Abstract
Forced expression of the transcription factor HoxB4 has been shown to enhance the self-renewal capacity of mouse bone marrow hematopoietic stem cells (HSCs) and confer a long-term repopulating capacity to yolk sac and embryonic stem (ES) cell–derived hematopoietic precursors. The fact that ES cell–derived precursors do not repopulate bone marrow without HoxB4 underscores an important role for HoxB4 in the maturation of ES-derived hematopoietic precursors into long-term repopulating HSCs. However, the precise molecular mechanism underlying this process is barely understood. In this study, we performed a genome-wide analysis of HoxB4 using ES cell–derived hematopoietic stem/progenitor cells. The results revealed many of the genes essential for HSC development to be direct targets of HoxB4, such as Runx1, Scl/Tal1, Gata2, and Gfi1. The expression profiling also showed that HoxB4 indirectly affects the expression of several important genes, such as Lmo2, Erg, Meis1, Pbx1, Nov, AhR, and Hemgn. HoxB4 tended to activate the transcription, but the down-regulation of a significant portion of direct targets suggested its function to be context-dependent. These findings indicate that HoxB4 reprograms a set of key regulator genes to facilitate the maturation of developing HSCs into repopulating cells. Our list of HoxB4 targets also provides novel candidate regulators for HSCs.
Introduction
All hematopoietic cells are continuously generated from hematopoietic stem cells (HSCs) to maintained blood homeostasis throughout the life span of a host. The crucial properties of HSCs rely on self-renewal and multipotency.1 Self-renewal is defined as entry into the cell cycle with restrictions of differentiation, apoptosis, and senescence. Strategies to expand HSC numbers ex vivo without losses of stemness have widespread clinical applications. Thus, elucidating the molecular mechanisms of self-renewal and lineage commitment is of great importance.2,3 Culturing HSCs with various combinations of cytokines or growth factors has achieved their expansion in vitro, although the effect was moderate.4-6 Several molecules are implicated in the regulation of HSC development and self-renewal, such as Runx1, Scl/Tal1, GATA2, Gfi1, c-Myc, HoxB4, and Bmi1, which are potential targets to manipulate for the expansion of HSCs.
HoxB4 is a member of the class 1 homeobox (Hox) gene family, characterized by a highly conserved homeobox domain with a helix-turn-helix structure important for the specific DNA binding. Many class 1 homeobox genes have been implicated in hematopoiesis.7 HoxB4 is predominantly expressed in hematopoietic stem and progenitor cells, and its forced expression in mouse bone marrow (BM) HSCs significantly enhanced their self-renewal and proliferation in ex vivo cultures.8-11 Forced expression of HoxB4 also allowed mouse yolk sac and embryonic stem (ES) cell–derived hematopoietic precursors to repopulate BM long-term in recipient mice. Of note, HoxB4 is the only gene that can confer a long-term repopulating capability to mouse ES cell–derived hematopoietic precursors.8-11 Thus, many attempts have been made to understand the function of HoxB4 in HSCs. Extensive analyses with microarrays implicated HoxB4 in modulating the response of BM HSCs to multiple extrinsic signals and acquisition of a set of transcripts closely associated with adult HSCs by ES-derived HSCs/hematopoietic precursor cells (HPCs).12-14 Recent studies defined Hemgn as a direct target of HoxB4 in the expansion of myeloid progenitor cells,12 and a chromatin immunoprecipitation (ChIP) on microarray (ChIP-Chip) analysis of a multipotent progenitor cell line, EML, presented a list of candidate target genes directly regulated by HoxB4.7,12-14 Nevertheless, no comprehensive analysis to identify direct targets of HoxB4 has been performed in ES-derived HSCs/HPCs; thus, the molecular mechanisms behind the HoxB4-mediated regulation of ES-derived HSCs remain obscure.
In this study, we expressed 3xFLAG-HoxB4 in mouse ES cell–derived hematopoietic precursors and cocultured the cells with OP9 stromal cells. We then purified HSCs/HPCs and carried out ChIP-chip and gene expression microarray assays. Through comparative analyses of the ChIP-chip and microarray data, we identified several candidate target genes, which are directly or indirectly regulated by HoxB4 in ES cell–derived HSCs/HPCs.
Methods
Cell culture
The mouse ES cell lines B6N-22UTR15 and tetracycline-inducible HoxB4 (iHoxB4; kindly provided by M. Kyba, University of Minnesota, Minneapolis, MN)11,16 were maintained on irradiated mouse embryonic fibroblasts in Dulbecco modified Eagle medium (Sigma-Aldrich)/15% fetal bovine serum (Invitrogen), 0.1mM nonessential amino acids (Invitrogen), 2mM glutamine, penicillin/streptomycin (Invitrogen), 0.1mM β-mercaptoethanol (Sigma-Aldrich), and 1000 U/mL leukemia inhibitory factor (Millipore).
In vitro differentiation of ES cells
B6N-22UTR or iHoxB4 ES cells were allowed to form embryoid bodies (EBs) as described previously.11 EBs were dissociated with 0.25% trypsin at day 6, and c-Kit+CD41+ cells were isolated by flow cytometry. Isolated cells were resuspended in HSC medium (Iscove modified Dulbecco medium, Sigma-Aldrich/15% fetal bovine serum, 2mM glutamine, penicillin/streptomycin, 5 ng/mL vascular endothelial growth factor, PeproTech; and 40 ng/mL thrombopoietin), and transferred onto semiconfluent OP9 cells. iHoxB4 cells were cultured in the presence or absence of 1 μg/mL doxycycline. The cultures were harvested and replated onto fresh OP9 cells every 4 days thereafter.
Cell isolation
Flow cytometric experiments were performed with FACSAria II (BD Biosciences), and the data were analyzed using FlowJo software (TreeStar). For magnetic cell sorting, cells were incubated with magnetic beads conjugated with anti-CD117 antibodies (Miltenyi Biotec), and the labeled cells were passed through magnetic-activated cell sorting separation columns to recover c-Kit+ cells.
Retrovirus vectors, virus production, and transduction
The 3xFLAG-mouse HoxB4 and human HoxB4 were subcloned into a pGCDNsam-ires-EGFP retrovirus vector kindly provided by M. Onodera (National Research Institute for Child Health and Development, Tokyo, Japan). The production and concentration of recombinant retroviruses and the transduction of cells have been described previously.16
ChIP-Chip experiment
ES-derived hematopoietic precursors (c-Kit+CD41+ cells) on day 6 EBs derived from B6N-22UTR ES cells were seeded on OP9 stromal cells and transduced with a HoxB4 retrovirus. The cells were further cultured on OP9 cells. c-Kit+ progenitors were collected at day 7 of culture by magnetic cell sorting using magnetic beads conjugated with anti-CD117 antibodies and subjected to a ChIP assay using an anti-FLAG antibody. ChIP was carried out as previously described.17 ChIP-on-chip analysis was carried out using the Mouse Promoter ChIP-on-chip Microarray Set (G4490A, Agilent Technologies). Purified immunoprecipitated and input DNA was subjected to T7 RNA polymerase-based amplification as described previously.18 Labeling, hybridization, and washing were carried out according to the Agilent mammalian ChIP-chip protocol (Version 9.0). Scanned images were quantified with Agilent Feature Extraction software under standard conditions. The assignment of regions bound by HoxB4 around transcription start sites (TSSs) was carried out using direct sequence alignment on the mouse genome database (National Center for Biotechnology Information Version 36). The location of HoxB4-bound regions was compared with a set of transcripts derived from the MGI database. We assigned bound regions that were within −5.5 kb to 2.5 kb of the TSS. Alignments on the mouse genome and TSSs of genes were retrieved from Ensembl (www.ensembl.org). Intensity ratios (IP/input: fold enrichment) were calculated, and the maximum value for each promoter region of a gene was used to represent the fold enrichment of the gene. Fold enrichment was calculated only for probes whose signals both from IP and input DNA were significant (P < .001). ChIP-chip data were submitted to Gene Expression Omnibus under accession number GSE26653.
Validation of HoxB4 binding by ChIP analysis
Cells were fixed with 1% formaldehyde in phosphate-buffered saline for 10 minutes at room temperature and washed twice with phosphate-buffered saline. Fixed cells were left to swell in a buffer (20mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.8, 1.5mM MgCl2, 10mM KCl, 0.1% NP-40, and 1mM dithiothreitol) for 10 minutes on ice, and nuclei prepared with a Dounce homogenizer were then lysed with RIPA (10mM Tris-HCl, pH 8.0, 0.5% sodium dodecyl sulfate, 140mM NaCl, 1mM ethylenediaminetetraacetic acid, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 0.1% sodium deoxycholate, and a proteinase inhibitor cocktail, Complete Mimi), and sonicated for 30 minutes using a Bioruptor (COSMO BIO). After centrifugation, the soluble chromatin fraction was precleared using a mixture of Protein A and G (Invitrogen) blocked with bovine serum albumin, and 300 μg of chromatin was immunoprecipitated overnight at 4°C using 25 μL of antibody-conjugated Protein A and G. The immunoprecipitates were washed extensively and subjected to a quantitative polymerase chain reaction (PCR) analysis with SYBR Premix Ex TaqTM II (Takara).
GO analysis
We performed gene ontology (GO) analysis using our own in-house programs written in Python and C++ and GO data retrieved from the Gene Ontology database (www.geneontology.org), KEGG (www.genome.jp/kegg), and others. The version of the dataset used was from October 27, 2006, submitted by Mouse Genome Informatics. We aligned microarray probes on mouse genes and assigned GO terms on all probes using these alignments. The significance of each term was determined using Fisher exact test and the Bonferroni adjustment for multiple testing. The P value reflects the likelihood that we would observe such enrichment by chance. Subsequent statistical examinations were also conducted using Fisher exact test.
