Abstract
In humans, embryonic, fetal, and adult hemoglobins are sequentially expressed in developing erythroblasts during ontogeny. For the past 40 years, this process has been the subject of intensive study because of its value to enlighten the biology of developmental gene regulation and because fetal hemoglobin can significantly ameliorate the clinical manifestations of both sickle cell disease and β-thalassemia. Understanding the normal process of loss of fetal globin expression and activation of adult globin expression could potentially lead to new therapeutic approaches for these hemoglobin disorders. Herein, we briefly review the history of the study of hemoglobin switching and then focus on recent discoveries in the field that now make new therapeutic approaches seem feasible in the future. Erythroid-specific knockdown of fetal gene repressors or enforced expression of fetal gene activators may provide clinically applicable approaches for genetic treatment of hemoglobin disorders that would benefit from increased fetal hemoglobin levels.
Introduction
Fetal hemoglobin (HbF, α2γ2) was first distinguished from adult hemoglobin (α2β2) by its resistance to alkaline denaturation by Korber1 and von Kruger2 almost 150 years ago. In 1934, Brinkman et al described experiments using alkaline denaturation, as measured by a spectrophotometer coupled to a photoelectric output, to differentiate fetal and adult hemoglobin by their dissimilar reaction kinetics.3 The differences in susceptibility to alkaline denaturation, along with what was known about the consistency of the heme moiety between umbilical cord and adult blood, led the authors to conclude that the difference between the entities must be attributable to the “globin” portion of the molecule.3 Now, nearly 80 years later, the field is making substantial progress in uncovering the molecular mechanisms that regulate the developmental expression of the fetal and adult globin genes. In the past several years, genetic approaches have helped identify 3 loci that may account for up to 50% of the variance in HbF expression in certain human populations.4-6 In addition, experimentation facilitated by murine transgenic, knockout, and vector knockdown technology has contributed to the identification of other potentially important regulators of fetal and adult globin gene expression.7,8 Herein, we will focus on the recent discoveries that have advanced the field.
A synopsis of hemoglobin switching: 1970 to 2000
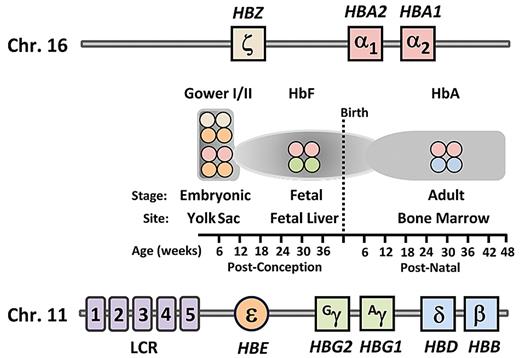
In their classic paper, Brinkman et al noted that “unsolved is the question whether human hemoglobin is changing from one form to the other, or whether there are 2, originally different forms (eg, the resistant fetal type and the adult form).”3 Much has been learned since that time, including the structure and sequence of both the α- and β-globin loci9-12 (Figure 1), the transcriptional regulation of the genes contained within these loci during development, and the clinical diseases that result from genetic changes to both the coding and noncoding portions of the genes.13-16
Schematic of genomic structural organization of the human α-globin and β-globin loci and temporal expression of the various hemoglobin types. The gene order of the α-globin locus on chromosome 16 and the β-globin locus on chromosome 11. Shown at each temporal stage, as indicated on the timeline at bottom, are the types of hemoglobin tetramers produced for each indicated site. LCR indicates locus control region; HbF, fetal hemoglobin; and HbA, adult hemoglobin.
Schematic of genomic structural organization of the human α-globin and β-globin loci and temporal expression of the various hemoglobin types. The gene order of the α-globin locus on chromosome 16 and the β-globin locus on chromosome 11. Shown at each temporal stage, as indicated on the timeline at bottom, are the types of hemoglobin tetramers produced for each indicated site. LCR indicates locus control region; HbF, fetal hemoglobin; and HbA, adult hemoglobin.
The hemoglobin molecule is a tetramer composed of 2 heterodimers. Each heterodimer is composed of one α-globin-like polypeptide chain and one β-globin–like chain. In humans, embryonic hemoglobin is expressed in primitive erythroblasts developing in the yolk sac during the first several weeks after conception. It is composed of the embryonic α-globin–like chain, ζ-globin, and the embryonic β-globin–like ϵ-chain (ζ2ϵ2; Figure 1). The first major hemoglobin switching event subsequently occurs as ζ- and ϵ-globin expression ceases and α- and γ-globin synthesis begins, leading to production of HbF (α2γ2). These events are coincident with the transition of the site of erythropoiesis from the yolk sac to the fetal liver. In humans, the γ-globin genes are duplicated and differ by one amino acid, leading to their designation as the Gγ and Aγ genes. The second switch in humans and Old World primates, which is the focus of this review, involves the perinatal decline of HbF synthesis (α2γ2) coupled with the increased synthesis of the adult form of hemoglobin (α2β2). After 2 years of age, HbF is present as a minor component of total hemoglobin17 in only a few percentage of mature red blood cells in healthy persons.18
Persistent, high-level expression of HbF was observed in the early 1960s in some persons from both black and Greek populations.19,20 Termed hereditary persistence of HbF (HPFH), this phenomenon has subsequently been shown to be the result of a variety of mutations, many of which are within the β-globin gene locus.21 Frequently, HPFH reflects a single nucleotide substitution in the upstream promoter region of a γ-globin gene leading to its up-regulation, suggesting the location of a regulatory element that interacts with transcription factors potentially relevant to the fetal to adult switch. An interesting aspect of these mutations is that they result in a nonuniform distribution of HbF in red cells, leading to their designation as heterocellular HPFH mutants. Deletion forms of HPFH, which usually result in a pancellular HPFH distribution, are thought to eliminate inhibitory sequences22 and/or result in juxtaposition of a remote enhancer(s) adjacent to the γ-globin genes.23 Persons homozygous for a mutant form of β-globin (βS) with sickle cell anemia or those homozygous for mutations that cause deficient β-synthesis with severe β-thalassemia who also inherit an HPFH mutation have a milder clinical phenotype with fewer manifestations of vaso-occlusion or milder anemia, respectively.15,21
The ameliorating effect of elevated levels of HbF on the clinical severity of sickle cell disease and β-thalassemia has been a significant driving force for the study of hemoglobin switching during the past 3 decades.24,25 Studies in the 1970s used sheep and baboon models to demonstrate the effect of anemic stress on synthesis of individual hemoglobins,26,27 whereas studies using primary human cells also began to uncover aspects of the control of HbF production.28 The cloning of the globin genes and the advent of first transgenic and later knock-in technology provided genetically altered mice for study of murine embryonic to adult globin switching as well as the fetal to adult switch of transferred human β-globin loci.29-35 Powerful chromatin opening and DNA enhancer elements, first identified as DNase-I hypersensitive sites located approximately 40 to 60 kb upstream of the globin genes, were shown to be necessary for high level globin gene expression.36,37 Collectively, these elements are called the locus control region (LCR). Tuan mapped the distribution of DNase-I hypersensitive sites in the entire human β-like-globin gene domain. Additional sites were associated with active transcription of the immediate downstream β-like globin gene.36 Others showed that, in transgenic mice, LCR elements can be used to obtain high level, copy number-dependent β-globin gene expression, which was relatively independent of the integration position of the transgene,37 whereas deletion of this region lead to silencing of transcription of the globin genes.38
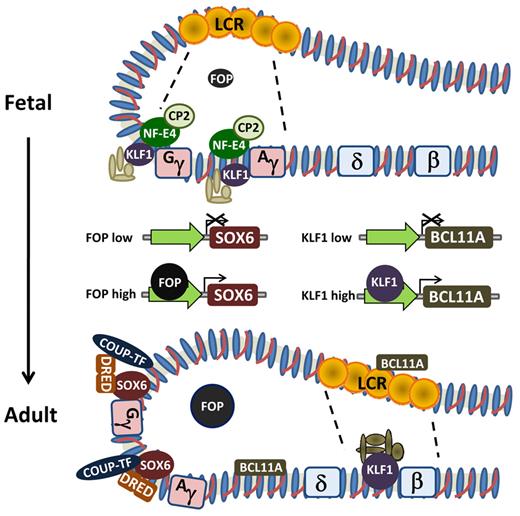
This body of work, coupled with studies using erythroid cell lines and primary human cells, resulted in the proposal of several models of hemoglobin switching. These include gene competition, chromosomal looping, gene silencing, and polymerase tracking.14,39 Wijgerde et al, using a then novel RNA fluorescence in situ hybridization technique, showed that transgenic human γ- and β-globin genes in day 12 mouse fetal erythroid cells were for the most part alternately, and not simultaneously, transcribed.40 Consistent with β-globin being the predominant mRNA, the transcription apparatus was associated with the β-globin gene for the majority of the time. However, some instances of flip-flop of transcription from one gene to the other at the single-cell level were also observed. Thus, the transcriptional process is dynamic and consistent with the idea that the LCR might activate each gene individually by direct interaction. Later work, using a technique called chromosomal conformational capture, confirmed direct interaction of sequences with the upstream β-globin LCR and the individual globin gene promoters, supporting the idea of chromosomal looping.41 Early studies in transgenic mice suggested that each hypersensitive site conferred a different developmental pattern on the individual globin genes42 ; but in the normal context, the individual sites are thought to interact with one another in selectively activating the individual globin genes during development40,41 (Figure 2). LCR interactions are not exclusive of autonomous gene silencing, which probably also occurs with both the embryonic and fetal genes.43-45 Finally, polymerase tracking may occur to open the chromatin structure and to fine-tune interactions.46 Thus, the various models that have been proposed are not mutually exclusive but rather complement one another.
Schematic of hemoglobin switching model based on looping and interaction of the LCR with the individual globin gene promoters. The various proteins demonstrated experimentally to be involved in regulating the change in expression from γ-globin to β-globin and individual effects of FOP and KLF1 on transcriptional regulation of SOX6 and BCL11A, respectively.
Schematic of hemoglobin switching model based on looping and interaction of the LCR with the individual globin gene promoters. The various proteins demonstrated experimentally to be involved in regulating the change in expression from γ-globin to β-globin and individual effects of FOP and KLF1 on transcriptional regulation of SOX6 and BCL11A, respectively.
Hemoglobin switching: 2000 and beyond
Inherited variation in HbF levels continues to provide many new insights into the fetal to adult hemoglobin switch.47 The common forms of heterocellular HPFH represent the upper tail of normal HbF variation and are inherited based on multiple quantitative genetic traits. As discussed in more detail in “Molecular control of the fetal to adult hemoglobin switch,” 3 major quantitative trait loci that influence HbF levels in otherwise normal persons as well as in patients with sickle cell disease or β-thalassemia have been identified.48,49 Similarly, study of thalassemia mutations continues to provide insights into the mechanisms of HbF regulation. Using the liquid culture system beginning from peripheral blood cells, Fraser's laboratory has studied heterozygotes and homozygotes with the Corfu thalassemia mutation.50 Two mutations are present on the Corfu chromosome: (1) a 7.2-kb deletion removes part of the δ-globin gene and upstream flanking sequences; and (2) a splice site mutation in intron 1 of the β-globin gene reduces splicing to approximately 20% of normal levels. Homozygotes for the Corfu deletion have very high levels of fetal hemoglobin and a mild clinical phenotype, whereas heterozygotes have low HbF levels. Quantitative analysis suggested that the γ-globin mRNA levels do not correspond to transcription levels but rather vary from patient to patient seemingly dependent on the level of β-globin mRNA. The proposed mechanism is a differential affinity of an mRNA-stabilizing ribonuclear protein complex, which leads to preferential accumulation of β-globin mRNA. Increased accumulation of γ-globin mRNA occurs only when β-globin mRNA is severely deficient as in homozygous persons.50 The Corfu deletion removes a chromatin boundary element located approximately 3 to 4 kb upstream of the δ-gene, which is thought to define 2 developmentally chromatin subdomains, fetal and adult. Deletion of the region containing the boundary element in transgenic mice results in a failure to open the adult subdomain, whereas silencing of the γ-genes is unperturbed.51 A 250-bp pyrimidine-rich DNA sequence located 1 kb upstream of the human δ-globin gene interacts with a multiprotein complex, which is thought to suppress LCR interactions with the γ-promoters and enhance interaction with the δ-promoter.52
Much has been learned during the past decade about hematopoiesis and transcriptional regulation that is ultimately relevant to the switching mechanisms. The origin of hematopoietic stem cells during embryonic life has recently been reviewed.53,54 Independent derivation of stem cells is attributed to the yolk sac and the aortic-gonad-mesonephrus with subsequent definitive erythropoiesis involving colonization of the fetal liver, thymus, spleen, and ultimately the bone marrow. The Wnt/β-catenin and Notch-δ signaling pathways have important roles in stem cell development. Several transcription factors, such GATA-2, RUNX1, TEL/ETV6, SCL/TAL1, and LM02, also have critical roles in stem cell development, whereas other transcriptional factors, namely, PU.1, GFIX, C/EBPα, and GATA-1, function more as lineage specific factors. However, many factors associated with lineage specificity are also expressed at low levels in more primitive cells, leading to the concept of lineage priming, which allows stem cells to go down one or another pathway dependent on other events within the cells or environmental stimuli. A recent study examined the genome-wide binding patterns and combinations of interactions for 10 key regulators of blood stem/progenitor cells.55 Direct protein interactions among multiple factors are involved in the formation of multiprotein complexes that bind regulatory elements within DNA and control stem cell differentiation. Transcription is highly dynamic with ongoing interactions among basic transcriptional machinery and regulatory components to modulate activities of RNA polymerase.56 Recent studies suggest that RUNX1 is a major player contributing to the opening of the chromatin surrounding the PU.1 gene.57 In this opening act, RUNX1 does not assemble into a detectable, stable transcription factor complex but instead takes the lead to begin the gradual changes in chromatin structure that, on reaching a threshold, trigger PU.1 expression to initiate a dramatic program of hematopoietic lineage differentiation.58
As recently reviewed, epigenetic mechanisms, such as DNA methylation and histone modifications, are important ancillary regulatory mechanisms.59 Histone modification by acetylation, methylation, phosphorylation, sumoylation, and ubiquitination and DNA methylation on CpG residues are among the most important epigenetic mechanisms. Primary regulation is thought to occur via sequence specific binding of multiprotein complexes to DNA, but such complexes have the capacity to recruit other multiprotein complexes that function to establish relevant epigenetic modifications on active or inactive loci. Alternatively, certain epigenetic marks may facilitate the recruitment of chromatin modifying complexes, which open chromatin to facilitate transcription factor complex binding. The ability to demethylate the CpG residues in the silenced γ-globin gene promoter with 5-azacytidine prompted demonstration of its ability to enhance fetal hemoglobin production in patients with thalassemia60 and sickle cell disease.61 Demethylating drugs, including decitabine, are potent activators of fetal hemoglobin production in adults, although clinical testing has been limited by largely unfounded fears regarding leukemogenesis. Recent use of a chemical genetic strategy to screen libraries of chemical probes coupled with RNA interference has identified 2 histone deacetylases (HDAC1 and HDAC2) as potential new molecular targets for mediating fetal hemoglobin induction.62
Studies in erythroid and other cells have shown that transcription occurs in a limited number of discrete sites or foci within nuclei termed “transcription factories.” A recent review describes a model indicating that polymerase localizes within these transcriptional factors and reels DNA strands through as they are transcribed.63 Both the mouse α- and β-globin genes are associated differentially with other actively transcribed, erythroid specific genes within transcriptional factories. Such factories are thought to boost the expression of clustered, coregulated genes by concentrating specific transcription factors required for their coordinate or increased transcription.64 Erythroid Kruppel-like factor is thought to have a critical role in gene recruitment to transcription factories in erythroid cells. Long-range gene control by remote enhancers and locus control regions is thought to involve direct interaction between chromatin at the enhancer and gene local regulatory elements within transcription factories.64
GATA-1 is a lineage-specific master transcriptional factor that is essential for erythroid and megakaryocytic development and antagonizes neutrophilic differentiation.65,66 A review of the interactive proteins, the distribution of GATA-1 binding sites, the changes in gene expression induced by GATA-1 and the potential role of posttranslational modifications in GATA-1 activity has recently been published.67 An integrated model is presented involving several multiprotein complexes containing GATA-1, which orchestrate gene expression or repression in erythroid cells. GATA-2 is important in early hematopoiesis; and during progression of erythropoiesis, there is a GATA-2 to GATA-1 switch.66 The GATA-2 to GATA-1 switch is reflected by replacement of one GATA factor by another at specific chromatin sites. Ongoing studies of the GATA proteins and their interactions with multiple transcriptional factors illustrate the complexity of transcriptional networks in controlling expression of particular genes.
Substantial evidence indicates that the globin switches that occur in the context of embryonic, fetal, and adult hematopoiesis occur within the various cell populations, coincidentally, according to a developmental clock rather than being based on development of separate, globin-specific lineages. The concept of the developmental clock mechanism for the control of hemoglobin switching arose from the study of γ- to β-switching in somatic cell hybrids between human fetal liver cells and mouse erythroleukemia cells.68 Hybrid cells formed with early-stage fetal liver cells initially made γ-globin but switched synchronously to β-globin during extended culture. Recent studies have shown the occurrence of globin switches in cultures of CD34+ cells developed from embryonic stem cells in which the switches occur within the cultured cell population over time.69 We have recently compared the pattern of hemoglobin production in erythroblasts derived from fetal liver, cord blood, or adult mobilized peripheral blood or bone marrow CD34+ cells in liquid culture.70 This system recapitulates expression of globin genes according to the developmental stage of the originating CD34+ cell source. Thus, the key molecular events that determine the pattern of hemoglobin production by developing erythroblasts have occurred in progenitors at a very early stage of hematopoiesis.
Molecular control of the fetal to adult hemoglobin switch
Substantial evidence suggests that the fetal to adult hemoglobin switch is influenced by multiple cis-acting elements and many transcriptional factors acting as multiprotein complexes. Evidence implicating a particular cis-acting element or a transcriptional factor in γ- to β-switching is strongest when there is both genetic evidence as well as functional and/or biochemical evidence supporting its role (Table 1). Several point mutations in one or the other of the γ-globin gene promoters are individually associated with increased expression of that γ-gene, as summarized in a recent review.47 Genetic linkage and genome-wide association studies have identified 3 major loci that account for genetic variation in HbF.4-6,48,49,71,72 One of these loci is the β-globin cluster on chromosome 11. Prior studies had suggested that a single nucleotide polymorphism at position −158 of the γ-promoter was associated with clinical phenotypic variation of HbF levels in patients with sickle cell disease, but recent studies indicate that the relevant single nucleotide polymorphism is approximately midway between the γ- and δ-globin gene.73 The second is a polymorphism on the long arm of chromosome 6 located in a non–protein-coding region between the genes HBS1L and MYB. The third and best characterized to date is the variant identified within intron 2 of the zinc finger transcription factor BCL11A; this polymorphism is associated with reduced expression of this gene.
Trans-acting factors involved in the transcriptional switch from fetal to adult hemoglobin production
| Factor . | Description . | Biologic role . | Mechanism of action/comment . | Method of identification as regulator of γ-globin expression . | References . |
|---|---|---|---|---|---|
| HBS1L-MYB | HBS1L: GTP-binding protein MYB: proto-oncogene | Unclear | Altered MYB expression influences γ-globin gene expression | Linkage analysis and GWAS | 71,72 |
| BCL11A | Zinc-finger transcription factor | Repressor of γ-globin gene expression | Binds to HS3 and γδ intergenic region; interacts with SOX6 | Linkage analysis and GWAS; β-YAC/BCL11A–deficient mice; shRNA knockdown in human erythroid cells | 5, 6, 49, 74, 75, 76 |
| SOX6 | SRY family of HMG box transcription factors | Involved in silencing of mouse embryonic globin genes; interacts with BCL11A to repress γ-globin gene expression | Directly interacts with BCL11A; binds to proximal γ-globin gene promoters to recruit BCL11A | Chromatin immunoprecipitation studies of murine ϵY-globin promoter; coimmunoprecipitates with BCL11A | 75, 77 |
| KLF1 (EKLF) | Erythroid-specific Kruppel-like factor | Repressor of γ-globin gene expression | Haploinsufficiency results in HPFH; directly interacts with CBP/p300 and BRG1 and recruits Sin3A and HDAC1 | Linkage analysis and GWAS; β-YAC/KLF1–deficient mice; shRNA knockdown in human erythoid cells | 7, 81 |
| p22NF-E4 | Fetal erythroid-specific transcription factor | Activator of γ-globin gene expression; interacts with CP2 to form SSP | Transcriptional activation of γ-globin gene expression | Biochemical found as component of SSP bound to γ-globin promoter; ectopic expression increased γ-globin expression in cord blood cells and delays fetal to adult Hb switch in β-YAC mice | 94, 96, 97 |
| COUP-TFII | Orphan nuclear transcription factor | Binds to DR-like sequences in proximal γ-globin gene promoters | Repressor of γ-globin gene expression | Binds DR-like sequence in γ-globin promoters; increased γ-globin expression observed when repressed by high levels of stem cell factor or shRNA knockdown in human erythroid cells | 90, 91 |
| DRED (TR2/TR4) | Orphan nuclear transcription factor | Binds to DR1 sequences in proximal γ-globin gene promoters | Repressor of γ-globin gene expression | Identified as repressor of murine embryonic globin genes; binds γ-globin promoters | 8, 85 |
| FOP | Chromatin-associated protein that directly interacts with and is methylated by PRMT1 | Regulator or repressor of γ-globin gene expression | shRNA-mediated knockdown of FOP reduces levels of Sox6 but does not alter levels of BCL11A in developing adult erythroid cells | Levels of FOP are reduced in erythroblasts derived from fetal liver compared with adult blood cells; increased γ-globin expression after shRNA knockdown in fetal liver cells of β-YAC mice and erythroid cells derived from normal and β-thalassemic donors | 95 |
| Factor . | Description . | Biologic role . | Mechanism of action/comment . | Method of identification as regulator of γ-globin expression . | References . |
|---|---|---|---|---|---|
| HBS1L-MYB | HBS1L: GTP-binding protein MYB: proto-oncogene | Unclear | Altered MYB expression influences γ-globin gene expression | Linkage analysis and GWAS | 71,72 |
| BCL11A | Zinc-finger transcription factor | Repressor of γ-globin gene expression | Binds to HS3 and γδ intergenic region; interacts with SOX6 | Linkage analysis and GWAS; β-YAC/BCL11A–deficient mice; shRNA knockdown in human erythroid cells | 5, 6, 49, 74, 75, 76 |
| SOX6 | SRY family of HMG box transcription factors | Involved in silencing of mouse embryonic globin genes; interacts with BCL11A to repress γ-globin gene expression | Directly interacts with BCL11A; binds to proximal γ-globin gene promoters to recruit BCL11A | Chromatin immunoprecipitation studies of murine ϵY-globin promoter; coimmunoprecipitates with BCL11A | 75, 77 |
| KLF1 (EKLF) | Erythroid-specific Kruppel-like factor | Repressor of γ-globin gene expression | Haploinsufficiency results in HPFH; directly interacts with CBP/p300 and BRG1 and recruits Sin3A and HDAC1 | Linkage analysis and GWAS; β-YAC/KLF1–deficient mice; shRNA knockdown in human erythoid cells | 7, 81 |
| p22NF-E4 | Fetal erythroid-specific transcription factor | Activator of γ-globin gene expression; interacts with CP2 to form SSP | Transcriptional activation of γ-globin gene expression | Biochemical found as component of SSP bound to γ-globin promoter; ectopic expression increased γ-globin expression in cord blood cells and delays fetal to adult Hb switch in β-YAC mice | 94, 96, 97 |
| COUP-TFII | Orphan nuclear transcription factor | Binds to DR-like sequences in proximal γ-globin gene promoters | Repressor of γ-globin gene expression | Binds DR-like sequence in γ-globin promoters; increased γ-globin expression observed when repressed by high levels of stem cell factor or shRNA knockdown in human erythroid cells | 90, 91 |
| DRED (TR2/TR4) | Orphan nuclear transcription factor | Binds to DR1 sequences in proximal γ-globin gene promoters | Repressor of γ-globin gene expression | Identified as repressor of murine embryonic globin genes; binds γ-globin promoters | 8, 85 |
| FOP | Chromatin-associated protein that directly interacts with and is methylated by PRMT1 | Regulator or repressor of γ-globin gene expression | shRNA-mediated knockdown of FOP reduces levels of Sox6 but does not alter levels of BCL11A in developing adult erythroid cells | Levels of FOP are reduced in erythroblasts derived from fetal liver compared with adult blood cells; increased γ-globin expression after shRNA knockdown in fetal liver cells of β-YAC mice and erythroid cells derived from normal and β-thalassemic donors | 95 |
GWAS indicates genome-wide association study; and SSP, stage selector protein.
BCL11A
The role of BCL11A as a regulator of γ-globin gene silencing was demonstrated experimentally by increased production of HbF in developing adult human erythroblasts after shRNA-mediated knockdown. BCL11A occupies the β-globin locus in developing erythroid cells and associates with distal regulatory elements with concentrated binding in the LCR (hypersensitive site 3) and the intergenic region between the Aγ-globin and δ-globin genes to reconfigure the β-globin cluster74-76 (Figure 2). Human γ-globin transgene and mouse embryonic globin gene expression is derepressed in the context of the Bcl11a−/− genotype, although red cell phenotype is normal.76 Recent evidence indicates that the mechanism of BCL11A-mediated silencing occurs through cooperation with the high-mobility group transcription factor SOX6, as BCL11A and SOX6 are coexpressed and physically interact. Lentiviral vector-mediated knockdown of both proteins results in synergistic up-regulation of HbF in developing erythroblasts.76,77 Thus, there are both substantial genetic and functional data supporting a key role for BCL11A in silencing of the γ-globin genes during developmental switching and its potential role in reactivation of HbF in adult erythroblasts.
KLF1
Also known as Erythroid Kruppel-like factor, KLF1, a zinc finger protein, binds the CACCC of the adult β-globin gene in mice and humans7,78 and is critical for its expression as ablation in mice caused embryonic lethality because of severe anemia.79,80 Although it was known that both endogenous mouse embryonic and human fetal globin gene expression was not properly down-regulated in the Eklf−/− mice, its role in hemoglobin switching was not apparent until 2 studies recently emerged.81,82 One study used a genome-wide single nucleotide polymorphism scan followed by linkage analysis of a family, with 10 of 27 members showing the HPFH phenotype. This led to identification of a nonsense mutation in KLF1, which ablated the DNA binding domain, resulting in haploinsufficiency in the heterozygous state.81 Of interest is that HbF levels varied among individual family members, suggesting the influence of other genetic and perhaps environmental factors. In addition to binding to the β-globin gene promoter, normal KLF1 was found to show strong binding to the BCL11A promoter, activating its expression in cultured, primary adult human erythroblasts (Figure 2). Thus, KLF1 may mediate its switching effects through a dual mechanism, acting both on the β-globin and BCL11A promoters. Zhou et al also found that decreased, but not absent, KLF1 levels in genetically altered mice led to markedly reduced Bcl11a mRNA and protein levels.82 They also found strong binding of KLF1 to the Bcl11a promoter. Mouse embryonic and human fetal globin transgene expressions were greatly increased in the setting of reduced KLF1. Similarly, γ-globin gene expression was up-regulated when KLF1 was knocked down in cultured adult human erythroblasts.82 Thus, there are both compelling genetic and functional data indicating that KLF1 has a significant role in the fetal to adult switch. Remaining to be solved is how a 2- to 3-fold increase in its expression in adult versus fetal cells results in KLF1's preferential association with the β-promoter in the former and the γ-promoters in the latter.
HBSL1-MYB DNA region
Variants in the intergenic region between the HBSL1 and MYB genes were identified in a genome-wide association study designed to uncover polymorphisms associated with the variability of HbF expression in nonanemic humans.71 Recent data suggest that the intergenic area most highly associated with HbF expression has properties of a regulatory element.72 The linkage to an equilibrium block, which is most closely associated with HbF levels, is between the HBS1L and MYB genes, which are found on opposite DNA strands. Three hypersensitive sites have been identified within that region. These sites exhibit the characteristic marks of active chromatin, including histone acetylation and RNA polymerase II binding in erythroid but not in nonerythroid cells.72 HBS1L is thought to be a housekeeping gene because it is ubiquitously expressed, whereas MYB has more of a restrictive pattern of expression and is crucial for erythroid development. Expression profiles of 5 genes within this region, including MYB and HBS1L, were compared in the erythroid cultures of 12 persons with elevated and 14 persons with normal HbF levels.83 Both cMYB and HBS1L were down-regulated in persons with elevated HbF levels, whereas the other 3 genes did not change in their level of expression. Overexpression of MYB inhibited γ-globin gene expression in human erythroleukemia cells, whereas overexpression of HBS1L had no effect. The investigators speculate that low levels of MYB result in fewer cell-cycle events early in erythropoiesis and that early maturation of erythroblasts yields red cells containing higher levels of HbF.83 Overall, MYB has an important role in erythropoiesis, and recent studies indicate that it acts, in part, by transactivation of KLF1 and LMO2 expression.84 Resequencing and genotyping studies have recently identified rare missense mutations that provide further evidence for the involvement of MYB in modulating HbF levels.73
SOX6
SOX6 was first observed to play a role in the silencing of the mouse embryonic globin genes and was shown by chromatin immunoprecipitation to bind to the murine ϵγ-globin promoter.77 Subsequently, it was shown that SOX6 interacts with BCL11A, although SOX6 strongly binds the γ-globin gene promoters (Figure 2). This suggested that the SOX6-BCL11A protein-protein interaction may involve chromatin looping.75 SOX6 binding to the γ-globin promoters also was found to require BCL11A. Finally, knockdown of SOX6 in cultured, primary human erythroblasts led to a small up-regulation of HbF but when combined with BCL11A knockdown, further enhanced HbF production, beyond that achieved with either knockdown alone.75
TR2/TR4 DR erythroid-definitive complex
Originally identified as a repressor of the human embryonic ϵ-globin gene,8,85 the direct repeat (DR) erythroid-definitive complex consists of a heterodimer of 2 orphan nuclear receptors, TR2 and TR4. This dimer, along with the nuclear factor COUP-TFII (next section, “COUP-TFII”) binds DRs in the ϵ-globin gene promoter in vitro and later was shown to also bind a similar repeat with the γ-globin gene promoters86 (Figure 2). Endogenous murine embryonic to adult globin gene and fetal to adult human β-globin transgene switching is delayed in mice that are null for TR2/TR4, whereas enforced expression of a dominant negative TR4 results in both loss of endogenous embryonic gene expression and human γ-globin transgene silencing. Interestingly, a mutation at −117 of the γ-globin promoter, associated with HPFH, affects a DR element and influences factor binding, adding to evidence that this heterodimer plays a role in stage-selective repression of the human γ-globin gene.87,88
COUP-TFII
Initially described as one of 2 factors necessary for transcription of the chicken ovalbumin gene,89 COUP-TFII was later found to bind to the γ-globin promoter DR in vitro as well90 (Figure 2). Mutation of DR elements results in derepression of γ-globin transgene expression in adult mice, suggesting that DR sites are involved in fetal globin gene silencing. An in vitro model of primary human erythroblast development was used to show that the cytokine stem cell factor, through activation of the Erk1/2 and/or p38 mitogen-activated protein kinase pathways, suppresses COUP-TFII expression at the mRNA and protein level, resulting in a large reduction in its binding to the γ-globin promoter, as assessed by chromatin immunoprecipitation assays.91 RNA polymerase II binding increased several-fold and correlated with increased γ-globin gene expression. Finally, knockdown of COUP-TFII with a short inhibitory RNA in cultured, primary human erythroblasts resulted in up-regulation of γ-globin expression. Similar to DR erythroid-definitive, repressor function of COUP-TFII is evidenced by the fact that 4 different HPFH mutations in the γ-globin gene promoter reduce COUP-TFII binding.92
FOP
Friend of protein arginine methyltransferase I (FOP) was recently described as a target of protein arginine methyltransferase 1 (PRMT1), which stably interacts with chromatin.93 Initially studied for its role in estrogen-dependent gene activation,93 FOP may also have a role in hemoglobin switching. Lentiviral-mediated knockdown of FOP in murine fetal liver cells transgenic for the β-globin locus significantly increased expression of both the mouse embryonic and human fetal globin genes.94 Similarly, knockdown of FOP in erythroblasts derived from human adult peripheral blood erythroid progenitors also resulted in up-regulation of γ-globin and increased production of HbF. HbF levels increased from background levels of 6% to 7% to an average of approximately 27%. Interestingly, BCL11A levels were not altered, but SOX6 protein appeared to be reduced. Thus, it is possible that FOP depletion increased γ-globin expression through reduction of SOX6 (Figure 2).
NF-E4
Originally identified biochemically as a component of the “stage selector protein,” which bound the stage selector element in the proximal γ-globin promoter in human K562 erythroleukemia cells, NF-E4, in conjunction with the ubiquitous transcription factor CP2, facilitates transcription of the γ-globin gene.95,96 The stage selector element is conserved in species having γ-globin genes but is not present in species lacking HbF. Biochemical data support the idea that p22 NF-E4 acts as an “activator” of γ-globin transcription by interacting with CP2 to recruit p45 NF-E2 and subsequently RNA polymerase II to the promoter (Figure 2). Further evidence supporting a role for NFE4 in HbF production is the demonstration that an HPFH mutation at −202 creates a new binding site for the NF-E4-CP2 complex.97 An alternate form, p14 NF-E4, apparently generated by translational initiation from an internal methionine, may play a role in γ-globin gene down-regulation or silencing. The smaller isoform is abundant in adult erythroid cells; and although it does not bind the γ-globin promoter, it appears to interact with CP2 and may function to sequester NF-E2 and prevent complexing of activating factors at the γ-globin promoter. Enforced expression of p22 NF-E4 in transgenic mice harboring the human β-globin locus delayed the switch, without increasing HbF expression in the adult.97
Genetic efforts to develop therapeutic blockade of the switch
These recent insights into the repressive factors that silence γ-globin gene expression have offered potentially novel approaches to “reactivate” HbF expression for therapeutic purposes in the context of sickle cell disease and β-thalassemia. Lentiviral vector delivery of shRNA designed to knock down these factors has been used to increase γ-globin and HbF expression in cultured adult human erythroblasts.74-77 Although knockdown of COUP-TFII, KLF1, BCL11A, SOX6, and FOP resulted in increased γ-globin mRNA levels, only the studies involving knockdown of BCL11A, SOX6, and FOP documented significant increases in HbF protein in cultured human adult erythroblasts, under conditions with minimal or modest baseline HbF levels. FOP knockdown resulted in an increase in HbF from approximately 6% to 7% to approximately 27%, whereas BCL11A knockdown resulted in HbF increasing from minimal levels of 1% or less to more than 30%. Recent work suggests that BCL11A knockdown in erythroblasts derived from the CD34+ cells of patients with β-thalassemia can significantly increase the level of HbF and raise total hemoglobin content to potentially therapeutic levels.98
An alternate approach put forward99 and recently evaluated in cultures of primary human normal and β-thalassemic erythroid cells involves delivery and expression of an artificial zinc finger transcriptional activator factor designed to interact with the γ-globin gene promoters at the −117 site. Lentiviral-mediated expression of this factor (GG1-VP64) resulted in HbF levels of up to 20% in erythroblasts derived from transduced, normal adult CD34+ cells, compared with background levels of HbF measuring less than or equal to 2%.70 Expression of GG1-VP64 in thalassemic erythroblasts, after lentiviral-mediated transfer into bone marrow CD34+ cells from transfusion-dependent, β+-thalassemic patients, resulted in high levels of HbF (54%) and, importantly, a doubling of the hemoglobin content, from 20% that of normal cells up to 39% of normal, a level that would probably be therapeutic.98 When combined with lentiviral-mediated transfer of a γ-globin gene, HbF levels in thalassemic erythroblasts approached 60%, with a similar further increase in total hemoglobin content. Coupling γ-globin gene addition with activation of the endogenous γ-globin gene loci may be therapeutically useful.
In conclusion, a vast body of scientific knowledge has been gained in the past 40 years from studying the expression of the globin genes. Although the critical events that establish the program of globin synthesis remain obscure in that they appear to be established in very early erythroid progenitors in accordance with a developmental clock, the factors that carry out the program of globin synthesis in maturing erythroblasts are emerging and are clearly amenable to manipulation to increase fetal hemoglobin production. The search for a single switching mechanism has given way to the realization that hemoglobin switching is indeed a very complex developmental phenomenon involving many players with various roles. Some, such as BCL11A, although having significant roles in lymphoid cell development, appears to have a very limited role in red cells.100 Thus, BCL11A is an attractive target for modulating HbF synthesis via lentiviral vector–mediated gene transfer and erythroid-specific expression of a BCL11A shRNA. Other factors that have equally important roles in the switch, such as KLF1 and MYB, have many other roles in red cell development and function, making it less obvious that they could be manipulated in the context of enhancing HbF production in adult cells without undesirable effects on other aspects of erythropoiesis. Several key players, such as BCL11A, KLF1, and MYB, have been identified and substantiated both by genetic and functional data, whereas other factors have less defined roles both functionally and quantitatively. New factors remain to be discovered, and their role in the emerging network of fetal hemoglobin regulation remains to be defined.100 Continued advances should facilitate the development of novel genetic approaches to therapy for patients with hemoglobin disorders.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Pat Streich for her assistance in the preparation of this manuscript.
This work was supported by the National Heart, Lung, and Blood Institute (PO1HL053749, A.W.N., D.A.P.; Basic and Translational Research Program Sickle Cell Disease Grant U54HL070590, D.A.P.), the Cooley's Anemia Foundation (D.A.P.), the Assisi Foundation of Memphis (A.W.N., D.A.P.), and the American Lebanese Syrian Associated Charities (A.W.N., D.A.P.).
National Institutes of Health
Authorship
Contribution: A.W., A.W.N., and D.A.P. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Derek A. Persons, Department of Hematology, St Jude Children's Research Hospital, 262 Danny Thomas Pl, MS 341, Memphis, TN 38105; e-mail: derek.persons@stjude.org.