancy.andrews@duke.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal