Abstract
Cytokine-mediated phosphorylation of Erk (pErk), ribosomal S6 (pS6), and Stat5 (pStat5) in CD34+/CD117+ blast cells in normal bone marrow from 9 healthy adult donors were analyzed over 60 minutes. Treatment with stem cell factor (SCF), Flt3-ligand (FL), IL-3, and GM-CSF and measurement by multiparametric flow cytometry yielded distinctive, highly uniform phosphoprotein kinetic profiles despite a diverse sample population. The correlated responses for SCF- and FL-stimulated pErk and pS6 were similar. Half the population phosphorylated Erk in response to SCF between 0.9 and 1.2 minutes, and S6 phosphorylation followed approximately a minute later (t½pS6 rise = 2.2-2.7 minutes). The FL response was equally fast but more variable (t½pErk rise = 0.9-1.3 minutes; t½pS6 rise = 2.5-3.5 minutes). Stat5 was not activated in 97% of the cells by either cytokine. IL-3 and GM-CSF were similar to each other with half of blast cells phosphorylating Stat5 and 15% to 20% responding through Erk and S6. Limited comparison with leukemic blasts confirmed universal abnormal signaling in AML that is significantly different from normal bone marrow blasts. These differences included sustained signals, a larger fraction of responding cells, and amplification of phosphorylation levels for at least one phosphoprotein. These data support the eventual use of this approach for disease diagnosis and monitoring.
Introduction
Hematopoiesis is regulated by a complex system of cytokines and receptors that transmit information on cellular demand through “cell signaling.” Current qualitative information is impressive and describes the system at cellular and subcellular levels with narrative/pictorial models derived from various venues. A quantitative viewpoint, particularly at the molecular level, does not exist. This is evident when the rigor of mathematic modeling is applied to cell signaling.1 Relative expression data at the protein and epitope level are not scarce but are scattered and buried in studies focused on other aspects. Aside from the lymphocytic branch of the human system, signaling dynamics are generally known through transformed cell line models. With this in mind, we measured cytokine stimulation of a limited set of pathways in human CD34+/CD117+ cells at closely spaced intervals in whole bone marrow aspirates (ie, without perturbation other than exposure to heparin, air, and loss of microenvironment). Recent whole blood fixation/permeabilization2 and cell-based methods to measure phosphoepitopes3-8 enabled this effort. In addition to a desire for pathway signal information that reflects natural systems, we also designed this study to provide baseline detail for the normal adult state to compare signaling in myeloid leukemias. This is preliminary to test the idea that cell signaling could be a leukemia “biomarker,” useful in hematopathology.
Leukemogenesis is an evolutionary process resulting in progressive, malignant disease. An oversimplified but attractive model for acute myeloid leukemia (AML) is that mutant transcription factors reduce the differentiation rate, and additional mutations in signaling genes promote proliferation and survival of hematopoietic precursor stages.9-12 Because the current classification schemes (French-American-British [FAB] and World Health Organization) rely on differentiation/maturation-related morphology and chromosomal translocations that often involve transcription factors, an inference from that model is that analysis of signaling would complement current diagnosis. Because significant targeted therapy efforts have been focused on kinase inhibitors, this enhanced information could affect therapeutic decisions.
An integrated, long-term study requires early, limiting decisions. First, we used whole bone marrow and flow cytometry to obtain quantitative, correlated data on samples that were minimally perturbed. Second, we focused on stem cell factor (SCF) and Flt3 ligand (FL) because the receptors are often mutated in AML.13-17 Third, we included both GM-CSF and IL-3 because both are important in myelopoiesis and the receptors are not often mutated in AML. Phosphorylation of Erk (pErk), ribosomal protein S6 (pS6), and Stat5 (pStat5) were chosen as endpoints because they have been well studied as cytometric signaling endpoints18-28 and antibodies were available as primary conjugates. We limited this study to a cell population defined by CD34, CD117, CD45, and side scatter that contains hematopoietic stem cells and multipotential myeloid precursors. With this reduced complexity, we could perform fine kinetics, defined as stimulation followed by assay at closely spaced intervals over a period that would allow us to capture the shape of the primary signal.
We stimulated unfractionated bone marrows from healthy adult volunteers with 4 cytokines to measure correlated signaling with 3 endpoints. This equates to 12 pathways, with a pathway defined as ligand → endpoint. In addition, we compared these results with SCF and FL stimulation of 4 AML patients of different FAB classes. With this limited sampling, we show distinct, classifiable signaling differences in the blast cells of each patient compared with normal bone marrow blasts.
Methods
Antibodies and reagents
Phospho-Erk1/2-Alexa 488 (pErk, clone E10), CD34-phycoerythrin (CD34, clone QBEnd10), CD117-PC7 (CD117, clone 104D2D1), and phospho-S235/S236-S6 ribosomal protein (pS6, clone D57.2.2E) from Cell Signaling Technologies conjugated to Pacific Blue (F/P = 4.0), were from Beckman Coulter. Phospho-Y694-STAT5-Alexa 647 (pSTAT5, clone 47) and CD45-peridinin chlorophyll protein (CD45, clone 2D1) were from BD Biosciences.
SCF (GenScript Corporation) and FL, IL-3, and GM-CSF (R&D Systems) stock solutions were prepared in phosphate-buffered saline (PBS) with 4% bovine serum albumin (Sigma-Aldrich). U0126 and rapamycin (Invitrogen) stock solutions were prepared in dimethyl sulfoxide and diluted with 10% dimethyl sulfoxide in PBS on use. Dilute Triton X-100 (eg, 0.127%) in PBS was prepared from 10% Triton X-100 (Thermo Scientific). Wash buffer was PBS with 4% bovine serum albumin or 4% heat-inactivated fetal bovine serum (Invitrogen). The Triton X-100 solution and wash buffer were supplemented with phosphatase inhibitors at final concentrations 0.2mM sodium orthovanadate (Sigma-Aldrich), 2mM sodium pyrophosphate decahydrate (Fisher Scientific), 2mM β-glycerophosphate (Sigma-Aldrich), and 10mM sodium fluoride (Sigma-Aldrich). A total of 80% methanol (Fisher Scientific) was prepared in PBS.
Bone marrow samples
Acquisition was approved by the Case Cancer Institutional Review Board of the Case Western Reserve University School of Medicine. Volunteers provided informed consent in accordance with the Declaration of Helsinki. Patient information was delinked. For healthy volunteers, 10 to 20 mL of normal bone marrow (NBM) was aspirated from the posterior iliac crest. Samples were collected and filtered through 40-μm nylon mesh. Elapsed time from aspiration to experiment was approximately 1 hour. Discarded BM from 4 AML patients was acquired within 24 hours of aspiration (supplemental Data, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Cytokine signaling and cytometry
A total of 100 to 200 μL of BM, containing approximately 2 million white blood cells, determined by a Hemavet 950 FS (Drew Scientific), were dispensed into 12 × 75 mm polystyrene round-bottom tubes (BD Biosciences) and incubated at 37°C (Thermolyne Dri-Bath) for 30 minutes. For inhibition, 100μM U0126, 1 μg/mL rapamycin, or both (final concentration) were added 20 minutes before cytokine addition. Signaling was initiated by addition of SCF (100 ng/mL) or FL, IL-3, or GM-CSF (25 ng/mL) and continued for 0, 1, 2, 4, 6, 8, 10, 15, 20, 25, 30, and 60 minutes (or a subset), and stopped by addition of formaldehyde to 4% (Polysciences) for 10 minutes at room temperature. Red blood cells were lysed with a dilute Triton X-100/PBS solution (37°C for 20 minutes), an equal volume of ice-cold wash buffer was added, and the suspension was centrifuged (300g, 6 minutes, 4°C). Cells were dehydrated with 80% methanol (prechilled to −20°C) and incubated on ice for 10 minutes. Then, cells were washed with ice-cold PBS, blocked with ice-cold wash buffer (4°C for 30 minutes), and incubated with an antibody cocktail (5 or 10 μL of pS6–Pacific Blue [0.25, 0.50 μg], 5 or 10 μL of pERK-Alexa 488, 10 μL of pSTAT5-Alexa 647, 20 μL of CD34-phycoerythrin, 10 μL of CD45–peridinin chlorophyll protein, and 10 μL of CD117-PC7 per tube) for 60 minutes at 4°C. Finally, samples were washed and resuspended in PBS. In some cases, cells were transferred to 96-well microplates (BD Biosciences) for antibody binding, washing, and cytometry. Samples were analyzed with a BD LSR II (BD Biosciences) or Gallios (Beckman Coulter) flow cytometer. A detailed protocol is available in Supplemental data.
Instrument settings and compensation were set with BD CompBeads (BD Biosciences). For Pacific Blue compensation, a single-stained, cellular control (ie, an aliquot of bone marrow treated with 0.5μM PMA for 10 minutes at 37°C) was used for the pS6-rabbit antibody which fails to react with anti-mouse CompBeads.
Data analysis
We analyzed cytometry data with WinList 6.0 3D (Verity Software House), and graphical analyses of signaling responses were analyzed in Excel (Microsoft) or GraphPad Prism, Version 5.03 (GraphPad Software). The level of phosphorylation was determined by calculating the median fluorescence intensity (MFI) on an entire subpopulation-specific dataset (both positive- and negative-stained cells). The purpose was to obtain the expression level for the population. MFIs were normalized to the stained BD CompBead controls (or, for pS6, to a cellular control), and backgrounds were subtracted. The resulting data were further transformed within each endpoint response by normalization to the average maximum or peak response of the volunteer samples. Response frequencies (fractions of responding cells) were calculated by Frequency = ([no. of phosphorylation-positive cells]/[total cells]) × 100 for the CD34+/CD117+ subpopulation.
Results
Healthy volunteer demographics
Samples from healthy donors (5 males, 4 females; age range, 26-49 years) contained 7 to 31 million white blood cells per milliliter (Table 1). Bone marrow samples were received and experiments were initiated within 1 hour of donation. Discarded AML samples from 4 females (29-76 years of age) were also acquired (Supplemental data). These samples were mixed 1:1 with RPMI 1640 with 10% fetal bovine serum and stored overnight at room temperature.
Demographics of healthy donor and AML BM
| BM . | Sex . | Age, y . | WBCs, 106/mL . |
|---|---|---|---|
| Donor 1 | Female | 33 | 16.2 |
| Donor 2 | Male | 31 | 15.0 |
| Donor 3 | Female | 26 | 10.6 |
| Donor 4 | Male | 48 | 30.8 |
| Donor 5 | Male | 43 | 10.1 |
| Donor 6 | Male | 47 | 9.9 |
| Donor 7 | Female | 31 | 27.1 |
| Donor 8 | Male | 49 | 26.3 |
| Donor 9 | Female | 26 | 7.4 |
| AML1 (M4) | Female | 71 | 40.7 |
| AML2 (M2) | Female | 29 | 11.5 |
| AML3 (M4Eo) | Female | 76 | 73.0 |
| AML5 (M1) | Female | 63 | 26.5 |
| BM . | Sex . | Age, y . | WBCs, 106/mL . |
|---|---|---|---|
| Donor 1 | Female | 33 | 16.2 |
| Donor 2 | Male | 31 | 15.0 |
| Donor 3 | Female | 26 | 10.6 |
| Donor 4 | Male | 48 | 30.8 |
| Donor 5 | Male | 43 | 10.1 |
| Donor 6 | Male | 47 | 9.9 |
| Donor 7 | Female | 31 | 27.1 |
| Donor 8 | Male | 49 | 26.3 |
| Donor 9 | Female | 26 | 7.4 |
| AML1 (M4) | Female | 71 | 40.7 |
| AML2 (M2) | Female | 29 | 11.5 |
| AML3 (M4Eo) | Female | 76 | 73.0 |
| AML5 (M1) | Female | 63 | 26.5 |
WBCs indicate white blood cells.
Blast cell population definition and phosphorylation measurements
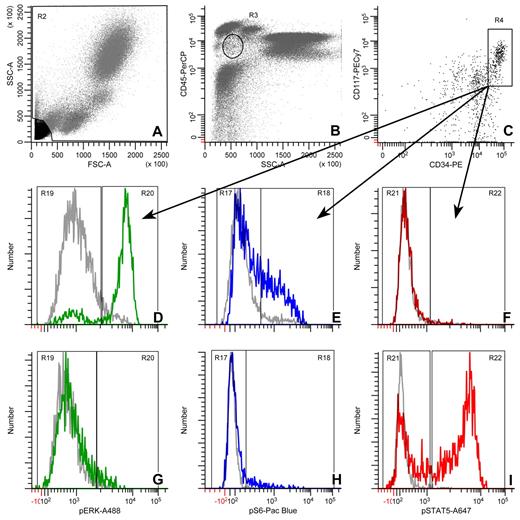
To restrict measurements to a specific, relevant population, we used the gating scheme shown in Figure 1 to identify the blast, CD34+, CD117+ cells. The single-parameter histograms show typical responses in these cells for SCF → pErk (2 minutes), pS6 (4 minutes), and pStat5 (2 minutes), and for IL-3 → pErk, pS6, and pStat5 (all at 8 minutes).
Gating strategy for pErk, pS6, and pStat5 in CD34+, CD117+ blast cells. (A) A region used to exclude debris. (B) The circle represents the blast region defined by CD45 expression and side scatter. (C) The box shows the CD34+/CD117+ region. (D,G) pErk histograms at t = 0 (gray) or 2 (green) minutes for SCF and t = 0 (gray) or 8 (green) minutes for IL-3, respectively. (E,H) pS6 histograms at t = 0 (gray) or 4 (blue) minutes for SCF and t = 0 (gray) or 8 (blue) minutes for IL-3, respectively. (F,I) pStat5 histograms at t = 0 (gray) or 2 (red) minutes for SCF and t = 0 (gray) or 8 (red) minutes for IL-3, respectively. The histograms are representative of all the flow cytometry data reported here.
Gating strategy for pErk, pS6, and pStat5 in CD34+, CD117+ blast cells. (A) A region used to exclude debris. (B) The circle represents the blast region defined by CD45 expression and side scatter. (C) The box shows the CD34+/CD117+ region. (D,G) pErk histograms at t = 0 (gray) or 2 (green) minutes for SCF and t = 0 (gray) or 8 (green) minutes for IL-3, respectively. (E,H) pS6 histograms at t = 0 (gray) or 4 (blue) minutes for SCF and t = 0 (gray) or 8 (blue) minutes for IL-3, respectively. (F,I) pStat5 histograms at t = 0 (gray) or 2 (red) minutes for SCF and t = 0 (gray) or 8 (red) minutes for IL-3, respectively. The histograms are representative of all the flow cytometry data reported here.
Assay specificity
The specificity of stimulated responses was demonstrated by U0126 and rapamycin inhibition of Erk and S6 phosphorylation (Mek and mTOR inhibition, respectively: supplemental Figure 1). U0126 completely inhibited SCF-stimulated Erk phosphorylation. Each inhibitor reduced S6 phosphorylation partially. Both inhibitors combined reduced S6 phosphorylation to background levels, confirming that both Ras/Erk and PI3K/mTOR pathways contribute. The specificity of SCF and FL in CD34+/CD117+ cells was further demonstrated by their inability to stimulate phosphorylation in lymphocytes (supplemental Figure 1).
Signaling in NBM
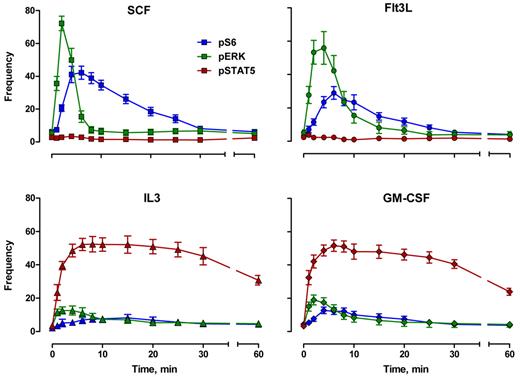
The CD34+/CD117+ cell response kinetics for 4 cytokines and 3 correlated phosphoprotein endpoints are shown in Figures 2 through 5. Figure 2 shows the timing and variation for each pathway response with respect to the fraction of the population that responded. Figures 3 and 4 show some of the same responses with each individual response normalized to itself. This eliminates constitutive signaling (by setting the untreated level to 0) and focuses on the timing and shape of the response (by setting each peak to 100). In these figures, deviation from 100 at the peak is an indication of the donor-to-donor variability for each signal.
Cytokine-mediated phosphoprotein kinetic profiles: frequency. Frequency indicates percentage of responding blast cells. Error bars represent SEM.
Cytokine-mediated phosphoprotein kinetic profiles: frequency. Frequency indicates percentage of responding blast cells. Error bars represent SEM.
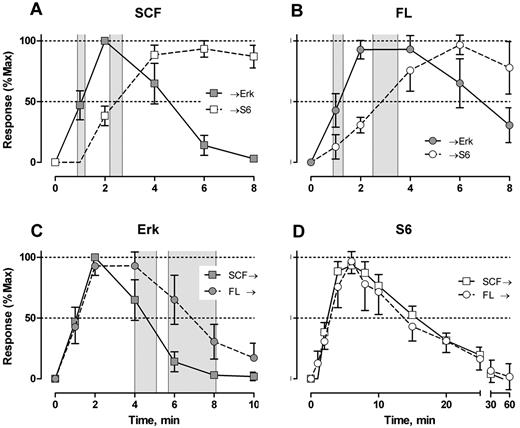
SCF- and FL-mediated Erk and S6 signal shape analysis. Response indicates percentage of responding cells with t = 0 level subtracted. Individual signaling profiles were normalized to t = 0 (equal to 0 response) and the largest value (maximum). t½Erk/S6 rise indicates time to 50% of maximum; and t½Erk/S6 decay, time to 50% loss of maximum. Gray zones represent 95% CI for t½Erk rise (A-B) and t½Erk decay (C). The response shapes for S6 phosphorylation were not significantly different (D). Error bars represent 95% CI.
SCF- and FL-mediated Erk and S6 signal shape analysis. Response indicates percentage of responding cells with t = 0 level subtracted. Individual signaling profiles were normalized to t = 0 (equal to 0 response) and the largest value (maximum). t½Erk/S6 rise indicates time to 50% of maximum; and t½Erk/S6 decay, time to 50% loss of maximum. Gray zones represent 95% CI for t½Erk rise (A-B) and t½Erk decay (C). The response shapes for S6 phosphorylation were not significantly different (D). Error bars represent 95% CI.
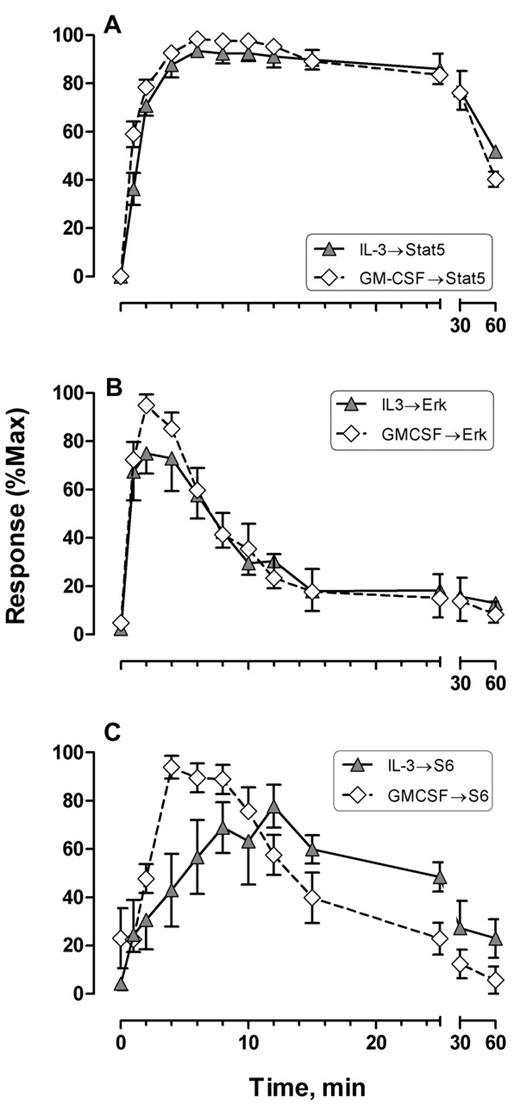
IL-3– and GM-CSF–mediated Stat5, Erk, and S6 signal shape comparisons. Response indicates percentage of responding cells with t = 0 level subtracted. Error bars represent SEM. Data were processed as described in the legend to Figure 3.
IL-3– and GM-CSF–mediated Stat5, Erk, and S6 signal shape comparisons. Response indicates percentage of responding cells with t = 0 level subtracted. Error bars represent SEM. Data were processed as described in the legend to Figure 3.
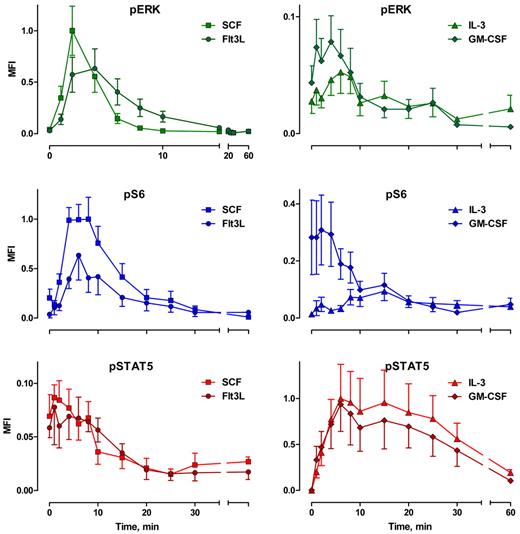
Cytokine-mediated phosphoprotein kinetic profiles: MFI. Error bars represent SEM.
Cytokine-mediated phosphoprotein kinetic profiles: MFI. Error bars represent SEM.
The overall response structure for each cytokine was essentially as expected from model systems (eg, Erk is phosphorylated before S6; the receptor tyrosine kinases signal strongly through Erk and S6; IL-3 and GM-CSF signal strongly through Stat5). However, these data provide the following new information. First, the responses were remarkably uniform in terms of timing and the populations that respond. Figure 2 represents the results from 9 persons of different ages and both sexes, randomly sampled. Second, although the correlated response structures for SCF and FL were similar (Figure 2), Stat5 was not activated in 97% of the cells, in contrast to interpretations of data derived from model systems.18 IL-3 and GM-CSF were quite similar to each other with at least half of the blast cells phosphorylating Stat5 and 15% to 20% responding through Erk and S6 (Figure 2). This must represent population heterogeneity. Third, the receptor tyrosine kinase responses were very fast. Half the population phosphorylated Erk in response to SCF between 0.9 and 1.2 minutes (95% confidence interval [CI]), and S6 phosphorylation followed approximately a minute later (t½pS6 rise = 2.2-2.7 minutes; Figure 3A). The FL/Flt3 response was equally fast but more variable (t½pErk rise = 0.9-1.3 minutes; t½pS6 rise = 2.5-3.5 minutes; Figure 3B). Fourth, the characteristic specificity of each cytokine, known by phenotype, was apparent biochemically. For example, the FL/Flt3 response through Erk was clearly different from SCF/c-Kit. CD34+/CD117+ cells phosphorylated Erk via Flt3 or c-Kit at the same rate, but the Flt3-mediated signal was held longer (t½pErk decay = 4.0-5.1 and 5.7-8.1 minutes for SCF and FL, respectively, Figure 3C). These timings cannot be true if the underlying biochemistries are exactly the same. In contrast, there were no differences in the rates for S6 phosphorylation/dephosphorylation for the 2 cytokines (Figure 3D). The simplest interpretation is that the 2 ligand/receptor systems signal differently to Erk but equivalently down the PI3K → mTor branch of the network (although the latter interpretation does not have to be true). The timing and fraction of cells responding to IL-3– and GM-CSF–mediated Stat5 phosphorylation responses rates were remarkably the same with a slight (but significant) difference in peak variability (Figure 4A). The rates of Stat5 phosphorylation and dephosphorylation were remarkably uniform between the 2 different receptor systems (Figures 2A, 4A, 5F). The IL-3– and GM-CSF–mediated Erk responses also displayed equivalent phosphorylation/dephosphorylation rates with more variation in the fraction responding to IL-3 (Figure 4B). Finally, the S6 signals were distinctly different with different response rates and different response intervals (Figures 2C-D, 4C). These responses provide distinct evidence for an individual character of each ligand/receptor response while using the same endpoints. This can be compared with contrast to canonical descriptions of the same pathways.
In Figure 5, we have plotted the MFIs of the CD34+/CD117+ population, which are proportional to the levels of phosphorylation. Here the responses have been normalized with the peak Erk and S6 signals in response to SCF and the IL-3-mediated Stat5 signal set to 1.0. This allows direct comparisons between cytokines. These measurements (Figure 5) are more variable than the fractional data (Figure 2) but demonstrated the same kinetic response patterns. The normalized data show that FL-driven Erk and S6 phosphorylation is approximately 40% lower than SCF-driven responses (Figure 5A). Further, while the Erk response to IL-3 or GM-CSF is measurable (10%-20% of the cells distinctly show a measurable response; Figure 2C-D), the phosphorylation level is low (5%-10% of the SCF-driven pErk levels). Equally, S6 phosphorylation in response to IL-3 is 9% of SCF-mediated peak response. GM-CSF is more complex with what appears to be a decaying moderate (30% of the SCF-mediated peak) constitutive response added to a weak (12% of SCF levels) GM-CSF mediated response. Increased cytokine concentrations may increase these phosphorylation responses; however, the doses used here are relatively “standard.” These weak Erk and S6 responses to GM-CSF can be contrasted within the same reactions to the response of monocytes, which can be equal to or much higher than SCF- or Flt3-driven Erk and S6 responses (supplemental Figure 2).
In summary, the fractions of responding cells provide signal shape, robust timing information, donor-to-donor variability, and an indication of the underlying population heterogeneity. The intensity of phosphorylation provides an indication of signal strength in terms of the average levels of phosphorylation of the endpoint proteins. In each case, the overall structure of the responses agrees with what we know from model systems and qualitative work. The details of the signaling responses, for each cytokine, display response specificity that should be related to the phenotypic specificity that is characteristic. This dataset and analysis represent the best sampling set that we could obtain and therefore are the response baseline or landscape for comparing other samples (eg, samples that are > 1 hour removed from the body, “hematologically normal” samples, such as staging marrows, for lymphomas, and discarded marrow from joint replacement surgeries) and provides guideposts for comparing abnormal samples (myeloproliferative and myelodysplastic disorders, and myeloid leukemias). In the following text, we compare 4 AML samples that are up to 24 hours old. Because we expect that samples change as a function of time after aspiration, signaling change may occur as well. A thorough investigation of these changes requires a large study. For the present, we have performed a limited analysis, comparing fresh to aged samples of normal and leukemia samples, and we observed small changes in NBM relative to the response range of NBM and overall small changes in AML samples relative to the differences displayed between NBM and AMLs (supplemental Data). Therefore, the following comparisons are valid. However, caution with respect to each detail in the comparison taken by themselves should be exercised.
Signaling in leukemic blast cells
Because of limited sample volumes for discarded material, we focused on SCF and FL signaling. Although the AML sample number is small, the data confirm abnormal signaling in the AML blasts and suggest that it is significantly different from NBM blasts in each case. The differences include (1) a larger fraction of responding cells, (2) sustained signals, and (3) AML phosphorylation levels are always amplified for at least one endpoint. Further, we see identical signaling patterns in 2 AMLs of different FAB class, which supports the idea that the relationship between signaling patterns and maturation is not tightly correlated and consequently measurements like this will enhance leukemia diagnosis. We report the results for 4 leukemias with 2 cytokines and 18 high-level endpoints (statistical analysis of the signaling is in Table 2).
Summary statistics: cytokine-stimulated signaling in blast cells
| Cytokine . | pERK . | pS6 . | pSTAT5 . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Amplitude, normalized MFI . | Peak, minutes . | Interval, minutes . | Amplitude, normalized MFI . | Peak, minutes . | Interval, minutes . | Amplitude, normalized MFI . | Peak, minutes . | Decay, MFI per minute . | |
| NBM | |||||||||
| SCF | 1.00 ± 0.24 | 2.0 ± 0.0 | 9.3 ± 1.4 | 1.00 ± 0.22 | 5.6 ± 0.7 | 40.0 ± 13 | 0.10 ± 0.02 | 2.3 ± 0.4 | ND |
| Flt3L | 0.66 ± 0.20 | 3.4 ± 0.4 | 17.0 ± 2.4 | 0.63 ± 0.25 | 6.0 ± 0.4 | 32.0 ± 5 | 0.10 ± 0.03 | 3.7 ± 1.2 | ND |
| IL3 | 0.06 ± 0.02 | 9.6 ± 1.9 | ND | 0.11 ± 0.03 | 15.0 ± 2.5 | ND | 1.00 ± 0.37 | 12.0 ± 3.2 | −0.013 ± 0.005 |
| GM-CSF | 0.09 ± 0.02 | 3.1 ± 1.0 | ND | 0.30 ± 0.11 | 12.0 ± 7.0 | ND | 0.94 ± 0.29 | 13.0 ± 3.1 | −0.014 ± 0.006 |
| AML1 (M4) | |||||||||
| SCF | 0.53 | 2.0 | 20.0* | 3.30 | 10.0* | 60.0* | 0.13 | 2.0 | ND |
| Flt3L | 0.27 | 4.0 | 20.0 | 0.93 | 10.0* | 60.0* | 0.03 | 2.0 | ND |
| IL3 | 0.02 | 6.0 | ND | 0.10 | 0.0 | ND | 0.27 | 6.0 | −0.004 |
| GM-CSF | 0.04 | 6.0 | ND | 0.07 | 0.0 | ND | 0.41 | 6.0 | −0.007 |
| AML2 (M2) | |||||||||
| SCF | 0.58 | 2 | 20.0* | 3.30* | 10.0* | 30.0 | 0.1 | 2.0 | ND |
| AML3 (M4Eo) | |||||||||
| SCF | 2.10* | 4.0* | 60.0* | 5.30* | 10.0* | 60.0* | 0.23 | 2.0 | ND |
| Flt3L | 2.10* | 6.0* | 60.0* | 4.60* | 10.0* | 60.0* | 0.21 | 6.0 | ND |
| AML5 (M1) | |||||||||
| SCF | 0.08 | 0.48 | |||||||
| Flt3L | 1.90* | 4.0 | 60.0* | 7.60* | 6.0 | 60.0* | ND | ND | ND |
| Cytokine . | pERK . | pS6 . | pSTAT5 . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Amplitude, normalized MFI . | Peak, minutes . | Interval, minutes . | Amplitude, normalized MFI . | Peak, minutes . | Interval, minutes . | Amplitude, normalized MFI . | Peak, minutes . | Decay, MFI per minute . | |
| NBM | |||||||||
| SCF | 1.00 ± 0.24 | 2.0 ± 0.0 | 9.3 ± 1.4 | 1.00 ± 0.22 | 5.6 ± 0.7 | 40.0 ± 13 | 0.10 ± 0.02 | 2.3 ± 0.4 | ND |
| Flt3L | 0.66 ± 0.20 | 3.4 ± 0.4 | 17.0 ± 2.4 | 0.63 ± 0.25 | 6.0 ± 0.4 | 32.0 ± 5 | 0.10 ± 0.03 | 3.7 ± 1.2 | ND |
| IL3 | 0.06 ± 0.02 | 9.6 ± 1.9 | ND | 0.11 ± 0.03 | 15.0 ± 2.5 | ND | 1.00 ± 0.37 | 12.0 ± 3.2 | −0.013 ± 0.005 |
| GM-CSF | 0.09 ± 0.02 | 3.1 ± 1.0 | ND | 0.30 ± 0.11 | 12.0 ± 7.0 | ND | 0.94 ± 0.29 | 13.0 ± 3.1 | −0.014 ± 0.006 |
| AML1 (M4) | |||||||||
| SCF | 0.53 | 2.0 | 20.0* | 3.30 | 10.0* | 60.0* | 0.13 | 2.0 | ND |
| Flt3L | 0.27 | 4.0 | 20.0 | 0.93 | 10.0* | 60.0* | 0.03 | 2.0 | ND |
| IL3 | 0.02 | 6.0 | ND | 0.10 | 0.0 | ND | 0.27 | 6.0 | −0.004 |
| GM-CSF | 0.04 | 6.0 | ND | 0.07 | 0.0 | ND | 0.41 | 6.0 | −0.007 |
| AML2 (M2) | |||||||||
| SCF | 0.58 | 2 | 20.0* | 3.30* | 10.0* | 30.0 | 0.1 | 2.0 | ND |
| AML3 (M4Eo) | |||||||||
| SCF | 2.10* | 4.0* | 60.0* | 5.30* | 10.0* | 60.0* | 0.23 | 2.0 | ND |
| Flt3L | 2.10* | 6.0* | 60.0* | 4.60* | 10.0* | 60.0* | 0.21 | 6.0 | ND |
| AML5 (M1) | |||||||||
| SCF | 0.08 | 0.48 | |||||||
| Flt3L | 1.90* | 4.0 | 60.0* | 7.60* | 6.0 | 60.0* | ND | ND | ND |
Data are mean ± SEM. Amplitude is defined as the maximum cytokine-stimulated response; amplitude data are normalized to the maximum response of each phosphoprotein in NBM. Peak is the time to maximum response. Interval is the duration of response from rise through decay, where normalized MFI ≥ 0.05. Decay is the slope from 15 to 30 minutes for the IL-3 and GM-CSF → pStat5 signal.
ND indicates not determined, generally because the data were either noisy or multimodal.
P < .05 compared with the corresponding value in NBM.
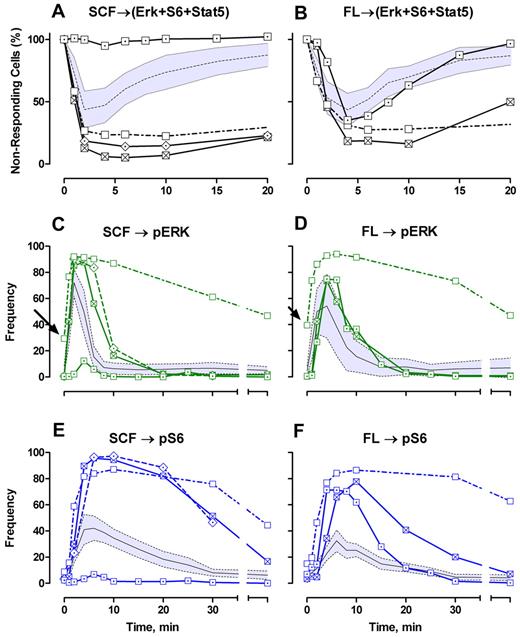
Three of the leukemias were CD117+ and 1 (AML5) was ∼ 99% CD117−. For each CD117+ leukemia, a higher fraction of blasts responded to SCF compared with NBM blasts. This is most easily seen by subtracting the constitutively active (phosphoepitope-positive) cells and then calculating the fraction of triple phosphoepitope-negative cells after stimulation (Figure 6A). Using this analysis, it is readily apparent that SCF signaling in these leukemic blasts is more rapid than the average normal blast, and the duration of signaling is prolonged (Figure 6A). Of the 3 leukemias stimulated with FL, 2 (AML1, AML5) responded abnormally but similar to SCF signaling (ie, rapid early responses and prolonged duration; Figure 6B). AML5 (the ∼ 99% CD117− leukemia) did not respond to SCF, as expected, and responded to FL more like normal but was abnormal in that the beginning response was delayed and more cells responded (Figure 6B). In Figures 6 and 7, the gray zones around the NBM responses are the 95% CI. Thus, from this global viewpoint, 7 of 7 signaling reactions were abnormal in the leukemias.
Phosphoprotein kinetic profiles of AML patient samples: frequency analysis. (A-B) The frequency of blast cells that are not positive for any of the 3 downstream markers are plotted with the frequency at t = 0 set to 100%. (C-F) Actual frequencies (in percentage) of cells positive for each marker at time t. Gray zones represent 95% CI for normal blast cell response; and black line, the center (mean) of that distribution. Arrows point to high constitutive activity for AML3.  represents AML2; □, AML3; ⊠, AML1; ⊡, AML5; and
represents AML2; □, AML3; ⊠, AML1; ⊡, AML5; and  , NBM.
, NBM.
Phosphoprotein kinetic profiles of AML patient samples: frequency analysis. (A-B) The frequency of blast cells that are not positive for any of the 3 downstream markers are plotted with the frequency at t = 0 set to 100%. (C-F) Actual frequencies (in percentage) of cells positive for each marker at time t. Gray zones represent 95% CI for normal blast cell response; and black line, the center (mean) of that distribution. Arrows point to high constitutive activity for AML3.  represents AML2; □, AML3; ⊠, AML1; ⊡, AML5; and
represents AML2; □, AML3; ⊠, AML1; ⊡, AML5; and  , NBM.
, NBM.
Phosphoprotein kinetic profiles of AML patient samples: MFI analysis. AMLs classified as 3 potential “phenotypes” by signaling patterns: AML3 (rows A and J); AMLs 1 and 2 (rows D and M); AML5 (rows G and P). Data have been normalized to NBM for SCF → Erk, SCF → S6, and IL-3 → Stat5 = 1.0 at max. Colored zones represent the 95% CI for the NBM blast response MFIs.
Phosphoprotein kinetic profiles of AML patient samples: MFI analysis. AMLs classified as 3 potential “phenotypes” by signaling patterns: AML3 (rows A and J); AMLs 1 and 2 (rows D and M); AML5 (rows G and P). Data have been normalized to NBM for SCF → Erk, SCF → S6, and IL-3 → Stat5 = 1.0 at max. Colored zones represent the 95% CI for the NBM blast response MFIs.
The fraction responding data can be examined at the individual pathway level (Figure 6C-F) and can subdivide the global information into abnormal and normal pathways. Each AML was abnormal with respect to SCF signaling through Erk or S6 (Figure 6C-E); AML1 and AML5 were normal for FL → Erk (Figure 6D) but abnormal for FL → S6 (Figure 6F). Each AML was abnormal with respect to S6 (Figure 6E-F). AML 3 stands out by itself as having the longest decay times for both cytokines and both endpoints (Figure 6C-F). Overall, the number of data points for which the AML responses are not within the normal 95% CI (Figure 6C-F) demonstrate that abnormality is measured at almost any time. However, the detailed signaling kinetic profiles provide the high-level characteristics that could be used to classify leukemias in a robust manner.