Gene expression microarray and quantitative RT-PCR
Total RNA was isolated using TRIZOL LS solution (Invitrogen). For gene expression analyses with microarrays, first-strand cDNA was synthesized and hybridized to Affymetrix GeneChip Mouse Gene Version 1.0 ST arrays (Affymetrix) to assess and compare overall gene expression profiles as described previously.17 Microarray data were submitted to Gene Expression Omnibus under accession number GSE26653. For quantitative reverse-transcribed (RT)-PCR, Total RNA was reverse-transcribed by the ThermoScript RT-PCR system (Invitrogen) with an oligo-dT primer. Real-time quantitative PCR was performed with an ABI Prism 7300 Thermal Cycler (Applied Biosystems) using FastStart Universal Probe Master (Roche Applied Science) and combinations of Universal Probe Library (Roche Applied Science) and primers according to the manufacturer's protocol.
Results
Genome-wide analysis of HoxB4
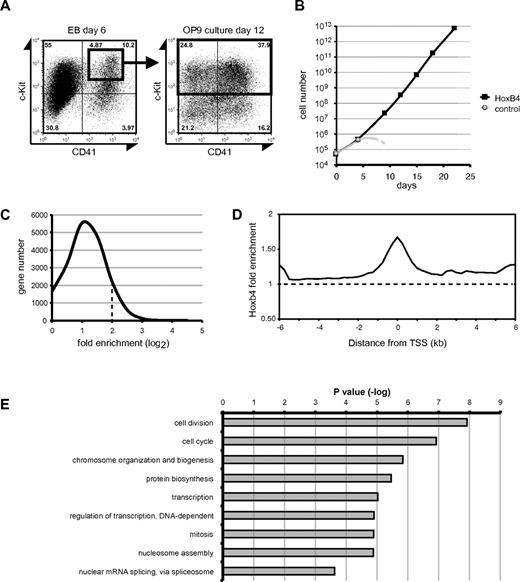
To identify the direct target genes of HoxB4 in HSCs/HPCs derived from ES cells, we conducted a ChIP-chip analysis. We differentiated hematopoietic progenitor cells from mouse ES cells. ES cells were differentiated for 6 days in EBs; then c-Kit+CD41+ hematopoietic precursor cells were isolated (Figure 1A), plated onto OP9 cells, and transduced with the GFP control or 3xFLAG-tagged HoxB4 retrovirus. Expansion of the control cells was limited, whereas cells overexpressing HoxB4 continuously proliferated as previously reported (Figure 1B).11,19 At day 12 of the coculture with OP9 cells, c-Kit+ progenitor cells overexpressing HoxB4 were collected by magnetic-activated cell sorting (Figure 1A) and subjected to a ChIP-chip analysis. The purity of the recovered c-Kit+ cells was more than 95% as judged by flow cytometry.
ChIP-chip analysis. (A-B) c-Kit+CD41+ cells in day 6 EBs were purified, seeded on OP9 stromal cells, and transduced with ether control or 3xFLAG-HOXB4 viruses. The cells were further cocultured with OP9 cells, and their growth was monitored. Control cells stopped growing after day 4 of culture (B). At day 12 of the OP9 coculture, c-Kit+ cells were purified by magnetic-activated cell sorting and subjected to a ChIP-chip analysis. (C) Distribution of genes in relation to the fold enrichment values (log2). The dotted line represents the ChIP-chip threshold (> 2). (D) The average distribution of HoxB4-bound probes. The dotted line represents the normalized average signal over the entire chip. (E) GO analysis of the ChIP-chip assay results. The P value of each GO term is indicated.
ChIP-chip analysis. (A-B) c-Kit+CD41+ cells in day 6 EBs were purified, seeded on OP9 stromal cells, and transduced with ether control or 3xFLAG-HOXB4 viruses. The cells were further cocultured with OP9 cells, and their growth was monitored. Control cells stopped growing after day 4 of culture (B). At day 12 of the OP9 coculture, c-Kit+ cells were purified by magnetic-activated cell sorting and subjected to a ChIP-chip analysis. (C) Distribution of genes in relation to the fold enrichment values (log2). The dotted line represents the ChIP-chip threshold (> 2). (D) The average distribution of HoxB4-bound probes. The dotted line represents the normalized average signal over the entire chip. (E) GO analysis of the ChIP-chip assay results. The P value of each GO term is indicated.
The ChIP-chip analysis was performed with mouse promoter microarrays containing approximately 17 000 probe sets covering from −5.5 kb upstream to 2.5 kb downstream of the TSS. 3xFLAG-HoxB4 was cross-linked to DNA and precipitated using the anti-FLAG M2 antibody. Gene promoters bound by HoxB4 were ranked according to fold enrichments calculated compared with signals obtained with the control antimouse IgG. Of the 18 216 gene promoter regions analyzed, 2292 showed high levels of HoxB4-binding with a fold enrichment greater than 4-fold (data listed in supplemental Data; available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The distribution of genes in relation to the fold enrichment is shown in Figure 1C. The peak of HoxB4-binding signals showed a typical binding pattern of transcription factors, which coincided with TSS region (Figure 1D). The functional annotation of the set of genes bound by HoxB4 was performed based on GO and showed significant enrichment for genes, which fell into the categories, such as “cell division,” “cell cycle,” and “chromosome organization and biogenesis” (Figure 1E).
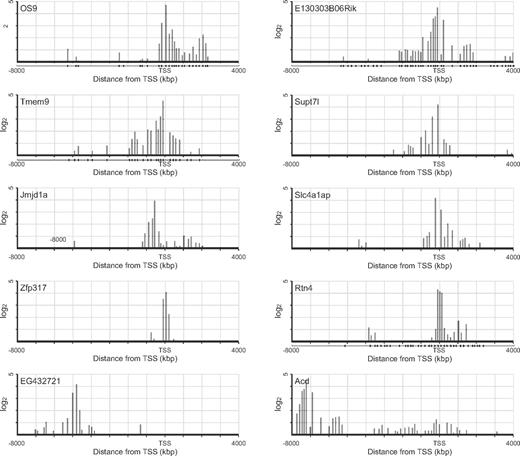
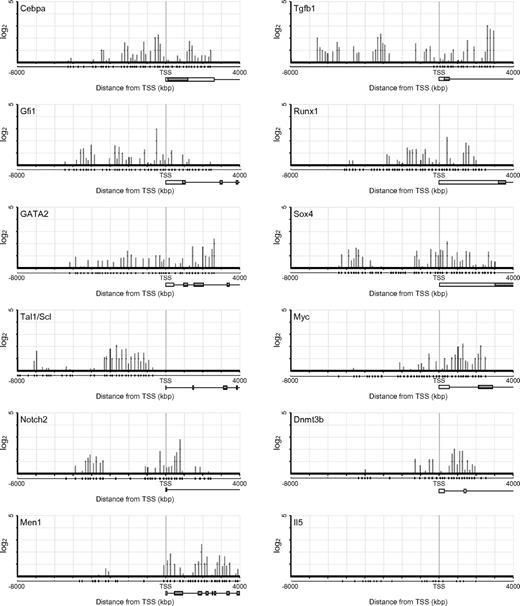
The list of top 20 genes ranked according to the ChIP-chip score showed fold enrichments more than 10-fold (Table 1). However, none of these genes has been characterized with respect to function in HSCs. The binding sites of HoxB4 sharply clustered around the TSS at these genes, except for EG432721 and Acd where HoxB4 mainly distributed in 5′-upstream regions (Figure 2). Notably, the listed genes with 4- to 8-fold enrichments included a number of genes well characterized as regulators of development, maintenance, or differentiation of HSCs (Table 2). Scl/Tal1, Runx1, and Gata2 encode transcription factors essential to the development of HSCs from hemoendothelial cells and also act in adult HSCs.20-26 Gfi1, Tgfb1, Sox4, E2f1, E2f4, and Cebpa have been implicated in HSC function.27-32 Although no significant binding of HoxB4 was detected at other HSC regulator genes, such as Myb, Erg, Gfi1b, Gata3, and Sox17, these findings indicate that HoxB4 simultaneously regulates a set of key HSC regulator genes. Contrary to the top 20 genes, the distribution of HoxB4 signals at these genes varied from the 5′-upstream region to gene bodies (Figure 3). The ChIP-chip data were confirmed by manual ChIP assays on selected genes, such as Os9, Runx1, Scl/Tal1, and Gata2 (supplemental Figure 1).
Top 20 genes ranked according to ChIP-chip scores
| Fold enrichment over input (log2) . | Fold difference HOXB4/control (log2) . | Gene symbol . | Gene name . |
|---|---|---|---|
| 4.72 | −1.124 | Os9 | Amplified in osteosarcoma |
| 4.51 | 1.566 | Tmem97 | Transmembrane protein 97 |
| 4.5 | ND | 4933405L10Rik | RIKEN cDNA 4933405L10 gene |
| 4.5 | 0.652 | Acd | Adrenocortical dysplasia |
| 4.5 | ND | E130303B06Rik | RIKEN cDNA E130303B06 gene |
| 4.34 | −1.947 | Rtn4 | Reticulon 4 |
| 4.2 | 2.659 | Slc4a1ap | Solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| 4.2 | 0.162 | Supt7 | Suppressor of Ty 7 (S cerevisiae)-like |
| 4.16 | 0.728 | Akr1e1 | Aldo-keto reductase family 1, member E1 |
| 4.16 | ND | Rpl29-ps2 | Ribosomal protein L29, pseudogene2 |
| 4.07 | 4.387 | Zfp317 | Zinc finger protein 317 |
| 4 | ND | 1110038F14Rik | RIKEN cDNA 1110038F14 gene |
| 3.94 | 0.993 | Arid1a | AT-rich interactive domain 1A (SWI-like) |
| 3.93 | ND | Jmjd1a | Jumonji domain containing 1A |
| 3.9 | −0.306 | 3200002M19Rik | RIKEN cDNA 3200002M19 gene |
| 3.73 | −0.487 | Arhgap1 | Rho GTPase activating protein 1 |
| 3.73 | 1.767 | Tmem48 | Transmembrane protein 48 |
| 3.73 | 1.745 | Zfp408 | Zinc finger protein 408 |
| 3.7 | ND | Map3k3 | Mitogen-activated protein kinase kinase kinase 3 |
| 3.63 | ND | Zc3h4 | Zinc finger CCCH-type containing 4 |
| 3.6 | 3.517 | 1700021C14Rik | RIKEN cDNA 1700021C14 gene |
| Fold enrichment over input (log2) . | Fold difference HOXB4/control (log2) . | Gene symbol . | Gene name . |
|---|---|---|---|
| 4.72 | −1.124 | Os9 | Amplified in osteosarcoma |
| 4.51 | 1.566 | Tmem97 | Transmembrane protein 97 |
| 4.5 | ND | 4933405L10Rik | RIKEN cDNA 4933405L10 gene |
| 4.5 | 0.652 | Acd | Adrenocortical dysplasia |
| 4.5 | ND | E130303B06Rik | RIKEN cDNA E130303B06 gene |
| 4.34 | −1.947 | Rtn4 | Reticulon 4 |
| 4.2 | 2.659 | Slc4a1ap | Solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| 4.2 | 0.162 | Supt7 | Suppressor of Ty 7 (S cerevisiae)-like |
| 4.16 | 0.728 | Akr1e1 | Aldo-keto reductase family 1, member E1 |
| 4.16 | ND | Rpl29-ps2 | Ribosomal protein L29, pseudogene2 |
| 4.07 | 4.387 | Zfp317 | Zinc finger protein 317 |
| 4 | ND | 1110038F14Rik | RIKEN cDNA 1110038F14 gene |
| 3.94 | 0.993 | Arid1a | AT-rich interactive domain 1A (SWI-like) |
| 3.93 | ND | Jmjd1a | Jumonji domain containing 1A |
| 3.9 | −0.306 | 3200002M19Rik | RIKEN cDNA 3200002M19 gene |
| 3.73 | −0.487 | Arhgap1 | Rho GTPase activating protein 1 |
| 3.73 | 1.767 | Tmem48 | Transmembrane protein 48 |
| 3.73 | 1.745 | Zfp408 | Zinc finger protein 408 |
| 3.7 | ND | Map3k3 | Mitogen-activated protein kinase kinase kinase 3 |
| 3.63 | ND | Zc3h4 | Zinc finger CCCH-type containing 4 |
| 3.6 | 3.517 | 1700021C14Rik | RIKEN cDNA 1700021C14 gene |
ND indicates no data.
ChIP-chip profile of HoxB4 occupancy of representative genes. HoxB4 signal maps are shown for representative top 20 genes with high ChIP-chip scores (y-axis in log2 scale). Plot under the x-axis shows the position of probe sets.
ChIP-chip profile of HoxB4 occupancy of representative genes. HoxB4 signal maps are shown for representative top 20 genes with high ChIP-chip scores (y-axis in log2 scale). Plot under the x-axis shows the position of probe sets.
Candidate target genes related to the HSC/HPC function
| Fold enrichment over input (log2) . | Fold difference HOXB4/control (log2) . | Gene symbol . | Gene name . |
|---|---|---|---|
| 3.05 | 2.861 | Tgfb1 | Transforming growth factor-β1 |
| 3 | −1.365 | Gfi1 | Growth factor independent 1 |
| 2.8 | 4.071 | Notch 2 | Notch gene homolog 2 (Drosophila) |
| 2.67 | 2.846 | Men 1 | Multiple endocrine neoplasia 1 |
| 2.54 | 1.972 | E2f4 | E2F transcription factor 4 |
| 2.42 | 1.906 | Gata 2 | GATA binding protein 2 |
| 2.39 | 1.723 | E2f1 | E2F transcription factor 1 |
| 2.35 | 0.281 | Runx 1 | Runt-related transcription factor 1 |
| 2.3 | 5.261 | Tle1 | Transducin-like enhancer of split 1, homolog of Drosophila E(spl) |
| 2.29 | 2.197 | Cebpa | CCAAT/enhancer binding protein (C/EBP), α |
| 2.27 | 3.204 | Myc | Myelocytomatosis oncogene |
| 2.22 | 5.874 | Sox4 | SRY-box containing gene 4 |
| 2.17 | 2.503 | Tal1/Scl | T-cell acute lymphocytic leukemia 1 |
| 2.02 | 13.147 | Dnmt3b | DNA methyltransferase 3B |
| 1.97 | 3.529 | Lmo2 | LIM domain only 2 (rhombotin-like 1) |
| 1.83 | −4.55 | Sfpi1 | SFFV proviral integration 1 |
| 1.75 | 23.626 | Nov | Nephroblastoma overexpressed gene |
| 1.69 | 2.706 | Meis1 | Meis homeobox 1 |
| 1.62 | 4.836 | Cd34 | CD34 antigen |
| 1.24 | 2.867 | Pbx3 | Pre-B-cell leukemia transcription factor 3 |
| 1.03 | 1.606 | Pbx1 | Pre-B-cell leukemia transcription factor 1 |
| 0.92 | 6.695 | Erg | Avian erythroblastosis virus E-26 (v-ets) oncogene related |
| 0.85 | −1.78 | Cdx4 | Caudal-type homeo box 4 |
| 0.84 | 4.508 | Hemgn | Hemogen |
| 0.47 | −5.725 | Ahr | Arylhydrocarbon receptor |
| Fold enrichment over input (log2) . | Fold difference HOXB4/control (log2) . | Gene symbol . | Gene name . |
|---|---|---|---|
| 3.05 | 2.861 | Tgfb1 | Transforming growth factor-β1 |
| 3 | −1.365 | Gfi1 | Growth factor independent 1 |
| 2.8 | 4.071 | Notch 2 | Notch gene homolog 2 (Drosophila) |
| 2.67 | 2.846 | Men 1 | Multiple endocrine neoplasia 1 |
| 2.54 | 1.972 | E2f4 | E2F transcription factor 4 |
| 2.42 | 1.906 | Gata 2 | GATA binding protein 2 |
| 2.39 | 1.723 | E2f1 | E2F transcription factor 1 |
| 2.35 | 0.281 | Runx 1 | Runt-related transcription factor 1 |
| 2.3 | 5.261 | Tle1 | Transducin-like enhancer of split 1, homolog of Drosophila E(spl) |
| 2.29 | 2.197 | Cebpa | CCAAT/enhancer binding protein (C/EBP), α |
| 2.27 | 3.204 | Myc | Myelocytomatosis oncogene |
| 2.22 | 5.874 | Sox4 | SRY-box containing gene 4 |
| 2.17 | 2.503 | Tal1/Scl | T-cell acute lymphocytic leukemia 1 |
| 2.02 | 13.147 | Dnmt3b | DNA methyltransferase 3B |
| 1.97 | 3.529 | Lmo2 | LIM domain only 2 (rhombotin-like 1) |
| 1.83 | −4.55 | Sfpi1 | SFFV proviral integration 1 |
| 1.75 | 23.626 | Nov | Nephroblastoma overexpressed gene |
| 1.69 | 2.706 | Meis1 | Meis homeobox 1 |
| 1.62 | 4.836 | Cd34 | CD34 antigen |
| 1.24 | 2.867 | Pbx3 | Pre-B-cell leukemia transcription factor 3 |
| 1.03 | 1.606 | Pbx1 | Pre-B-cell leukemia transcription factor 1 |
| 0.92 | 6.695 | Erg | Avian erythroblastosis virus E-26 (v-ets) oncogene related |
| 0.85 | −1.78 | Cdx4 | Caudal-type homeo box 4 |
| 0.84 | 4.508 | Hemgn | Hemogen |
| 0.47 | −5.725 | Ahr | Arylhydrocarbon receptor |
ChIP-chip profile of HoxB4 occupancy at genes related to the HSCs/progenitor cell function. HoxB4 signal maps are shown for representative HSC/progenitor-related factor genes (y-axis in log2 scale). Plot under the x-axis shows the position of probe sets. Schematic gene structures are also shown.
ChIP-chip profile of HoxB4 occupancy at genes related to the HSCs/progenitor cell function. HoxB4 signal maps are shown for representative HSC/progenitor-related factor genes (y-axis in log2 scale). Plot under the x-axis shows the position of probe sets. Schematic gene structures are also shown.
Comparison of gene lists between the ChIP-chip and microarray assays
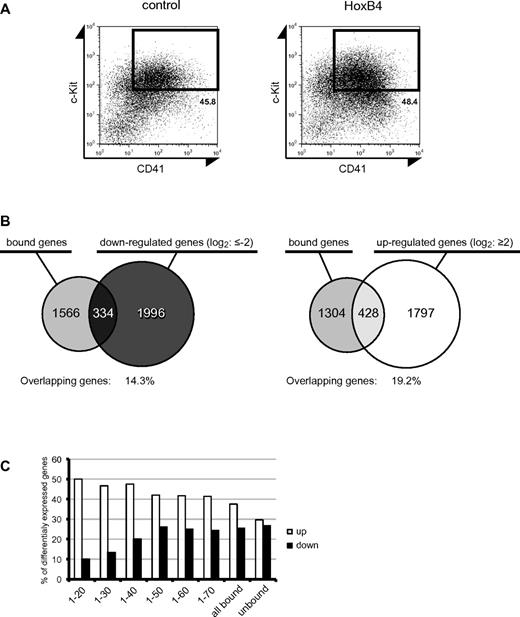
We next analyzed the comprehensive gene expression changes induced by HoxB4 using gene expression microarrays. ES cells genetically engineered for tetracycline-inducible expression of HoxB4 (iHoxB4 ES cells) have been broadly used to overexpress HoxB4 in ES cell–derived hematopoietic cells. Because a retroviral system induced a much higher expression of HoxB4 than a tetracycline-inducible system (data not shown), we induced HoxB4 expression using both tetracycline-inducible and retroviral systems. EBs were formed with iHoxB4 ES cells in the presence of the tetracycline analog doxycycline, and then c-Kit+CD41+ precursor cells were purified at day 6 of EB culture. The precursors were seeded on OP9 stromal cells and cultured in the presence and absence of doxycycline. At day 8 of the coculture with OP9 stromal cells, c-Kit+CD41+ cells were isolated by cell sorting and subjected to a microarray analysis. In parallel, purified c-Kit+CD41+ precursor cells from EB cultures at day 6 were seeded on OP9 stromal cells and immediately transduced with the GFP control or HoxB4 retrovirus in the absence of doxycycline. At day 8 of culture, c-Kit+CD41+ cells were similarly isolated (Figure 4A) and subjected to a microarray analysis. As expected, both the iHoxB4 and retroviral systems established very similar gene expression profiles in c-Kit+CD41+ cells during 8-day coculture with OP9 cells, and the changes in gene expression were generally greater with retrovirally expressed HoxB4 than with the iHoxB4 system (supplemental Data). Therefore, we focused on the data from the retroviral system in the subsequent analyses. The microarray results were validated by quantitative RT-PCR of selected representative genes (supplemental Figure 2).
Comparison between ChIP-chip and expression microarray datasets. (A) At day 8 of OP9 coculture, c-Kit+CD41+ cells were flow cytometrically isolated and used for microarray analyses of gene expression. (B) Venn diagrams showing the number of genes bound by HoxB4, the number of down-regulated genes (left) or up-regulated genes (right) on HoxB4 expression, and the overlap between the 2. (C) Graph showing the percentage of HoxB4-bound genes when the fold change threshold stringency of ChIP-chip enrichment is changed. The x-axis shows the ranking of genes in descending order in terms of the ChIP-chip score. Each number corresponds to fold enrichment values as follows: 1, 4.72; 20, 3.45; 30, 3.35; 40, 3.28; 50, 3.21; 60, 3.13; and 70, 3.11.
Comparison between ChIP-chip and expression microarray datasets. (A) At day 8 of OP9 coculture, c-Kit+CD41+ cells were flow cytometrically isolated and used for microarray analyses of gene expression. (B) Venn diagrams showing the number of genes bound by HoxB4, the number of down-regulated genes (left) or up-regulated genes (right) on HoxB4 expression, and the overlap between the 2. (C) Graph showing the percentage of HoxB4-bound genes when the fold change threshold stringency of ChIP-chip enrichment is changed. The x-axis shows the ranking of genes in descending order in terms of the ChIP-chip score. Each number corresponds to fold enrichment values as follows: 1, 4.72; 20, 3.45; 30, 3.35; 40, 3.28; 50, 3.21; 60, 3.13; and 70, 3.11.
The microarray analysis identified 2330 down-regulated and 2225 up-regulated genes with more than 2-fold changes compared with the GFP control (supplemental Data). Of these, 334 down-regulated and 428 up-regulated genes showed more than a 4-fold enrichment in the ChIP-chip analysis (Figure 4B). To understand the role of HoxB4 in transcriptional regulation, we analyzed the relationship between the mode of HoxB4-binding and transcriptional status. The percentage of up-regulated and down-regulated genes among the unbound genes was comparable (30% and 27%, respectively). In contrast, among the HoxB4-bound genes, the percentage of up-regulated genes became substantially higher than that of down-regulated genes as the ChIP-chip score increased. This tendency was evident with the top 20 genes (50% and 10%, respectively; Figure 4C). These results indicate that HoxB4 acts as a transcriptional activator and repressor but prefers to activate the transcription of its target genes.
Most of the key regulator genes defined as direct targets of HoxB4 were differentially expressed in HoxB4-overexpressing cells: Scl/Tal1, Gata2, Tgfb1, and Sox4 were up-regulated and Gfi1 was down-regulated (Table 2). However, some of them did not show significant differences in expression levels compared with the control, including Runx1, E2f1, and E2f4. Among the differentially expressed genes, those with marginal (2- to 4-fold) enrichments and no binding of HoxB4 included the HSC regulator genes Lmo2 and Nov, and Erg, Meis1, Pbx1, Hemgn, and Ahr, respectively, and were considered indirect targets of HoxB4 (Table 2). Of these, Hemgn was previously shown to be directly up-regulated by HOXB4 in mouse BM HSCs/HPCs.12 In our ChIP-chip analysis, however, HoxB4 did not show significant binding to Hemgn in developing HSCs/HPCs from ES cells.
Comparison of differentially expressed HoxB4 targets with other databases
Recently, several groups enumerated candidate target genes of HoxB4.12-14 We then compared our list of 762 differentially expressed target genes (genes with > 4-fold enrichments and differentially expressed by more than 2-fold) with other comprehensive analyses of HoxB4 target genes. Schiedlmeier et al performed expression profiling of lineage marker−Sca-1+c-Kit+ cells from BM HSCs/HPCs expanded by inducible HoxB4 ex vivo and hematopoietic precursors developed in EBs under inducible HoxB4 expression.14 They identified 52 genes as differentially expressed in both KSL and EB cells, and 13 of which were on our list of differentially expressed HoxB4 target genes (supplemental Table 1). Lee et al performed ChIP-chip and gene expression analyses of HoxB4-overexpressing EML cells, a mouse immature hematopoietic cell line established by transducing mouse BM cells with a dominant-negative form of retinoic acid receptor-α.13,33 Among 71 genes identified as differentially expressed targets of HoxB4 in EML cells, 7 were on our list including Sox4 (supplemental Table 2).
Discussion
In this study, we performed ChIP-chip and gene expression analyses using HSCs/HPCs differentiated from ES cells. Of note, this is the first report of a ChIP-chip analysis of ES cell–derived HSCs/HPCs. Thus, comparison of the 2 datasets provided a new list of HoxB4 target genes quite distinct from those reported before.
The ChIP-chip analysis clearly demonstrated that HoxB4 mainly binds around the TSS of target genes, suggesting that HoxB4 typically acts on promoters. However, HoxB4 was also located in the 5′-upstream regions of the genes or in the middle of gene bodies. Thus, HoxB4 would also mediate enhancer or silencer activity. These binding profiles correspond well to those of general transcription factors. The GO analysis revealed that HoxB4 is involved in the transcriptional regulation of genes with various biologic functions, highlighting a global role for HoxB4 in the reprogramming of ES cell–derived embryonic hematopoietic precursors into mature HSCs with self-renewal capability. The ChIP-chip data combined with expression profiling indicated that HoxB4 tends to transactivate its target genes. However, the down-regulation of a significant portion of HoxB4-bound genes, including Os9 and Rtn4 listed among the top 20 genes in the ChIP-chip analysis, suggests that its function is promoter context-dependent. Unexpectedly, none of the genes to which HoxB4 showed the strongest binding has been characterized in HSCs. Given that HoxB4 regulates genes with a broad range of biologic functions, the characterization of these novel targets should unveil new aspects of HoxB4 function in HSCs. In this regard, the list from the ChIP-chip analysis could be a useful database for HSC research. Some of the highly ranked target genes of HoxB4 were not differentially expressed compared with the control cells. Nonetheless, we might simply have missed the differentially expressed time points of these genes in the maturation of ES cell–derived precursors.
In contrast to the targets of HoxB4 reported by other groups, we successfully identified several genes essential for the formation of HSCs/HPCs during embryonic development, such as Runx1, Tal1/Scl, and GATA2.20,24 The different profiles of target genes might depend on the cells characterized. We analyzed HSCs/HPCs that had differentiated from ES cells, whereas other groups used adult BM-derived cells or immortalized lymphohematopoietic progenitors established from adult BM cells. The identification of essential regulator genes as direct targets of HoxB4 clearly indicates that HoxB4 regulates the expression of a broad range of regulator genes simultaneously to reprogram them into adult-type HSCs/HPCs that can repopulate the BM.
Our list also included many regulators for adult HSCs. Although numerous regulating genes were up-regulated by HoxB4, Gfi1 was slightly down-regulated (Table 2). Gfi1 is reported to restrict the proliferative capacity of adult BM HSCs to keep them in a quiescent state.28 Down-regulation of Gfi1 by HoxB4 in developing HSCs/HPCs might allow fetal HSCs to undergo cell cycling to expand their numbers. A previous report demonstrated that the overexpression of Nov or addition of a recombinant Nov protein enhanced human HSC/HPC activity,34 Although binding of HoxB4 was not detected in our study, Nov was significantly up-regulated on HoxB4 expression. Recently, small compound antagonists of the Aryl hydrocarbon receptor (AhR) were demonstrated to promote the expansion of human HSCs.35 AhR is a ligand-dependent transcriptional activator originally identified as an intracellular mediator of xenobiotic signaling pathways, mediates toxic effects of dioxin, and also regulates lymphocyte or vascular development and the reproductive ability of females. Our results showed that the expression of AhR and its direct targets AhRR and CYP1B1 were all significantly decreased on HoxB4 expression. Although the binding of HoxB4 was not detected on AhR or AhRR and CYP1B1 (Table 2; and data not shown), these genes might be regulated downstream of the direct targets of HoxB4. These findings suggest that HoxB4 also promotes the acquisition of a set of transcripts closely associated with adult HSCs by ES-derived HSCs/HPCs. Among target gene candidates shared between our study and other studies, only Sox4 has been reported to promote the proliferation of HSCs,30 although other shared genes of unknown function have potential importance, which should be analyzed further.
In conclusion, we presented a list of newly identified direct targets of HoxB4. Our list provides new insight into the HoxB4-mediated regulation of HSCs. Further study is needed to understand the effect and mechanism of the regulation of regulator gene sets by HoxB4, and should unveil an approach to manipulating HSCs for clinical applications.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Michael Kyba for providing iHoxB4 ES cells, Dr Masafumi Onodera for the pGCDNsam vector, and Haruna Takagi for technical support.
This work was supported in part by the Global Center for Education and Research in Immune System Regulation and Treatment, MEXT, Japan (Grants-in-Aid for Scientific Research and for the Global COE Program), the Japan Science and Technology Corporation (Grant-in-Aid for Core Research for Evolutional Science and Technology), the Takeda Science Foundation and National Institutes of Health grant R01HL081186.
National Institutes of Health
Authorship
Contribution: M. Oshima performed the experiments, analyzed results, made the figures, and actively wrote the manuscript; M.E. and H.K. performed the ChIP-chip and microarray analyses; Y.N.-T. assisted with the experiments, including the cell culture and flow cytometric analysis; T.A.E. and T.T. performed the database analysis; F.S. provided B6N-22UTR ES cells; M.K. provided iHOXB4 ES cells; and A.I. and M. Osawa conceived of and directed the project, secured funding, and actively wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Atsushi Iwama, 1-8-1 Inohana, Chuo-ku, Chiba 260-8670, Japan; e-mail: aiwama@faculty.chiba-u.jp; and Mitsujiro Osawa, 1-8-1 Inohana, Chuo-ku, Chiba 260-8670, Japan; e-mail: mitsujiro.osawa@faculty.chiba-u.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal