Abstract
Notch signaling critically mediates various hematopoietic lineage decisions and is induced in mammals by Notch ligands that are classified into 2 families, Delta-like (Delta-like-1, -3 and -4) and Jagged (Jagged1 and Jagged2), based on structural homology with both Drosophila ligands Delta and Serrate, respectively. Because the functional differences between mammalian Notch ligands were still unclear, we have investigated their influence on early human hematopoiesis and show that Jagged2 affects hematopoietic lineage decisions very similarly as Delta-like-1 and -4, but very different from Jagged1. OP9 coculture experiments revealed that Jagged2, like Delta-like ligands, induces T-lineage differentiation and inhibits B-cell and myeloid development. However, dose-dependent Notch activation studies, gene expression analysis, and promoter activation assays indicated that Jagged2 is a weaker Notch1-activator compared with the Delta-like ligands, revealing a Notch1 specific signal strength hierarchy for mammalian Notch ligands. Strikingly, Lunatic-Fringe– mediated glycosylation of Notch1 potentiated Notch signaling through Delta-like ligands and also Jagged2, in contrast to Jagged1. Thus, our results reveal a unique role for Jagged1 in preventing the induction of T-lineage differentiation in hematopoietic stem cells and show an unexpected functional similarity between Jagged2 and the Delta-like ligands.
Introduction
The Notch pathway is composed of a highly conserved signaling mechanism that mediates cell fate decisions in numerous developmental systems.1 Notch receptor-ligand interactions induce proteolytic cleavages of the Notch receptor, resulting in the translocation of the Notch intracellular domain to the nucleus. There, Notch intracellular domain interacts with CBF-1, Su(H), Lag-1 (CSL), resulting in the recruitment of transcriptional coactivators and induction of Notch target gene expression. In Drosophila, the 2 Notch ligands Delta and Serrate induce different signaling outcomes on activation of a unique Notch receptor, and Fringe further modifies these events by potentiating Delta-induced Notch signaling and weakening Serrate-induced activation. In mammals, the pathway is composed of 4 Notch receptors (Notch1-4) that can be activated through 5 canonical Notch ligands. Based on structural homology with Drosophila, these ligands are subdivided into 2 classes: the Serrate-like ligands Jagged1 (JAG1) and -2 (JAG2), and the Delta-like ligands (DLL) Delta-like-1, -3, and -4. Delta-like-3 seems incapable of activating Notch signaling, but it is unclear what the precise differences are between the other Notch ligands with respect to their Notch activation potentials.2 Because of their structural homology, parallels are drawn between Serrate-like ligands on the one hand and Delta-like ligands on the other hand. In agreement, different roles for Jagged versus Delta-like ligands have been illustrated,3,4 but few studies have investigated all mammalian Notch ligands in the same developmental model. Notch signaling in mammals is further modulated by 3 different Fringe proteins: Lunatic, Manic, and Radical. Lunatic and Manic generally strengthen Delta-mediated Notch signaling and reduce Jagged-induced Notch activation.5,6 However, exceptions have been reported,7,8 suggesting that Fringe-mediated effects might be context-dependent.
Notch signaling has been well studied in the hematopoietic system where it is involved in the developmental and functional maturation of a variety of blood cell types.9-12 Especially T cells are highly dependent on Notch signaling during their development in both mouse and humans. In contrast to other blood cell lineages that develop in the bone marrow, T lymphocytes develop in the thymic microenvironment after migration of hematopoietic progenitor cells (HPCs) with multilineage potential from the bone marrow into the thymus.13 T lymphopoiesis involves a series of discrete differentiation steps that gradually restricts this multipotency toward more restricted T-lineage precursors, and these stages can be identified through specific surface markers. In human, the earliest intrathymic progenitors express high levels of CD34 and are triple-negative for mature T-cell markers CD4, CD8, and CD3. Notch-mediated T-lineage specification results in the sequential up-regulation of CD7 and CD5,14,15 whereas subsequent CD1 expression marks irreversible T-cell commitment. During these stages, TCR rearrangements of the TCR-δ, TCR-γ, and TCR-β loci are initiated that will determine the developmental outcome.16 In-frame TCR-δ and TCR-γ rearrangements will result in the generation of CD3+ TCR-γδ+ T cells, whereas a TCR-β chain will pair with a surrogate TCR-α chain, pre-Tα, and this pre-TCR complex drives further development of αβ-lineage cells into CD4+CD8+ double positive cells and finally into CD3+TCR-αβ+ T cells.
Intrathymic Notch signaling is essential to induce T-cell development in HPCs, as well as to inhibit differentiation into alternative lineages, such as B and myeloid fates.17-19 Although Delta-like-4 is essential in vivo at the earliest stages of T-cell specification,20,21 both Delta-like-1 and -4, but not Jagged1, can induce T-cell development in HPCs in vitro when expressed by OP9 stromal cells.3,4,22,23 Jagged2 has been poorly studied in early hematopoiesis, and its role during T-lineage development is still unclear. Jagged2-deficient mice die embryonically and have a 2-fold reduction in γδ T cells.24 Furthermore, because it has only been shown that Delta-like-4 is essential at the earliest stage of T-cell development in adult mice to generate early thymic progenitors, the role of the various Notch ligands during fetal thymopoiesis as well as during adult intrathymic and Notch-dependent extrathymic T-cell development is still unclear. The amount of Notch signal strength induced by the various Notch ligands most probably determines their influence on hematopoietic lineage decisions,15,25 but this has not been well characterized. Because hematopoiesis in conjunction with the well-defined OP9 coculture system provides a good model system to study Notch signaling, we performed a comparative study to gain novel insights into the mechanisms through which Jagged1, Jagged2, Delta-like-1, and Delta-like-4 activate Notch signaling. Unexpectedly, we found functional and biochemical similarities between Delta-like ligands and Jagged2, in contrast to Jagged1 that acted as a classic Serrate-like ligand.
Methods
Reverse-transcription PCR
CD34+lin− cord blood (CB) cells were cocultured on Notch ligand-coated tissue culture plates or on OP9 stromal cells expressing the different Notch ligands. Plates were coated for 2 hours with human Notch ligand Fc proteins at 10 μg/mL (DLL1-Fc; DLL4-Fc from Alexis; Jagged1- and Jagged2-Fc from R&D Systems), and cells were harvested after 3 days of culture. For OP9 cocultures, cells were harvested at indicated time points and stained with CD45-phycoerythrin (Miltenyi Biotec) to sort CD45+ human leukocytes from the OP9 stromal cells. Cells were resuspended in RLT buffer (QIAGEN) and stored at −70°C. RNA was extracted using RNeasy RNA isolation kit (QIAGEN) and reverse-transcribed into cDNA using Superscript RT II (Invitrogen). Real-time PCRs were performed using quantitative PCR Core kit for SYBR Green I (Eurogentec) on a 7300 Real-time PCR system (Applied Biosystems) and with primers as described.15,26 Relative expression levels were calculated for each gene using the ΔCt or ΔΔCt method.
Luciferase reporter assay
U2OS Tet-on flp-in cells bearing isogenic transgenes encoding Notch1-Gal4 or Notch2-Gal4 chimeric receptors27 were stably transduced with control or LFng. Subsequently, cells were transfected with 5 μg of Gal4-firefly luciferase and 500 ng pRL-TK-Renilla (Promega) reporter plasmid, using calcium phosphate precipitation protocol (Invitrogen) according to the guidelines of the manufacturer. After 24 hours, K562 cells expressing Notch ligands were added to the transfected cells in the presence of tetracycline (2 μg/mL). After 24-hour coculture, luciferase activity was measured in cell lysates using a dual luciferase reporter assay system (Promega), also according to the guidelines of the manufacturer.
Isolation of TECs
Thymus samples were obtained from children undergoing heart surgery (Department of Cardiac Surgery, Medical School of the University of Heidelberg and approved by its Ethic Committee and with informed consent of participating subjects, in accordance with the Declaration of Helsinki) and enzymatically digested as described28 with the following modifications. For medullary thymic epithelial cell (mTEC) isolation, pooled trypsin-ethylenediaminetetraacetic acid fractions were depleted of CD45+ cells using CD45-Microbeads (Miltenyi Biotec). Enriched stromal cells were stained with biotinylated EpCAM (HEA125; DKFZ), CDR2-Alexa488 (CDR-2; DKFZ), Alexa680-conjugated HLA-DR (L243; DKFZ), and CD45-PerCP (BD Biosciences), followed by sav-phycoerythrin (BD Biosciences). The mTECs were sorted as CD45−CDR2−EpCAMhigh cells and thymocytes as CD3high (BD Biosciences) from the medium fraction. Dead cells were excluded with propidium iodide, and populations were sorted on a FACSAria (BD Biosciences). RNA isolation and cDNA synthesis were performed as described previously.28 Before quantitative PCR, cDNA was purified using MicroSpin G-50 columns (GE Healthcare). For the Notch-ligand staining, the TEC staining panel was slightly modified: anti–EpCAM-bio (purified and conjugated at the DKFZ) was detected with anti–sav-allophycocyanin (BD Biosciences).
Results
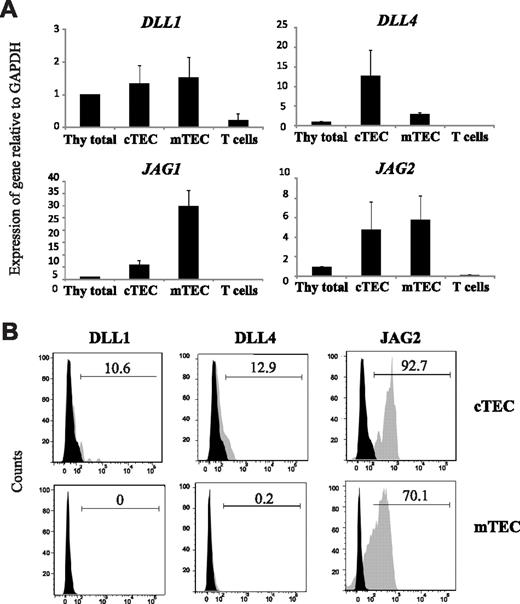
Jagged2 is abundantly expressed by human TECs
Within the hematopoietic system, Notch is well known for its role in mediating the T- versus B-cell lineage decision and the paradigm is that Delta-like, but not Jagged ligands, can initiate T-cell development and inhibit B-lineage fate. However, Jagged2 has not been thoroughly studied in this system, although Jag2 has been shown to be expressed in the mouse thymus. Because our work focuses on human hematopoiesis, we first determined Notch ligand (DLL1, DLL4, JAG1, and JAG2) expression in cortical and medullary TECs (cTECs and mTECs, respectively) of the human postnatal thymus through gene expression analysis (Figure 1A) and flow cytometry (Figure 1B). mRNA for all 4 ligands was detected in both epithelial subsets, in contrast to mature thymic T cells that virtually lacked Notch ligand expression. Interestingly, whereas JAG2 and DLL1 expression seemed equally distributed over cTECs and mTECs, DLL4 and JAG1 showed opposing expression patterns as DLL4 was primarily expressed by cTECs, in accordance with its requirement during early stages of T development, whereas JAG1 was preferentially expressed by mTECs (Figure 1A). Protein analysis confirmed that Delta-like-1 is very weakly expressed in human TECs, whereas Delta-like-4 was only detected in approximately 10% to 20% of cTECs (Figure 1B), comparable with the expression pattern in mouse.29 Interestingly, Jagged2 was very abundantly expressed in both cTECs and mTECs (Figure 1B).
Jagged2 is abundantly expressed by human TECs. (A) Quantitative RT-PCR analysis of human TECs for the expression of Notch ligands, DLL1, DLL4, JAG1, and JAG2. Expression levels are normalized using the ΔΔCt method, relative to GAPDH and relative to levels in total thymus. Results show mean of 3 independent sets of samples. Errors bars represent SEM. (B) Flow cytometric analysis of Delta-like-1, Delta-like-4, and Jagged2 expression in human cTECs (top row) and mTECs (bottom row). Histogram plots show isotype control (black) versus specific Notch ligand antibody staining (gray). Data are representative for at least 2 independent donors.
Jagged2 is abundantly expressed by human TECs. (A) Quantitative RT-PCR analysis of human TECs for the expression of Notch ligands, DLL1, DLL4, JAG1, and JAG2. Expression levels are normalized using the ΔΔCt method, relative to GAPDH and relative to levels in total thymus. Results show mean of 3 independent sets of samples. Errors bars represent SEM. (B) Flow cytometric analysis of Delta-like-1, Delta-like-4, and Jagged2 expression in human cTECs (top row) and mTECs (bottom row). Histogram plots show isotype control (black) versus specific Notch ligand antibody staining (gray). Data are representative for at least 2 independent donors.
Jagged2, like Delta-like-1 and Delta-like-4 but in contrast to Jagged1, can mediate the T- versus B-cell lineage decision
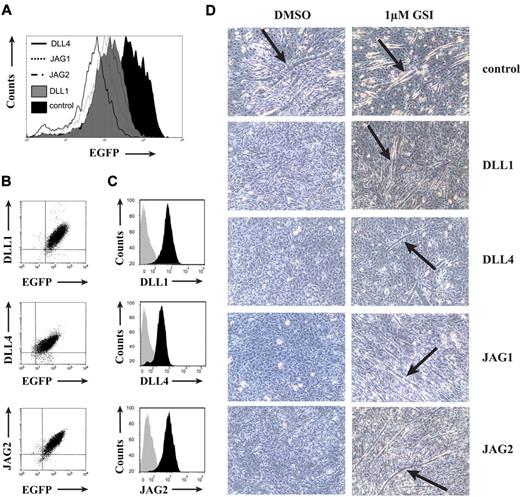
Because all 4 ligands are expressed at the site where in vivo T-cell development occurs, we investigated their impact on early T-cell development using OP9 cocultures. Therefore, we generated OP9 cells expressing human DLL1, DLL4, JAG1, and JAG2 through retroviral transduction using a bicistronic vector that also encodes the marker gene EGFP.15 EGFP transduced OP9 cells served as control (named “control” throughout the manuscript). After transduction, OP9 cells were fluorescence-activated cell sorter sorted for EGFP to obtain a homogeneous population that expresses similar levels of EGFP protein (Figure 2A) and mRNA for each particular Notch ligand (data not shown). Consistent with the use of a bicistronic vector, EGFP expression levels correlated with protein levels for each Notch ligand (Figure 2B), and comparison of protein expression with control transduced cells revealed a similar increase in Notch ligand expression in each OP9 cell line (Figure 2C). PCR analysis of murine Notch ligand expression indicated that the retroviral transduction procedure did not alter the endogenous expression pattern of the Notch ligands in OP9 cells (data not shown). To show that the generated OP9 cells expressed a functional human Notch ligand, the C2C12 myoblast differentiation assay was used.30 All Notch ligands repressed C2C12 differentiation into myotubes compared with the control, and this was reversible by adding a γ-secretase inhibitor (GSI) that inhibits Notch signaling (Figure 2D), indicating that each ligand could activate Notch signaling.
Characterization of OP9 stromal cells expressing different human Notch ligands. (A) Flow cytometric analysis of OP9-control, OP9-DLL1, OP9-DLL4, OP9-JAG1, or OP9-JAG2 stromal cells for EGFP expression. (B) Flow cytometric analysis of EGFP and Notch ligand expression in OP9-DLL1, OP9-DLL4, and OP9-JAG2 cells, as indicated. Isotype and OP9-control stainings were used as negative controls, and ligand specificity was determined through the absence of antibody staining on OP9 cells that express a different Notch ligand. (C) Flow cytometric analysis of Notch ligand expression in OP9-DLL1, OP9-DLL4, and OP9-JAG2 cells (black histograms), as indicated, versus OP9-control cells (gray histograms). (D) Differentiation of Notch1-expressing C2C12 myoblast cells (C2C12N1) after 5-day coculture with OP9-control, OP9-DLL1, OP9-DLL4, OP9-JAG1, or OP9-JAG2 in the presence of 1μM GSI or 0.1% dimethyl sulfoxide as a control. Arrows indicate myotube formation. Images were acquired from cells in culture medium with a Leica DM IL microscope (20×/0.3 objective) using a Leica DCF420 camera with Leica 3.1.0 Application Suite software.
Characterization of OP9 stromal cells expressing different human Notch ligands. (A) Flow cytometric analysis of OP9-control, OP9-DLL1, OP9-DLL4, OP9-JAG1, or OP9-JAG2 stromal cells for EGFP expression. (B) Flow cytometric analysis of EGFP and Notch ligand expression in OP9-DLL1, OP9-DLL4, and OP9-JAG2 cells, as indicated. Isotype and OP9-control stainings were used as negative controls, and ligand specificity was determined through the absence of antibody staining on OP9 cells that express a different Notch ligand. (C) Flow cytometric analysis of Notch ligand expression in OP9-DLL1, OP9-DLL4, and OP9-JAG2 cells (black histograms), as indicated, versus OP9-control cells (gray histograms). (D) Differentiation of Notch1-expressing C2C12 myoblast cells (C2C12N1) after 5-day coculture with OP9-control, OP9-DLL1, OP9-DLL4, OP9-JAG1, or OP9-JAG2 in the presence of 1μM GSI or 0.1% dimethyl sulfoxide as a control. Arrows indicate myotube formation. Images were acquired from cells in culture medium with a Leica DM IL microscope (20×/0.3 objective) using a Leica DCF420 camera with Leica 3.1.0 Application Suite software.
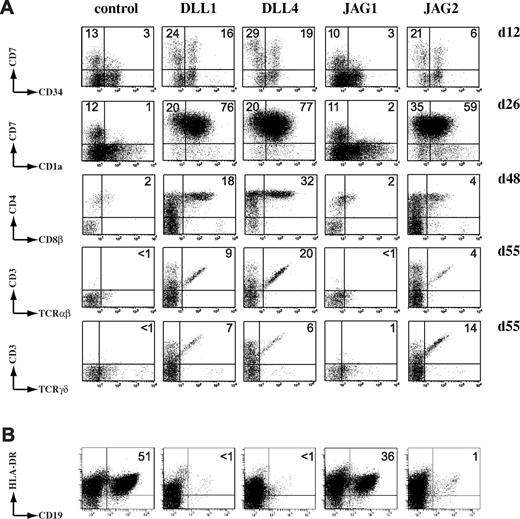
To investigate how Notch ligands mediate human T- and B-cell development, we initiated cocultures of human CD34+CD38−lin− CB HPCs onto the different OP9 stromal cells. As expected, both DLL1 and DLL4, but not JAG1 or control transduced OP9 cells, supported the generation of early human CD34+CD7++ T-/NK-cell precursors (day 12, Figure 3A) that further differentiated into CD7+CD1+-committed T-lineage precursors (day 26, Figure 3A), a process that is Notch-dependent.15 Surprisingly, also JAG2-transduced OP9 cells supported the development of early CD34+CD7++ T/NK and CD7+CD1+ committed T-cell precursors (Figure 3A). Consistently, only DLL1-, DLL4-, and JAG2-transduced OP9 cells allowed further development into CD4+CD8β+ double-positive thymocytes and CD3+TCR-αβ+ and CD3+TCR-γδ+ T cells (day 48 and day 55, Figure 3A). In agreement, NK cells that express cytoplasmic CD3E, a Notch-dependent and T-lineage specific protein,31 were preferentially generated after coculture on Delta-like-1, Delta-like-4, and Jagged2, not on Jagged1 or the control (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). As well as initiating T-lineage differentiation, Delta/Notch signaling also inhibits B-cell development. Analysis of B-lineage differentiation showed that, consistent with the results on early T-cell development, Delta-like-1, Delta-like-4, and Jagged2, but not Jagged1, could efficiently inhibit B-cell development (day 48, Figure 3B). Similar results on T- and B-cell development were obtained with murine c-Kit+Sca-1+Lin− fetal liver-derived HPCs, showing that these results are not a peculiarity of human HPCs (supplemental Figure 2).
Jagged2 mediates the T- versus B-cell lineage decision. (A) Jagged2, like Delta-like ligands, induces T-cell differentiation in human HPCs. Kinetic flow cytometric analysis of human CD34+CD38−lin− CB precursors after coculture on OP9 stromal cells expressing the different human Notch ligands, as indicated above the dot plots. Numbers in quadrants indicate the frequency of the corresponding populations. Data shown are representative for 4 independent experiments. (B) Jagged2, Delta-like-1, and Delta-like-4 inhibit B-cell development. Dot plots show analysis of CD19 versus HLA-DR staining of human CD34+CD38−lin− CB precursors cultured on OP9 stromal cells for 48 days. Dot plots shown are representative of 3 independent experiments.
Jagged2 mediates the T- versus B-cell lineage decision. (A) Jagged2, like Delta-like ligands, induces T-cell differentiation in human HPCs. Kinetic flow cytometric analysis of human CD34+CD38−lin− CB precursors after coculture on OP9 stromal cells expressing the different human Notch ligands, as indicated above the dot plots. Numbers in quadrants indicate the frequency of the corresponding populations. Data shown are representative for 4 independent experiments. (B) Jagged2, Delta-like-1, and Delta-like-4 inhibit B-cell development. Dot plots show analysis of CD19 versus HLA-DR staining of human CD34+CD38−lin− CB precursors cultured on OP9 stromal cells for 48 days. Dot plots shown are representative of 3 independent experiments.
Thus, besides the Delta-like ligands, Jagged2 is also capable of inducing T-lineage commitment in HPCs at the expense of B-lineage differentiation, in contrast to Jagged1-induced Notch signaling.
Dose-dependent inhibition of Notch signaling reveals a signal strength hierarchy
Although these findings revealed large similarities between the Delta-like ligands and Jagged2, it was unclear whether the Notch ligands induced similar or different Notch signal strengths in the signal-receiving cells. To gain further insights, we performed OP9 cocultures with human hematopoietic stem cells (HSCs) to which increasing amounts of GSI were added, thereby allowing dose-dependent inhibition of Notch signaling as illustrated by changes in HES1 expression (supplemental Figure 3). With respect to inducing early T-/NK-cell differentiation, Jagged2 generated a lower absolute number of CD34+CD7+ T-/NK-cell precursors compared with both Delta-like ligands, suggesting that Jagged2 is a weaker Notch ligand (Figure 4A). The addition of increasing dosages of GSI, however, resulted in a similar decrease in both the frequency and absolute number of early human CD34+CD7+ T-/NK-cell precursors, independent of whether the cells were cultured on OP9-DLL1, -DLL4, or -JAG2 (Figure 4A). B-cell development is strongly repressed by Delta-like-1, Delta-like-4, and Jagged2 (Figure 3B). Remarkably, the addition of GSI resulted in a much faster recovery of B-lineage differentiation in OP9-JAG2 cocultures compared with OP9-DLL1 and OP9-DLL4, both in terms of frequency and absolute cell number (Figure 4B). Similarly, differentiation into CD4+CD14+ monocytes (Figure 4C) and CD123+CD303+ plasmacytoid dendritic cells (Figure 4D), 2 lineages that are repressed by Notch signaling during early stages of T-cell development,17,18,32 was less profoundly repressed by Jagged2 compared with both Delta-like ligands and was faster restored on OP9-JAG2-cultured cells when GSI was added. In contrast, Jagged1 had very little effect on the absolute number of B, monocytic, and plasmacytoid dendritic cells that were generated in these culture conditions, and the addition of GSI had no obvious effect. Although presumably not relevant in vivo,33 Jagged1-mediated Notch signaling has also been suggested to be involved in the expansion of HSCs.34-36 In OP9 coculture experiments, Jagged1 did promote, more efficiently than other Notch ligands, the expansion of human CD34+ HPCs (supplemental Figure 4A), and experiments with increasing GSI amounts showed that this was indeed a Notch-dependent process (supplemental Figure 4B).
Dose-dependent Notch inhibition reveals a signal strength hierarchy. Effects of dose-dependent Notch inhibition on the induction of T-lineage differentiation (A), B-cell development (B), monocyte development (C), and plasmacytoid differentiation (pDC; D). Left panels: Flow cytometric analysis of human CD34+CD38−lin− CB HPCs after OP9 coculture in the presence of 0 or 1μM GSI (7 N-[N-(3,5-difluorophenyl-L-alanyl]-S-phenylglycine t-butyl ester). Numbers in quadrants indicate the percentage of CD34+CD7+ T-cell precursors (A), CD19+HLA-DR+ B cells (B), CD14+CD4+ monocytes (C), and CD123+CD303+ pDCs (D). Results shown are representative of 3 independent experiments. Right panels: Corresponding average of the absolute number of CD34+CD7+ T-cell precursors (A), CD19+HLA-DR+ B cells (B), CD14+CD4+ monocytes (C), and CD123+CD303+ pDCs (D) for cultures depicted in the corresponding left panel. Error bars represent SEM.  indicates an increasing dosage of GSI, corresponding to 0, 0.1, 0.3, and 1μM GSI.
indicates an increasing dosage of GSI, corresponding to 0, 0.1, 0.3, and 1μM GSI.
Dose-dependent Notch inhibition reveals a signal strength hierarchy. Effects of dose-dependent Notch inhibition on the induction of T-lineage differentiation (A), B-cell development (B), monocyte development (C), and plasmacytoid differentiation (pDC; D). Left panels: Flow cytometric analysis of human CD34+CD38−lin− CB HPCs after OP9 coculture in the presence of 0 or 1μM GSI (7 N-[N-(3,5-difluorophenyl-L-alanyl]-S-phenylglycine t-butyl ester). Numbers in quadrants indicate the percentage of CD34+CD7+ T-cell precursors (A), CD19+HLA-DR+ B cells (B), CD14+CD4+ monocytes (C), and CD123+CD303+ pDCs (D). Results shown are representative of 3 independent experiments. Right panels: Corresponding average of the absolute number of CD34+CD7+ T-cell precursors (A), CD19+HLA-DR+ B cells (B), CD14+CD4+ monocytes (C), and CD123+CD303+ pDCs (D) for cultures depicted in the corresponding left panel. Error bars represent SEM.  indicates an increasing dosage of GSI, corresponding to 0, 0.1, 0.3, and 1μM GSI.
indicates an increasing dosage of GSI, corresponding to 0, 0.1, 0.3, and 1μM GSI.
Thus, these dose-dependent Notch inhibition experiments reveal a signal strength hierarchy among the different Notch ligands, whereby both Delta-like ligands appear to be the strongest Notch ligands, followed by Jagged2 and Jagged1. Because, besides an increase in progenitor cell expansion, virtually no changes in developmental outcome were observed between control and OP9-JAG1-cocultured HSCs, both in the presence of absence of GSI, this suggests that Jagged1-induced Notch signaling is very weak in HSCs.
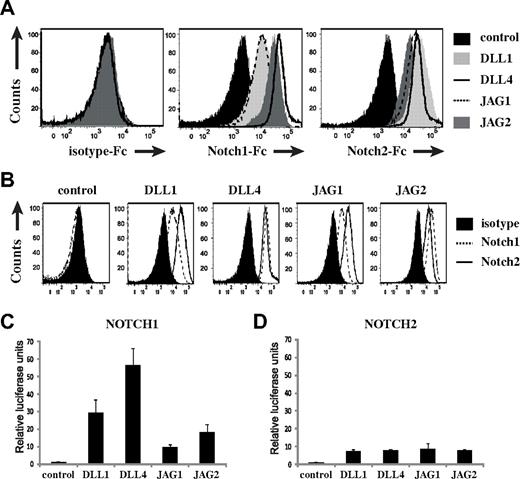
Notch ligands display differential receptor binding and signal strength induction
T-cell development critically depends on the Notch1 receptor, but CB HSCs express both Notch1 and Notch2.15,37 Therefore, we investigated the capacity and efficiency of Notch1 and Notch2 to bind to Delta-like-1, Delta-like-4, Jagged1, and Jagged2 to determine whether this could account for the previously observed developmental differences. In agreement, Notch1 displayed the lowest binding efficiency to Jagged1 (Figure 5A-B). Binding to Jagged2, however, was stronger, even stronger compared with Delta-like-1, but lower compared with Delta-like-4. Thus, all Notch ligands are capable of binding Notch1, although with different efficiencies. Notch2 displayed a slightly different binding pattern (Figure 5A-B). However, because Notch receptor/ligand binding efficiencies do not always correlate with an equivalent induction of Notch signal strength, we carried out specific Notch reporter assays using U2OS Tet-on flp-in cells bearing isogenic transgenes encoding Notch1-Gal4 or Notch2-Gal4 chimeric receptors,27 in which Notch was activated by coculture with K562 cells that each express a particular Notch ligand (supplemental Figure 5). As shown in Figure 5C, Notch1 specific activation confirmed the signal strength hierarchy that was observed in the coculture experiments. Jagged1 induced the lowest amount of Notch1 signaling, followed by Jagged2, Delta-like-1, and Delta-like-4. In contrast, Notch2-dependent Notch activation was very similar for all for Notch ligands (Figure 5D).
Ligand-specific Notch1 and Notch2 binding and signal strength transmission. (A) Staining of control-Fc, Notch1-Fc, and Notch2-Fc fusion proteins to K562 cells expressing the different Notch ligands as indicated. (B) Ligand-specific representation of the same data as in panel A. (C) Luciferase reporter assay of U2OS Tet-on flp-in-Notch1 and U2OS Tet-on flp-in-Notch2 cells cotransfected with CBF-luciferase reporter plasmid pGL2-Gal4-luciferase and the normalizing plasmid pRL-TK-expressing Renilla luciferase. After plasmid transfection, cells were cocultured with K562 expressing DLL1, DLL4, JAG1, JAG2, or control for 24 hours, and thereafter luciferase activity was measured. Bar graphs represent the average of 3 independent experiments. Errors bars represent SEM.
Ligand-specific Notch1 and Notch2 binding and signal strength transmission. (A) Staining of control-Fc, Notch1-Fc, and Notch2-Fc fusion proteins to K562 cells expressing the different Notch ligands as indicated. (B) Ligand-specific representation of the same data as in panel A. (C) Luciferase reporter assay of U2OS Tet-on flp-in-Notch1 and U2OS Tet-on flp-in-Notch2 cells cotransfected with CBF-luciferase reporter plasmid pGL2-Gal4-luciferase and the normalizing plasmid pRL-TK-expressing Renilla luciferase. After plasmid transfection, cells were cocultured with K562 expressing DLL1, DLL4, JAG1, JAG2, or control for 24 hours, and thereafter luciferase activity was measured. Bar graphs represent the average of 3 independent experiments. Errors bars represent SEM.
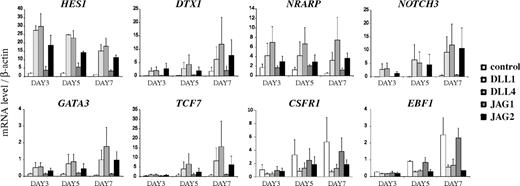
Jagged2, like Delta-like-1 and Delta-like-4, induces a T-lineage specific gene program
To gain insights into the molecular mechanisms that account for the differences in HSC lineage outcome on Notch ligand exposure, the impact on gene expression changes for known important mediators of hematopoietic lineage decisions was determined in human HSCs after OP9 coculture using quantitative RT-PCR (Figure 6).38 In agreement with their expression profile during early human T-cell development, the Notch1 target genes DTX1, HES1, NRARP, and NOTCH3 were strongly up-regulated in OP9-DLL1-, OP9-DLL4-, and OP9-JAG2-cocultured HSCs compared with the control, although slightly weaker in case of Jagged2-mediated activation, consistent with the results from the differentiation cultures. In contrast, Jagged1 only induced a small increase in HES1 and NRARP expression, but no increase in DTX1 or NOTCH3 was observed. In agreement with the capacity of Jagged2, Delta-like-1, and Delta-like-4 to induce and support T-cell development, the T-lineage transcription factors TCF7 and GATA3 were only up-regulated in HSCs exposed to these ligands, but not in cells cultured on OP9-JAG1 or control. These findings were also confirmed with human HSCs that were exposed to coated Notch ligand Fc fusion proteins (supplemental Figure 6). In contrast, the myeloid specific gene CSFR1 (coding for MCSF-R) and B-lineage specific transcription factor EBF1 were exclusively up-regulated in HSCs cultured on OP9-JAG1 and OP9-control, consistent with their developmental lineage outcome.
Jagged2 induces a T-lineage specific gene program. Kinetic and quantitative RT-PCR analysis of human CD34+lin− CB cells after coculture on OP9-control, OP9-DLL1, OP9-DLL4, OP9-JAG1, or OP9-JAG2 stromal cells. Cells were analyzed at different time points, as indicated in the x-axis. Units of expression are given relative to β-actin and relative to levels in the purified starting population. Data show the average of 2 sets of independent samples. Errors bars represent SEM.
Jagged2 induces a T-lineage specific gene program. Kinetic and quantitative RT-PCR analysis of human CD34+lin− CB cells after coculture on OP9-control, OP9-DLL1, OP9-DLL4, OP9-JAG1, or OP9-JAG2 stromal cells. Cells were analyzed at different time points, as indicated in the x-axis. Units of expression are given relative to β-actin and relative to levels in the purified starting population. Data show the average of 2 sets of independent samples. Errors bars represent SEM.
Thus, similar to Delta-like ligands, Jagged2 induces a strong enough Notch signal to impose a T-lineage program in human HSCs at the expense of other hematopoietic lineages. In contrast, Jagged1 is not capable of reaching the required Notch signal strength threshold to induced T-lineage differentiation and is not capable of blocking genes important for driving myeloid and B-cell fates.
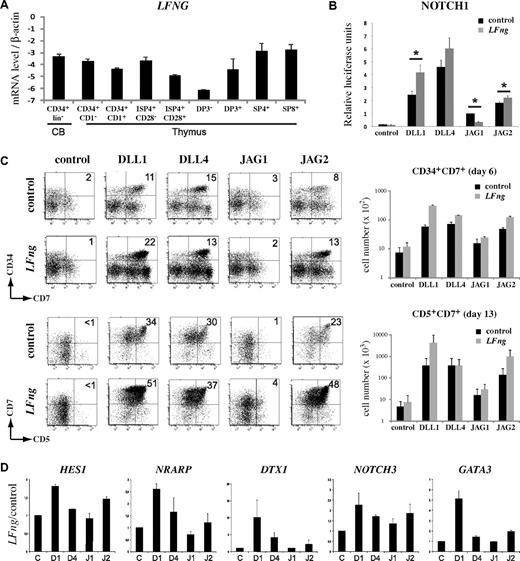
Fringe-mediated glycosylation increases Jagged2-induced Notch1 signaling
The differentiation cultures and gene expression analysis revealed clear functional differences between the Serrate-like ligands Jagged1 and Jagged2, with Jagged2 behaving very similarly as the Delta-like ligands. One important biochemical difference between the Serrate-like and Delta-like ligands is their behavior toward Fringe-glycosylated Notch receptors. The paradigm is that Fringe-mediated addition of N-acetylglucosamine to O-fucosylated EGF repeats on Notch receptors potentiates Delta-mediated Notch signaling but weakens Serrate-mediated Notch activity. Consistent with murine expression data,39 human CB CD34+CD38− HSCs express LFNG, the most important Fringe-family member during early T-cell development, to a similar level as early developing postnatal human thymocytes (Figure 7A). To investigate the effect of Fringe-mediated glycosylation on Jagged2-induced Notch signaling, we performed the same Notch reporter assay as described earlier, but this time with control or LFng transduced U2OS-Notch1 cells. As expected, LFng transduction increased Notch activation by both Delta-like ligands and reduced activation by Jagged1. Surprisingly, LFng also slightly increased Jagged2-mediated Notch activation in a significant manner (Figure 7B). In contrast, Lfng had no impact on Notch2-mediated activation by any of the 4 ligands (supplemental Figure 7). Next, we examined the effect of Fringe-mediated glycosylation on the developmental lineage potential of human HSCs on exposure to the various Notch ligands. Human CB CD34+lin− HSCs were transduced with either control or LFng and subsequently seeded onto OP9 cells expressing the different Notch ligands. Consistent with the reporter assays, LFng increased the frequency and absolute number of early CD34+CD7+ and CD7+CD5+ T-cell precursors derived from CB HSCs when cultured on OP9-DLL1, -DLL4 and -JAG2 (Figure 7C). Gene expression analysis further confirmed that this was mediated by increased Notch activation in LFng-transduced HSCs that were cultured on these 3 Notch ligands, as the Notch target genes HES1, DTX1, NRARP, and NOTCH3 were all up-regulated compared with control-transduced HSCs (Figure 7D).
Fringe-mediated glycosylation increases Jagged2-induced Notch1 signaling. (A) Quantitative RT-PCR of LFNG expression during human T-cell development.15,26 Data show the average of 3 independent sets of samples. Errors bars represent SEM. (B) Reporter assay of control and LFng-transduced U2OS Tet-on flp-in-Notch1 cells cotransfected with CBF-luciferase reporter plasmid pGL2-Gal4-luciferase and the normalizing plasmid pRL-TK expressing Renilla luciferase. After transfection, cells were cocultured with K562 expressing DLL1, DLL4, JAG1, JAG2, or control for 24 hours, and thereafter luciferase activity was measured. Bar graphs represent the average of 8 independent experiments. Errors bars represent SEM. (C) Left panels: Flow cytometric analysis of CD34+ Lin− CB cells transduced with either control or LFng after coculture on OP9 cells expressing different Notch ligands (indicated above dot plot). Numbers in the dot plots indicate the frequency of CD34+CD7+ and CD7+CD5+ T-cell precursors. Right bar graphs: Average of the corresponding absolute cell numbers. Errors bars represent SEM. (D) Quantitative RT-PCR analysis of gene expression in human CD34+lin− CB cells transduced with control or LFng, after 5 days of coculture on OP9-control (C), OP9-DLL1 (D1), OP9-DLL4 (D4), OP9-JAG1 (J1), or OP9-JAG2 (J2) stromal cells. Data show the ratio of expression in LFng versus control-transduced HPCs, and expression levels are normalized to β-actin. Data are the average from 2 sets of independent samples. Errors bars represent SEM.
Fringe-mediated glycosylation increases Jagged2-induced Notch1 signaling. (A) Quantitative RT-PCR of LFNG expression during human T-cell development.15,26 Data show the average of 3 independent sets of samples. Errors bars represent SEM. (B) Reporter assay of control and LFng-transduced U2OS Tet-on flp-in-Notch1 cells cotransfected with CBF-luciferase reporter plasmid pGL2-Gal4-luciferase and the normalizing plasmid pRL-TK expressing Renilla luciferase. After transfection, cells were cocultured with K562 expressing DLL1, DLL4, JAG1, JAG2, or control for 24 hours, and thereafter luciferase activity was measured. Bar graphs represent the average of 8 independent experiments. Errors bars represent SEM. (C) Left panels: Flow cytometric analysis of CD34+ Lin− CB cells transduced with either control or LFng after coculture on OP9 cells expressing different Notch ligands (indicated above dot plot). Numbers in the dot plots indicate the frequency of CD34+CD7+ and CD7+CD5+ T-cell precursors. Right bar graphs: Average of the corresponding absolute cell numbers. Errors bars represent SEM. (D) Quantitative RT-PCR analysis of gene expression in human CD34+lin− CB cells transduced with control or LFng, after 5 days of coculture on OP9-control (C), OP9-DLL1 (D1), OP9-DLL4 (D4), OP9-JAG1 (J1), or OP9-JAG2 (J2) stromal cells. Data show the ratio of expression in LFng versus control-transduced HPCs, and expression levels are normalized to β-actin. Data are the average from 2 sets of independent samples. Errors bars represent SEM.
Thus, Fringe-mediated glycosylation increases Jagged2-induced Notch signaling, similar as the Delta-like ligands, but in contrast to expected for Serrate-like ligands.
Discussion
Notch signaling is critical for various cell fate decisions, and activation of the pathway in mammals can be mediated by several different Notch ligands that have been classified into 2 different classes based on structural homology: Delta-like and Jagged (Serrate in Drosophila). Because very few studies have investigated the 4 activating canonical Notch ligands in a single model system, the nature of the difference between the various Notch ligands is still unclear. During hematopoiesis, Delta, but not Jagged, family members have been thought to induce T-cell and inhibit B-cell differentiation as a result of specific activation of Notch1. Here, we unexpectedly found Jagged2 able to activate Notch1 in addition to Notch2 and induce T-cell differentiation. We further show a hierarchy of Notch signaling strengths induced by Delta family members, followed by Jagged2 and Jagged1. Previous studies have shown that quantitative differences in Notch activation by varied densities of Delta-like-1 led to differences in cellular outcomes, with relatively lower amounts of Notch signaling induced in cells cultured with lower densities of Delta-like-1 leading to self-renewal of progenitors with primarily B-lymphoid and myeloid potential, whereas higher amounts of Notch signaling inhibited B-cell differentiation and promoted differentiation toward the T-cell lineage.25 Studies reported herein extend these findings, demonstrating quantitative differences in activation of Notch signaling induced by specific Notch ligands, a finding that may result from selective activation of Notch1 or Notch2 in HSCs. Our observation that glycosylation events regulate this hierarchy of Notch signaling strengths is also probably the result of glycosylation events regulated by genes, such as those mediated by Fringe family members in determining the selectivity of Notch ligands for specific Notch paralogs. Taken together, our findings clearly illustrate that each Notch ligand imposes a different signaling outcome by inducing quantitative differences in the strength of activation and by selective activation of specific Notch paralogs as a result of glucose-mediated modifications. This system provides the Notch pathway with a broad diversity of signaling outcomes, consistent with its essential role in a variety of developmental systems that demand differential signaling outcomes through this common pathway.
The hematopoietic system has been well studied and is therefore a suitable developmental model to study the functional and biochemical differences between the different Notch ligands, particularly because Notch signaling has been so extensively studied in this system.10-12,40 Especially the role of Notch1 in mediating the T- versus B-cell lineage decision has been well established both in mouse and human.19,41,42 Yet, the consensus thus far, based on studies that included both Delta-like ligands and Jagged1, but not Jagged2, has been that Delta-like ligands, but not Jagged ligands, can initiate T-cell development in HPCs at the expense of B-cell differentiation via activation of Notch1. Our results unambiguously show that Jagged2 can also mediate these cell fate decisions in both mouse and human, similar to Delta-like ligands. As a consequence, this indicates that, among the Notch ligands, Jagged1 has a unique role within the hematopoietic system. Jagged1 is the main Notch ligand that is expressed in the HSC niche.35 Although a role for Notch signaling in HSC has been controversial, recent data have indicated that Notch2 enhances the rate of formation of short-term repopulating multipotent progenitor cells as well as long-term repopulating HSCs, while delaying myeloid differentiation in bone marrow during nonhomeostatic conditions, including after chemotherapy induced injury or during marrow regeneration after stem cell transplantation.43 Within the thymus, Jagged1 could be involved in reducing and/or preventing Notch activation during the later stages of T-cell development by antagonizing the other Notch ligands, similar to its function during angiogenesis.44 Consistently, Jagged1 is primarily expressed in the medulla, the site where thymocytes undergo the final stages of T-cell maturation, a process that is Notch1 independent.45 Such a mechanism could explain the lack of Notch target gene expression in post-β selection thymocytes,26,46 despite the presence of Notch1 protein on the surface of these cells.47
Through several experiments, we show that the different Notch ligands each possess a different level of activation strength that is specific for signaling through Notch1, with Delta-like-4 being the strongest Notch1 activator, followed by Delta-like-1, Jagged2, and Jagged1, respectively. We did not observe such ligand-specific activation events for Notch2, indicating that the main developmental effects of quantitative differences in ligand strength seen in HPC differentiation are mediated through Notch1, and this is consistent with its essential nonredundant role during early T-cell development,48 in contrast to Notch2.49 Because these ligand-specific differences in Notch1 activation are observed in primary cells through both differentiation cultures and gene expression analysis, as well as in a cell line model in which reporter gene expression is directly regulated by the amount of cleaved Notch receptor protein, these findings suggest that Notch ligands induce quantitative differences, through the amount of ICN protein that is released, that ultimately lead to qualitative differences. With respect to the induction of early T-cell development, gene expression analysis shows a clear up-regulation of Notch target genes by both Delta-like ligands and Jagged2, suggesting that Jagged2 also induces a sufficient strong Notch signal to impose a T-lineage program in human HSCs at the expense of other hematopoietic lineages. In agreement, genes that are critical for driving alternative lineages are repressed. In contrast, Jagged1 is not capable of reaching the required Notch signal strength threshold to induce T-lineage differentiation and is not capable of blocking genes that are important for driving myeloid and B-cell fates. One factor that mediates this threshold is LRF as loss of this gene results in T-cell development in the bone marrow under normal Notch signaling conditions,50 but we observed no significant differences in LRF mRNA expression in response to the different Notch ligands (data not shown), suggesting that protein-protein interactions might be responsible for passing the LRF-mediated threshold. This might involve the activity of Deltex1, Nrarp, and/or Notch3. Although their role during early T-cell development is still unclear, it is probable that at least one of these genes mediates the specific hematopoietic lineage decisions that result from the differential Notch activation because these genes are not up-regulated in response to Jagged1, in contrast to when Notch activation is induced by the other ligands. Alternatively, Jagged1 may specifically lack or possess the potential to activate so-called noncanonical Notch signaling pathways that interfere with the canonical cascade in a positive or negative manner, respectively.
For several reasons, we are convinced that the observed differences in Notch activation capacity between the different Notch ligands are not the result of variations in expression levels but instead reflect the true biologic differences between these ligands. First, each of the OP9 cells that were generated was sorted based on similar EGFP expression levels, and fluorescence-activated cell sorter analysis with antibodies against Notch ligands revealed that the amount of Notch ligand protein that is expressed indeed correlates with the amount of EGFP protein, as also illustrated previously.31 Second, we confirmed the expression of similar mRNA levels for each ligand through quantitative RT-PCR. Third, similar differences in activation capacity were obtained in 2 different models systems (HPCs and U2OS cells), in which the Notch ligands were expressed by 2 different cell lines (OP9 and K562 cells) or through coating of an equal amount of protein (Notch ligand-Fc proteins). The observed signal strength hierarchy seems specific for Notch1 as we could not detect any significant difference in Notch activation when signaling occurred exclusively through Notch2. Our findings are also in agreement with a recent report that showed that Jagged1-induced Notch1 signaling is most sensitive to blocking Notch1 antibodies, followed by Delta-like-1, Jagged2, and Delta-like-4.51 The difference in Notch1 activation potential is at least in part correlated with the capacity of each ligand to bind to the Notch1 receptor. However, as also observed by others,8,52 this correlation is not absolute because Jagged2 displays a higher Notch1-binding capacity compared with Delta-like-1, yet Jagged2 is a weaker activator. Consistently, other mechanisms, such as ligand endocytosis and glycosylation, might also contribute to the signaling outcome.53
One aspect that contributes to the potential of Jagged2 to induce a sufficient strong Notch signal that supports the induction of T-lineage differentiation is the observation that Fringe-mediated glycosylation of Notch1 slightly increases Jagged2-mediated activation, as illustrated with experiments both in primary cells and in a cell line model. Indeed, both mouse and human HPCs express Lunatic-Fringe, implying that their Notch receptors are modified with N-acetylglucosamine residues, and it is well established that these modifications alter ligand-induced Notch activation. In contrast to Jagged1 whose activity is strongly weakened by Lunatic-Fringe-modified Notch1, Jagged2 induced Notch1 signaling was not reduced but in contrast even slightly increased, suggesting that Jagged2 might not behave as a classic Serrate-like Notch1 ligand. Although Fringe-deficient knockout mice will provide more direct evidence for this, our results suggest that the differential capacity of Jagged1 and Jagged2 to induce T-cell development is potentially mediated by a different sensitivity toward Fringe-modified Notch1. To our knowledge, an increase in Jagged-mediated Notch activation by Lunatic-Fringe has thus far only been illustrated for Jagged1-mediated Notch2 activation.8 Together, these findings suggest that the effects of Fringe-mediated glycosylation on Notch activation depend on the specific receptor-ligand interaction, indicating that Fringe proteins play a critical role in mediating Notch signaling effects.
For the hematopoietic system, the high diversity of signaling outcomes that is created by the various components of the Notch signaling pathway seems critical to discriminate between numerous developmental and differentiation events that are Notch-dependent in many different blood cell types, as recently reviewed.11,12,54 In particular, T cells depend on various Notch signaling events during their life span, from their earliest point of development until they reach functional competence.9,11,12,54,55 A variety of T-lineage subsets are generated intrathymically and extrathymically, with further differences between fetal and postnatal thymopoiesis. At present, it is clear that postnatal intrathymic early thymic progenitor formation depends on the DLL4/Notch1 interaction20,21,48 ; however, it is still unknown which Notch ligands are responsible for inducing extrathymic and fetal intrathymic T-cell development. It also remains to be determined which Notch ligands drive the further Notch-dependent differentiation of uncommitted early thymic progenitors into committed T-cell precursors, and which ligands control their further diversification into the various T-cell subsets. Because of its abundant expression in the thymus, its efficient binding to Notch1, and its potential to increase Notch1-dependent activation after Lfng-mediated glycosylation, Jagged2 is a good candidate to be involved in some of these processes. Furthermore, Jagged2-deficient mice display a reduction in fetal TCR-γδ T-cell development,24 suggesting an important role for this ligand in these processes. Site-specific conditional deletion of Jagged2 will be of critical importance to address these questions.
In each case, our results significantly advance our understanding of the Notch ligands. Although evolutionary derived from either a Serrate or a Delta ligand in Drosophila, our functional and biochemical data suggst that the mammalian Jagged and Delta-like Notch ligands have evolved independently to broaden the scope of Notch signaling effects. Therefore, it will also be of great interest to determine whether our observations also pertain to other developmental systems.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Weinmaster (University of California-Los Angeles, Los Angeles, CA) for the C2C12N1 cells, Dr Zúñiga-Pflücker (University of Toronto, Toronto, ON) for OP9 stromal cells, Dr Parreira for the LZRS-DLL1 and LZRS-JAG1 constructs (Instituto de Histologia e Embriologia, Lisboa, Portugal), and Dr Harris (University of Oxford, Oxford, United Kingdom) and Dr Tosato (National Institutes of Health, Bethesda, MD) for the LZRS-DLL4 construct.
This work was supported by the Odysseus program of the Fund for Scientific Research Flanders, the Odysseus program of the Fund for Scientific Research Flanders, the Flemish Institute for the advancement of Scientific-Technological Research in the Industry, and the Concerted Research Action of Ghent University. T.T. is a postdoctoral researcher of the Odysseus program of the Fund for Scientific Research Flanders. I.V.d.W. is a PhD student supported by a grant from the Flemish Institute for the advancement of Scientific-Technological Research in the Industry. M.G. and B.K. are supported by the German Cancer Research Center and the EU Consortium “Tolerage.”
Authorship
Contribution: I.V.d.W. and T.T. performed and designed research, analyzed and interpreted data, and wrote the manuscript; G.D.S. and M.G. performed research; M.D.S. performed research and interpreted data; E.W. performed and designed research and analyzed and interpreted data; B.V., G.L., and J.C.A. provided critical reagents; J.P. provided critical reagents, designed research, and interpreted data; and I.D.B., C.J.G., and B.K. provided critical reagents and interpreted data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tom Taghon, Department of Clinical Chemistry, Microbiology and Immunology, Ghent University, University Hospital Ghent, 4BlokA, De Pintelaan 185, B-9000 Ghent, Belgium; e-mail: Tom.Taghon@ugent.be.

![Figure 4. Dose-dependent Notch inhibition reveals a signal strength hierarchy. Effects of dose-dependent Notch inhibition on the induction of T-lineage differentiation (A), B-cell development (B), monocyte development (C), and plasmacytoid differentiation (pDC; D). Left panels: Flow cytometric analysis of human CD34+CD38−lin− CB HPCs after OP9 coculture in the presence of 0 or 1μM GSI (7 N-[N-(3,5-difluorophenyl-L-alanyl]-S-phenylglycine t-butyl ester). Numbers in quadrants indicate the percentage of CD34+CD7+ T-cell precursors (A), CD19+HLA-DR+ B cells (B), CD14+CD4+ monocytes (C), and CD123+CD303+ pDCs (D). Results shown are representative of 3 independent experiments. Right panels: Corresponding average of the absolute number of CD34+CD7+ T-cell precursors (A), CD19+HLA-DR+ B cells (B), CD14+CD4+ monocytes (C), and CD123+CD303+ pDCs (D) for cultures depicted in the corresponding left panel. Error bars represent SEM. indicates an increasing dosage of GSI, corresponding to 0, 0.1, 0.3, and 1μM GSI.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/17/10.1182_blood-2010-06-290049/4/m_zh89991170050004.jpeg?Expires=1765905992&Signature=XdCnl4~UWUTh3mmrW-5pL6xh6c03vfzkQHvN-vniO6ZjOb8JdmQAm89Uh0CPZCieiUcwVlyN~eAD2vbQ8W9L86mFm0PQn1UE~4ekn9hkUn9g~-7ocKMN-sdzMUlKzc35JaocySQgxBSw9o9wxEbG0VasYk-bc4FHeMN6YA2Hu5GnBBmIZejAN-ryhpfzRzOafQeM1mlwH0X9Mxp-6HDP9G1HBDNYegOdfioTaspsE-u4Ou3sU867nHp5Rw~9Nb7CrKhu4ZDuPX0Jv7TckNk38CP48FTcIaG4GsPxr8~TP1fTEAymwls4y7iPDTW1fmsOptXYLRl4bt2r2CBDkaFmag__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)