Signaling by phosphorylation intensity (MFI).
Figure 7 shows the phosphorylation kinetics for 4 AMLs as measured by the median intensity of phosphorylation. The shaded bands represent the 95% CI for NBM. Here, again we begin to see the beginnings of a classification system. The 3 rows for SCF and the related rows for FL contain the “classes.”
The top rows (AML3, Figure 7A-C,J-L) contain the “strongest” phenotype with both Erk and S6 phosphorylation dramatically elevated above normal for both cytokines and responses that were elevated for significantly longer time than normal. In addition, even Stat5 phosphorylation is above the normal range; albeit the signal is a fraction of IL-3 stimulated signal of normal cells.
The middle rows (Figure 7D-F,M-O) contain 2 leukemias (AML1, AML2) that demonstrated identical responses to SCF (only AML1 was tested for FL). However, the early SCF → Erk phosphorylations were in the normal range early on but reached peak levels represented by the bottom end of the distribution of normal signaling and shifted from the normal range because of an extended decay time. This evaluation is supported by the cell frequency responses (Figure 6C). The SCF → S6 signals for AML1 and AML2 were also normal for early time points but rose to 3-fold more than normal (Figure 7E). The FL → Erk signal for AML1 was also suppressed compared with both normal (but within normal limits) and with the other leukemias (AML3, AML5). The SCF and FL → Stat5 signals for AML1 were within normal limits but at the low end of the normal distribution, whereas AML2 showed stimulation of SCF → Stat5 at the upper end of the NBM distribution. Neither Stat5 signal was remarkable, and each would be considered normal with respect to Stat5 without other evidence.
The third and sixth rows show AML5, an ∼ 99% CD117− leukemia. Signaling through SCF was absent (Figure 7G-I). AML5 showed Stat5 phosphorylation above the NBM response limits (Figure 7I), that, in view of the absence of c-Kit and pErk/PS6 signals (Figure 7G-H), probably represents constitutive signaling through Stat5 throughout the time course. However, like the other AMLs examined here, the signals were detected in very few blasts (1%-2%) and were not very intense (on average) relative to IL-3 → Stat5 in NBM. The absence of an SCF signal is readily apparent as abnormal. In contrast, signaling through FL → Erk and FL → S6 are dramatically elevated, especially S6 (Figure 7Q), which is the strongest signal of the entire set at 8-fold more than the NBM SCF → S6. Subtleties include a distinct delayed beginning of the FL → Erk response in this leukemia.
High-end analytical endpoints.
We have used the term “endpoints” to refer to markers at the downstream ends of pathways, conventionally defining a pathway as ligand → endpoint. There is another, higher-level set of endpoints. These are derived from analysis of signaling waveforms. Although this could be more complicated and/or extensive, we chose the following set of MFI-based endpoints: (1) peak height (amplitude), which is the phosphorylation level at the peak of response; (2) peak time, which is the time of the highest phosphorylation value measured during the time course; and (3) response interval, which was measured as the time between departure from baseline (constitutive, untreated) and return to 5% of the peak value. These results are presented in Table 2. At a glance, the ability to detect abnormality is readily apparent. These statistics represent one possible approach to classification of AMLs by signaling.
Constitutive expression.
We did not set out to study constitutive signaling. However, the fractions of positive cells at t = 0 provide some information. NBM exhibited a mean of 5.0% plus or minus 4.0% (SD) Erk-positive and 4.9% plus or minus 3.9% (SD) S6-positive cells before stimulation. Somewhat surprising, AML1, AML2, AML3, and AML5 were significantly more quiet with respect to Erk (range, 0.3%-0.7% positive cells) and within the lower part of the normal range (0.3%-12%) for S6 (3%-5%). AML3 (M4eo) stood out with 35% Erk-positive cells (Figure 6C arrow); constitutive S6 phosphorylation was at the high end of normal (12%) and significantly elevated above the other AMLs (Figure 6D arrow). In this case, this high constitutive activity was matched by equally strong elevations in response to both cytokines (Figure 7A-B,J-K). Interestingly, common mutations in KIT or FLT3 were not detected in AML3.
Correlated analysis
We have focused on simplified analyses of the data presented in this paper. However, because the signaling data reported here are correlated, there is significantly more information that can be obtained from extended analyses. For example, supplemental Figure 3 shows correlated kinetic plots of SCF and Flt3-stimulated Erk and S6 phosphorylation. These types of correlated plots demonstrate an integrated ability to distinguish abnormal from normal signaling responses as opposed to side-by-side 2 pathway comparisons. In this analysis, there is really only one region of conjunction between normal and abnormal, a period from just after stimulation to just after 1 minute of stimulation. From this plot, it is also apparent how similar the responses of AML1 and AML2 are to each other and how markedly different is AML3. We think that the power that this approach potentially brings to AML diagnosis and patient monitoring is novel and additive rather than substitutive.
Discussion
AML is characterized by uncontrolled proliferation, impaired differentiation, and increased leukemic blast survival.29,30 In part, these dysfunctions are related to altered signal transduction. At the receptor level, altered signaling has been associated with increased activation of c-Kit or Flt3, through increased expression, gain-of-function mutations, and autocrine/paracrine stimulation. At the pathway level, the available data suggest that much of the aberrant signaling is confined to a few common pathways with PI3K → Akt, Ras → MAPK, and JAK → STAT prominent among them. In addition, effector molecules within these pathways are subject to activating mutations.16
In this report, we presented well-defined, high-resolution signaling profiles for SCF, FL, IL-3, and GM-CSF stimulation of NBM CD34+/CD117+ cells of 9 healthy, adult donors and 4 AML patients. The experiments were performed using whole, unfractionated bone marrow aspirates. The measured endpoints included the frequency of responding cells and median level of phosphorylated Erk, ribosomal S6, and Stat5. As far as we are aware, data with this level of resolution have not been published for NBM CD34+/CD117+ cells.
We have focused on blast/CD34+/CD117+ cells because they include hematopoietic stem cells and multipotential myeloid precursors in NBM and leukemia stem cells in AML samples; all are targets of ongoing research and developing therapies.31 A companion paper32 explores signaling along differentiation/maturation paths in normal and AML samples. The primary conclusions of each paper are in agreement despite the use of different sources of NBM.
Our results demonstrate remarkably uniform timing of Erk, S6, and Stat5 phosphorylation between donors for each cytokine. This was particularly evident for analysis of the responding fraction of CD34+/CD117+ cells. An example of this precision is that the relative position of the first 3 time points (1, 2, and 4 minutes) for the SCF → Erk signal was uniform in relative amplitudes for all 9 donors (by frequency or MFI). Thus, the peak timing statistic had an SD of 0 (Table 2). To distinguish between SCF/FL and IL-3/GM-CSF signaling, we did not need finely measured timing because the pathways are qualitatively quite different (based on activation of the Stat family). However, the literature does not lend itself to an easily distinguished difference in signaling output between SCF and FL. Here, the signal profiles were distinguishable and characteristic for each cytokine based on timing and levels of phosphorylation in the responding cell population.
Few studies focus on the kinetics of signaling responses in normal human hematopoietic precursor cells, and fewer yet explore biologic variance. Irish et al19 did explore biologic variance (between 3 normal donors) for B-cell receptor signaling in CD20+ B cells treated with cross-linking antibodies and H2O2, added to inhibit phosphatases that amplified and extended the signal. Without H2O2, a single example demonstrated that Erk phosphorylation peaked at 2 minutes, in close agreement with our results. The phosphatase-inhibited data for 3 donors (Erk peak = 60 minutes) also showed remarkable uniformity.21 Caruana et al33 and Montero et al34 showed differences in signaling kinetics between 2 isoforms of c-Kit that differ by the absence or presence of 4 amino acids (GNNK) in the juxtamembrane region. Using transfected NIH-3T3 or MMS1 (human myeloma) cells, respectively, both studies showed kinetic differences (slower or faster) for Erk phosphorylation via these Kit isoforms (but in contradiction to each other). In addition, one study showed large differences in the amount of phosphorylated Erk, whereas the other study did not show a difference. Taken together, these studies suggest (as expected) that the context in which signaling is measured has strong effects on the outcome. However, almost all studies support the generally held concepts for the structure of signaling data: upstream molecules signal before downstream molecules. Kalaitzidis and Neel23 presented kinetic signaling data on SCF stimulation of mouse Lin−/Sca+/Kit+ (LSK) and Lin−/Sca−/Kit+ cells. The LSK cells are most analogous to the cells studied here. The results are similar to the results of our study. LSK cells showed very uniform Erk response (peak = 5 minutes, but only 0, 5, and 10 minutes data were reported). The S6 response peaked at 5 minutes with a response duration of approximately 30 minutes, and a very weak, early Stat5 response. Marvin et al,32 using normal bone marrow from human femurs (from hip replacements), also showed similar kinetics for the 4 cytokines explored here. Thus, CD34+/CD117+ cells and mouse LSK cells display evidence of tight control over the SCF → Erk response, more variation in the S6 response, and a weak signal through Stat5.
There is significant evidence for c-Kit signaling through Jak2, Stat1, Stat3, and Stat5 and FL signaling through Stat5A; however, there are also c-Kit studies wherein this was not observed.35,36 The underlying cause of the discrepancy is not clear. Most work used transformed or transfected cell lines, but fetal liver and bone marrow preparations have been used also. One consistent feature is that, when Stat activation has been detected, signaling peaked early, timing that is more consistent with Erk phosphorylation than Stat5 phosphorylation via IL-3. We detected Stat5 signaling as an endpoint for SCF and FL in 3% of normal and 1% to 2% of leukemic CD34+/CD117+ cells. The signals were real and phosphorylation levels were approximately as strong as IL-3 stimulation for some fraction of individual cells (supplemental Figure 4). At present, our data (and those of Marvin et al32 ) indicate that a very small fraction of normal blast cells signal through Stat5 in response to SCF or FL. For the 4 AMLs studied here, Stat5 signaling via SCF or FL was not very different from normal.
The data reported here are consistent with current models for genetic abnormalities in AML. SCF- and FL-stimulated S6 phosphorylation was universally and dramatically elevated above normal NBM blast levels. Although it is speculative, because KIT and FLT3 mutations appear to be mutually exclusive, we can guess that none of the 4 AMLs we have analyzed harbored receptor mutations (because both SCF and FL stimulated positive, though abnormal, signals). We do not have genetic data for each AML, but we do know that AML3 was negative for KIT and FLT3 mutations. It is reasonable to presume that elevated pS6 is the consequence of downstream pathway dysfunction(s). Although speculative, it is possible that the signaling patterns as we have defined them here will be classifiable (eg, AML1 and AML2 are identical), that they will not be highly concordant with the FAB classification (eg, AML1 and AML2 are M2 and M4), and that signaling patterns will be associated with specific genetic alterations in the kinases, scaffolding proteins, and/or phosphatases that control these pathways.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank David Hedley and Sue Chow (Princess Margaret Hospital, Toronto) and James Marvin (Northwestern University) for a longstanding and continuous discussion of the underlying principles, technology, and significance of this research effort; Tammy Stefan and Mike Sramkoski (Case Western Reserve University) for early technical input into the work; Meryl Forman (Beckman Coulter) for significant input and associated work on matching fluorochromes; Steve Koester, George Quintana, Patti Grom, and Chris Wilkins (Beckman Coulter) for the logistics of coordinating cooperation and work between the 2 laboratories (J.W.J. and C.L.G.) and Beckman Coulter; and Cindy Collins and Brad Calvin for supporting this work far above the contractual agreements between Beckman Coulter and the universities.
This work was supported by Beckman Coulter, the Hematopoietic Stem Cell Core Facility (PA30 CA43703), and the Cytometry and Imaging Microscopy Core Facility (P30 CA43703) of the Case Comprehensive Cancer Center of Case Western Reserve University and University Hospitals of Cleveland.
National Institutes of Health
Authorship
Contribution: P.G.W. designed and executed all experiments, analyzed all data, wrote the first draft of the manuscript, prepared a first draft set of figures, and wrote and edited the final versions of the manuscript and figures; L.A.S. was responsible for acquisition of normal donor bone marrow samples and participated in and implemented the design of sample acquisition and processing logistics; H.J.M. was responsible for acquisition of leukemia samples, advised on the design of immunophenotyping markers, and wrote or edited sections of the manuscript; T.V.S. advised on fluorochrome/antibody selection and fixation and wrote and edited the manuscript; C.L.G. advised on immunophenotype markers, choice of cytokines, fixation, and experimental design, and edited the manuscript; and J.W.J. was responsible for the overall project, approved the final project design, advised P.G.W., analyzed data, and wrote and edited the manuscript.
Conflict-of-interest disclosure: T.V.S. owns shares in Beckman Coulter, which markets some of the reagents used in this study. The other authors declare no competing financial interests.
Correspondence: James W. Jacobberger, Case Comprehensive Cancer Center, Wolstein Research Bldg, Case Western Reserve University, 2103 Cornell Rd, Cleveland, OH 44106; e-mail: jwj@case.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal