Abstract
Evi1 (ecotropic viral integration site 1) is essential for proliferation of hematopoietic stem cells and implicated in the development of myeloid disorders. Particularly, high Evi1 expression defines one of the largest clusters in acute myeloid leukemia and is significantly associated with extremely poor prognosis. However, mechanistic basis of Evi1-mediated leukemogenesis has not been fully elucidated. Here, we show that Evi1 directly represses phosphatase and tensin homologue deleted on chromosome 10 (PTEN) transcription in the murine bone marrow, which leads to activation of AKT/mammalian target of rapamycin (mTOR) signaling. In a murine bone marrow transplantation model, Evi1 leukemia showed modestly increased sensitivity to an mTOR inhibitor rapamycin. Furthermore, we found that Evi1 binds to several polycomb group proteins and recruits polycomb repressive complexes for PTEN down-regulation, which shows a novel epigenetic mechanism of AKT/mTOR activation in leukemia. Expression analyses and ChIPassays with human samples indicate that our findings in mice models are recapitulated in human leukemic cells. Dependence of Evi1-expressing leukemic cells on AKT/mTOR signaling provides the first example of targeted therapeutic modalities that suppress the leukemogenic activity of Evi1. The PTEN/AKT/mTOR signaling pathway and the Evi1-polycomb interaction can be promising therapeutic targets for leukemia with activated Evi1.
Introduction
Evi1 (ecotropic viral integration site 1) is a nuclear transcription factor that is indispensable for proliferation of hematopoietic stem cells (HSCs) both during embryogenesis and in the adult.1,2 Aberrant expression of Evi1 is implicated in the development of myeloid disorders, including acute myeloid leukemia (AML), myelodysplastic syndrome, and chronic myelogenous leukemia (CML).3-5 High Evi1 expression occurs in ∼ 10% of cases of AML and genetically defines one of the largest clusters in AML.6 The patients classified into this cluster show an extremely poor outcome.6,7 Evi1 possesses diverse functions as an oncoprotein, including suppression of transforming growth factor-β–mediated growth inhibition,8 negative regulation of the c-Jun N-terminal kinase pathway,9 and stimulation of cell growth by activator protein-1 (AP-1).10 Aberrant expression of Evi1 affects hematopoietic differentiation in various lineages. Several groups reported that Evi1 blocks myeloid differentiation.11-13 Evi1 also affects differentiation of erythroid and megakaryocytic cells.11,14-16 In addition to its DNA-binding activity, Evi1 has the potential to recruit diverse proteins for transcriptional regulation. We and others have identified several target genes that are activated by Evi1, including globin transcription factor 2 (GATA2)2,17 and pre-B-cell leukemia homeobox 1 (PBX1).18 However, there are no reports of genes that are directly repressed by Evi1, despite that Evi1 interacts with several transcriptional corepressors, such as C-terminal binding protein (CtBP),19 SUV39H1, and G9a.20-22
Phosphatase and tensin homologue deleted on chromosome 10 (PTEN) plays critical roles in cell growth, migration, and death.23 It is mutated or deleted with high frequency in various human cancer tissues to promote tumorigenesis.24 The primary target of PTEN in cancer is phosphatidylinositol 3,4,5-triphosphate,25 and the loss of PTEN leads to constitutively high expression of phosphatidylinositol 3,4,5-triphosphate, which induces AKT kinase activation.26 AKT, in turn, phosphorylates a plethora of targets. In particular, AKT has a remarkable effect on cellular growth by activating the mammalian target of rapamycin (mTOR). Deletion of PTEN in murine adult HSCs has been studied by 2 groups.27,28 Both groups observed the rapid onset of myeloproliferative disorders within 4-6 weeks after PTEN knockout, which rapidly progressed to acute leukemia. Importantly, these phenotypes could be rescued by mTOR inhibitor rapamycin.27
Epigenetic perturbations such as altered DNA methylation, misregulation of chromatin remodeling by histone modifications have emerged as common hallmarks of tumors.29,30 Polycomb group (PcG) proteins are one of such epigenetic regulators. PcG proteins catalyze the addition of a methyl group at lysine 27 of histone H3 (H3K27me) and function as transcriptional regulators that silence specific sets of genes through chromatin modification.31,32 PcG proteins comprise 2 functionally and biochemically distinct multimeric polycomb repressive complexes (PRCs), called PRC1 and PRC2/3/4.
This study shows a novel function of Evi1 to regulate the PTEN/AKT/mTOR signaling pathway and involvement of PRCs in PTEN down-regulation by Evi1. These results provide a possibility of overcoming the poor prognosis of patients with leukemia with high Evi1 expression, which is supported by our newly established mouse model and the analysis of human samples.
Methods
Subjects
Studies that involved human subjects were done in accordance with the ethical guidelines for biomedical research involving human subjects, which was developed by the Ministry of Health, Labor, and Welfare, Japan, the Ministry of Education, Culture, Sports, Science, and Technology, Japan, and the Ministry of Economy, Trade, and Industry, Japan, and enforced on March 29, 2001. This study was approved by ethical committee of Tokyo University. Written informed consent was obtained from all patients whose samples were collected after the guideline was enforced in accordance with the Declaration of Helsinki. All animal experiments were approved by the University of Tokyo Ethics Committee for Animal Experiments.
Reporter assay
Analysis of luciferase activities was performed as described previously.8 Briefly, cells were seeded in 12-well culture plates at a density of 2 × 104/well and were transfected with the use of FuGENE6 (Roche). The transfected cells were harvested 48 hours after transfection and assayed for luciferase activity with the use of dual luciferase kit (Piccagene). Firefly luciferase activity was measured as relative light units. The relative light units from individual transfection were normalized by measurement of Renilla luciferase activity in the same samples. Relative PTEN promoter activity was presented as the ratio of normalized luciferase activity of mock-transfected cells.
Retrovirus production and bone marrow transplantation assays
These procedures were performed as described previously.1,33-35 Briefly, Plat E packaging cells were transiently transfected with each retrovirus vector, and supernatant containing retrovirus was collected 48 hours after transfection and used immediately for infection. Fluorouracil-primed bone marrow (BM) cells isolated from C57/B6 mice were used for retroviral transduction. To establish Evi1-induced murine AML models, pMYs-Evi1–internal ribosome entry site (IRES)–green fluorescent protein (GFP) or empty retrovirus was used, and 0.2-1.2 × 106 of Evi1-IRES-GFP35,36 or GFP-transduced BM cells (Ly5.1) were injected through the tail vein into C57/B6 (Ly5.2)–recipient mice that had been administered a sublethal irradiation (5.25 Gy). To test the sensitivity of rapamycin in vivo, 1 × 106 of Evi1-, translocated ets leukemia (TEL)/platelet-derived growth factor β receptor (PDGFβR)–AML1/ETO–, or AML1_S291fsX300-induced leukemic cells were injected to sublethally irradiated (7.5 Gy) secondary recipient mice. Diagnosis of AML was made according to the Bethesda proposals.37
Colony-forming assays
For short hairpin RNA (shRNA)–mediated knockdown assays, transformed BM cells from the third to fourth round of in vitro replating were subsequently infected with retrovirus encoding shRNAs. After retroviral transduction, BM cells were resuspended in Methocult3434 (StemCell Technologies) at a concentration of 4 × 104 cells/mL before selection or 1 × 104 cells/mL after selection and seeded at 1 mL/35-mm petri dish in duplicate. Colony number of each dish was scored weekly. G418 (1.0 mg/mL) or puromycin (1.0 μg/mL) was added to the Methocult for the purpose of selection. We defined a colony as a cluster of ≥ 100 cells. For in vitro inhibitor assays, rapamycin (Cell Signaling), LY294002 (Cell Signaling), BMS345541 (Calbiochem), or DAPT (Calbiochem) was reconstituted with dimethyl sulfoxide (Sigma-Aldrich) and added to methylcellulose.
siRNA interference
Specific siRNA oligos targeting EZH2, SUZ12, and EED mRNAs were designed as indicated by Clontech and cloned into pSIREN-RetroQ vectors. Control shRNA is a nonfunctional construct provided from Clontech. See supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article) for more information.
Quantitative real-time PCR
Real-time polymerase chain reaction (PCR) was carried out with the LightCycler480 (Roche) or the ABI PRISM 7000 Sequence Detection System (Applied Biosystems) according to the manufacturer's instructions. The results were normalized to β-actin levels. See supplemental Methods for more information.
ChIP
Detailed protocols for chromatin immunoprecipitation (ChIP) assays are presented in supplemental Methods. Immunoprecipitated DNA fragments were quantified by real-time PCR with the use of the following primers (Figure 1D); PCR primers 1 and primers 3 amplify sequences, including putative Evi1 binding sites (5′-AGACAGGTGAGGAAA-3′ fragment at position −4257/−4243 and 5′-AAAATAGAA-3′ fragment at position 1935/1943, respectively), which were identified by the rVISTA2.0 tool (http://rvista.dcode.org/) with the use of a matrix similarity threshold of 0.80 and 0.85, respectively, and PCR primers 2 amplify a sequence, including possible murine Egr1 binding site (5′-CCGCCCACTCGC-3′ fragment at position −1907/−1896 upstream of the initiation codon ATG [+1]). Primers for human samples correspond to the primers 3 in Figure 1D, and they amplify a sequence, including 5′-AGAAGATAA-3′ fragment.
EMSA
Protein lysates were obtained from 293T cells transfected with plasmids encoding Flag-tagged Evi1 or its mutants, immunoprecipitated with EZview Red ANTI-FLAG M2 Affinity Gel (Sigma-Aldrich), and eluted with 3× Flag peptide (Sigma-Aldrich) according to the manufacturer's recommendation. The procedures for electromobility shift assay (EMSA) were performed with the EMSA “Gel Shift” Kit (Panomics) according to the manufacturer's recommendation. Biotin-labeled probes were added. A competition control was set up by adding non–biotin-labeled cold probes to the reaction. See supplemental Methods for more information.
Microarray analysis
Gene expression analysis was carried out as previously described1 with the use of the Mouse Genome 430 2.0 Array (Affymetrix). All microarray data have been deposited in National Center for Biotechnology Information's Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through accession no. GSE22434. See supplemental Methods for more information.
Statistical analysis
Statistical significance of differences between parameters was assessed with a 2-tailed unpaired t test. The correlation between Evi1 and PTEN expression was estimated with both the Pearson product-moment correlation coefficient and the Spearman rank-correlation coefficient. The overall survival of mice in BM transplantation assays was calculated according to the Kaplan-Meier method.
Results
PTEN is a direct target of Evi1
To identify new target genes of Evi1, we first carried out genomewide gene-expression analysis. BM cells derived from wild-type C57/B6 mice were transduced with Evi1-GFP or GFP-expressing retroviruses, and GFP+ cells were sorted and subjected to gene expression profiling (Figure 1A). To narrow down the candidate genes, we assessed the correlation between expression of Evi1 and the potential target genes in AML samples with the use of published gene expression data of 285 persons with AML.6 When we looked at the genes that showed both ≥ 1.3-fold decrease in the expression value of Evi1-GFP–transduced cells (P < .05; n = 8) and the inverse correlation with Evi1 in human AML (P < .01; n = 285) (see supplemental Methods for detailed information), we noted that 3 probe sets for PTEN appeared in the list (supplemental Table 1). PTEN transcription was inversely correlated with that of Evi1 in 285 AML samples (supplemental Table 2). A similar trend was observed when we analyzed another published dataset of 43 patients with AML with lower statistical power38 (supplemental Table 2). We confirmed the above data by quantitative real-time PCR analysis (Figure 1B). We also found that PTEN expression was higher in Evi1-deficient lineage−, c-Kit+, Sca-1+ cells than in Evi1+/+ lineage−, c-Kit+, Sca-1+ cells, which was recovered by forced expression of Evi1 (Figure 1C).
Evi1 down-regulates PTEN expression in BM cells. (A) Schematic representation of gene expression analysis. Evi1-GFP– or GFP-transduced BM cells (n = 4 for each) were sorted and subjected to gene expression analysis. Representative fluorescence-activated cell sorting data show the gene transfer efficiencies of 78% and 90% for Evi1-GFP and GFP-expressing retrovirus, respectively. (B) Real-time PCR for PTEN expression. Error bars indicate SD (n = 6; *P < .0001). (C) PTEN mRNA expression in lineage−, c-Kit+, Sca-1+ cells derived from Mx-Cre Evi1flox/flox mice. Error bars indicate SD (n = 3; *P < .05). Evi1 was retrovirally added back. The Cre-mediated Evi1 deletion and the expression of wild-type Evi1 were evaluated by PCR with the use of genome DNA and cDNA, respectively, and representative figures are shown. Detailed experimental methods are shown in supplemental Data. (D) Schematic representation of mouse PTEN promoter region, possible Evi1 binding site predicted by rVISTA 2.0 (http://rvista.dcode.org/), constructs cloned into pGL4.10[Luc2], mutagenesis strategy, probe sets used for EMSAs, and primers for ChIP assays. (E) Relative luciferase activity of Evi1 on each PTEN promoter. Error bars indicate SD (n = 6; *P < .01). Jurkat cells were used. Error bars indicate SD. (F) Protein expression of Flag-tagged wild-type (WT) Evi1 and Evi1_R205N used for EMSAs. Western blot analysis was done with anti-Flag antibody. (G) Results of EMSAs are shown. Probe sets 3 or 3_mut are indicated in Figure 1D. Corresponding cold-specific competitors or nonspecific competitors were added as indicated. KO indicates knockout.
Evi1 down-regulates PTEN expression in BM cells. (A) Schematic representation of gene expression analysis. Evi1-GFP– or GFP-transduced BM cells (n = 4 for each) were sorted and subjected to gene expression analysis. Representative fluorescence-activated cell sorting data show the gene transfer efficiencies of 78% and 90% for Evi1-GFP and GFP-expressing retrovirus, respectively. (B) Real-time PCR for PTEN expression. Error bars indicate SD (n = 6; *P < .0001). (C) PTEN mRNA expression in lineage−, c-Kit+, Sca-1+ cells derived from Mx-Cre Evi1flox/flox mice. Error bars indicate SD (n = 3; *P < .05). Evi1 was retrovirally added back. The Cre-mediated Evi1 deletion and the expression of wild-type Evi1 were evaluated by PCR with the use of genome DNA and cDNA, respectively, and representative figures are shown. Detailed experimental methods are shown in supplemental Data. (D) Schematic representation of mouse PTEN promoter region, possible Evi1 binding site predicted by rVISTA 2.0 (http://rvista.dcode.org/), constructs cloned into pGL4.10[Luc2], mutagenesis strategy, probe sets used for EMSAs, and primers for ChIP assays. (E) Relative luciferase activity of Evi1 on each PTEN promoter. Error bars indicate SD (n = 6; *P < .01). Jurkat cells were used. Error bars indicate SD. (F) Protein expression of Flag-tagged wild-type (WT) Evi1 and Evi1_R205N used for EMSAs. Western blot analysis was done with anti-Flag antibody. (G) Results of EMSAs are shown. Probe sets 3 or 3_mut are indicated in Figure 1D. Corresponding cold-specific competitors or nonspecific competitors were added as indicated. KO indicates knockout.
To determine whether Evi1 regulates the transcription of PTEN, we next performed luciferase reporter assays. The genomic region of ∼ 10-kb base pairs around the transcription start site (TSS) of PTEN is well conserved across human and mouse, and 2 putative binding sequences for the first zinc finger (ZF1-7) domain of Evi1 were predicted by rVISTA 2.0 with the use of a matrix similarity threshold of 0.80 (http://rvista.dcode.org/). The one is located at position −4257/−4243 and has the [GAC/TA] N0-6 [GAT/CA]–like motif2 (matrix similarity = 0.80), and the other one is located at position 1935/1943 and contains the GACAAGATA-like motif39 (matrix similarity = 0.85). Thus, we divided this region of murine PTEN promoter into 2 fragments (PTEN1 and PTEN2; Figure 1D) and inserted them into pGL4.10 [luc2]. Each reporter plasmid was transiently transfected into Jurkat cells, in which endogenous Evi1 expression is low (data not shown), with or without Evi1 expression plasmid. Evi1 achieved an ∼ 0.25-fold decrease of luciferase activity over basal levels with the PTEN2 constructs (Figure 1E). Stepwise deletion constructs of PTEN reporter (PTEN3 and PTEN4_wt) showed that Evi1 represses PTEN transcription through the region between 1.8 and 3.8 kb downstream of the TSS. Then we created a promoter construct carrying mutated Evi1-binding sites (PTEN4_mut) placed at position 1935/1943, which harbors the evolutionarily conserved GACAAGATA-like sequence (Figure 1D). As shown in Figure 1E, the mutated promoter was insensitive to Evi1 expression, indicating that this sequence is responsible for the effect of Evi1 on PTEN regulation. We obtained essentially the same results in other cell lines, THP-1 and COS7 (supplemental Figure 1A-B).
We next performed EMSAs to test whether Evi1 binds to the GACAAGATA-like motif located at position 1935/1943 downstream of the PTEN TSS. Murine Evi1_R205N harbors a single amino acid change within the ZF1-7 domain and was previously reported to be incapable of binding to DNA containing the GACAAGATA-like motif.39 Flag-tagged wild-type Evi1 and Evi1_R205N were transfected in 293T cells, purified with immunoprecipitation with the use of anti-Flag affinity gel (Figure 1F), and applied to EMSA. Wild-type Evi1 formed a specific DNA-protein complex with the probe set 3 that contains murine PTEN promoter sequence with the AAAAGATAA motif, and it was weakened by cold-specific competitors but not by nonspecific competitors (Figure 1G). Wild-type Evi1 did not bind to the probe set 1 or 2 that was set upstream of the probe set 3 (supplemental Figure 2). However, Evi1_R205N failed to bind to the probe set 3, and wild-type Evi1 did not bind to the probe set 3_mut, whose sequence was mutated in the same way as PTEN4_mut that was used in the reporter assays. These results indicate that Evi1 specifically interacts with the GACAAGATA-like motif within the PTEN promoter via its ZF1-7 domain.
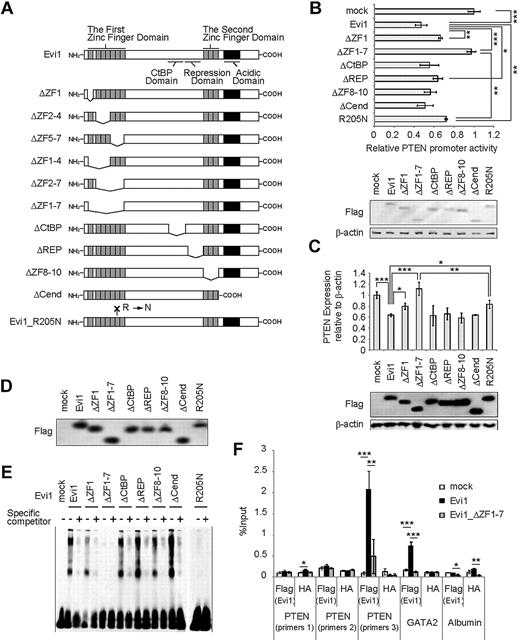
Next, we analyzed transcriptional activities of a series of Evi1 mutants40 (Figure 2A). The ZF1-7 domain is a DNA-binding domain and is essential for interaction with several proteins, including Sma and Mad related protein 3 (SMAD3)8 and c-Jun N-terminal kinase.9 The second zinc finger (ZF8-10) domain is another DNA-binding domain and is essential for activator protein-1 activation.10 The repression domain is required for the efficient repression of transforming growth factor-β signaling.8 The region containing CtBP-binding motif-like sequences is responsible for the interaction with CtBP1.19 In addition, Evi1 contains a highly acidic domain at the C-terminus, which is required for Evi1-mediated P-Sp hematopoiesis.17 The deletion of ZF1-7 almost completely abolishes the repressive activity of Evi1 in Jurkat cells, (Figure 2B), THP-1 cells (supplemental Figure 1C), and COS7 cells (supplemental Figure 1D). Evi1_ΔZF1 and Evi1_R205N have partially lost repressive effects on the PTEN promoter. The murine homologue of these mutants (Evi1_ΔZF1, Evi1_ΔZF1-7, or Evi1_R205N) did not fully repress PTEN transcription when it was retrovirally transduced in primary BM cells (Figure 2C). Moreover, EMSAs suggested that Evi1_ΔZF1-7 did not bind to the PTEN promoter (the probe set 3) (Figure 2D-E). Thus, ZF1-7 domain of Evi1 plays a major role for PTEN repression, although there is a possibility that the other domains of Evi1 and/or the other sites within ZF1-7 make additional contribution.
Evi1 represses PTEN expression via its first zinc finger domain. (A) Schematic representation of Evi1 and its mutants. (B) Relative luciferase activity of Evi1 and its mutants on PTEN4_wt promoter (n = 4) together with immunoblot of assayed Jurkat cells transfected with Flag-tagged wild-type Evi1, its mutants, and mock. Error bars indicate SD; *P < .05, **P < .01, and ***P < .001. (C) PTEN mRNA expression in Flag-tagged Evi1-, its mutants-, or mock-transduced BM cells together with protein expression of Evi1 and its mutants (n = 3). Error bars indicate SD; *P < .05, **P < .01, and ***P < .001. (D) Protein expression of Flag-tagged wild-type Evi1 and its mutants used in EMSAs. Western blot analysis was done with anti-Flag antibody. (E) EMSAs used mutants of human Evi1 and the probe set 3. (F) ChIP analysis for Flag-Evi1, Flag-Evi1_ΔZF1-7, or mock-expressing BM cells that used the indicated antibodies (n = 3). Error bars indicate SD; *P < .05, **P < .01, and ***P < .001. REP, repression domain; Cend, domain at the C-terminus.
Evi1 represses PTEN expression via its first zinc finger domain. (A) Schematic representation of Evi1 and its mutants. (B) Relative luciferase activity of Evi1 and its mutants on PTEN4_wt promoter (n = 4) together with immunoblot of assayed Jurkat cells transfected with Flag-tagged wild-type Evi1, its mutants, and mock. Error bars indicate SD; *P < .05, **P < .01, and ***P < .001. (C) PTEN mRNA expression in Flag-tagged Evi1-, its mutants-, or mock-transduced BM cells together with protein expression of Evi1 and its mutants (n = 3). Error bars indicate SD; *P < .05, **P < .01, and ***P < .001. (D) Protein expression of Flag-tagged wild-type Evi1 and its mutants used in EMSAs. Western blot analysis was done with anti-Flag antibody. (E) EMSAs used mutants of human Evi1 and the probe set 3. (F) ChIP analysis for Flag-Evi1, Flag-Evi1_ΔZF1-7, or mock-expressing BM cells that used the indicated antibodies (n = 3). Error bars indicate SD; *P < .05, **P < .01, and ***P < .001. REP, repression domain; Cend, domain at the C-terminus.
We further tested whether Evi1 binds to the PTEN promoter in primary BM cells. ChIP assays that used Flag-Evi1, Flag-Evi1_ΔZF1-7, or mock-transduced BM cells showed that Evi1 was significantly enriched in the region containing Evi1-binding sequence, which was amplified with primers 3 (Figure 1D) but not in the other regions of the PTEN promoter (amplified with primers 1 or 2) or in the irrelevant albumin promoter (Figure 2F). The association of Evi1 with the PTEN promoter was stronger than that detected with GATA2 promoter, a well-established target of Evi1.2 In contrast, we observed little to no enrichment of Evi1_ΔZF1-7 in the promoter of PTEN or GATA2. No enrichment was detected with the use of an unrelated antibody (anti-HA). Taken together, these data suggest that Evi1 binds to the PTEN promoter through ZF1-7 domain to repress its transcription.
Evi1 activates the AKT/mTOR pathway
We then asked whether Evi1 represses protein expression of PTEN and induces the activation of the downstream AKT/mTOR signaling pathway in primary BM cells. We prepared Evi1- or mock-transduced BM cells and investigated the status of this pathway. As shown in Figure 3A, Evi1 decreased the protein level of PTEN and increased the phosphorylation of AKT as well as mTOR. In contrast, Evi1 had little to no effect on the phosphorylation of extracellular signal-regulated kinase 1/2, signal transducer and activator of transcription 3, or signal transducer and activator of transcription 5 (Figure 3B), suggesting that Evi1 selectively activates the PTEN/AKT/mTOR pathway. Next, we compared the status of PTEN/AKT/mTOR signaling in several murine BM cells that were transformed by various oncogenes. Evi1 is known to enhance self-renewal potential,11 and we could replate Evi1-transduced BM cells > 15 times (data not shown). AML1/ETO, E2A/HLF, and PML/RARα are chimeric genes generated in t(8;21), t(17;19), and t(15;17) leukemias, respectively, all of which are known to transform murine BM cells. With the use of Evi1-, AML1/ETO-, E2A/HLF-, and PML/RARα-, or mock-transduced BM cells, we assessed the status of the PTEN/AKT/mTOR pathway. As shown in Figure 3C and 3D, Evi1-transduced BM cells showed decreased PTEN expression at both mRNA and protein levels and increased phosphorylation of AKT/mTOR compared with AML1/ETO-, E2A/HLF-, PML/RARα-, or mock-transduced cells, indicating that the PTEN/AKT/mTOR pathway is activated in Evi1-transduced BM cells.
Evi1 represses PTEN protein level and activates downstream AKT/mTOR signaling. (A-B) Analysis of indicated protein levels in Evi1 or mock-transduced BM cells. BM cells (1-2 × 106) were used in these assays. Experiments were repeated for > 2 times, and the representative figures are presented. (C) Comparison of PTEN mRNA expression between various oncogene-transduced BM cells. Mock, AML1/ETO, E2A/HLF, PML/RARα, or Evi1-transduced BM cells were prepared and were analyzed after 1 week of G418 selection. Error bars indicate SD (n = 4; *P < .01). Results were represented as the averages of 4 independent experiments performed in duplicate. (D) Comparison of PTEN/AKT/mTOR signaling of the indicated cells analyzed by Western blotting. BM cells (1-2 × 106) were used in these assays, and 4 independent experiments were performed and representative figures are shown (n = 2 for each). (E) Rapamycin was added to each oncogene- or mock-transduced BM cells with the indicated concentrations in semisolid medium. The average colony counts were converted to percentages, defining the colony number without rapamycin as the cell viability of 100%. P values were calculated by comparing percentages of viable cells at 0.4nM rapamycin. Error bars indicate SD (n = 8 from 4 independent experiments; *P < .00001). (F) Cell cycle analysis of Evi1- or mock-transduced BM cells with/without addition of rapamycin. Representative fluorescence-activated cell sorting data (left), and average percentage of cells in G2/M phase (right) were shown. Error bars indicate SD (n = 3; *P < .01). (G) A BM smear of Evi1-induced AML, stained with Wright-Giemsa, showed an increase of myeloblasts. Slides were examined by Olympus BH-2 microscope with 40×/0.75 NA oil objective. Picture was taken with Olympus DP20-E camera and analyzed with Adobe Photoshop 7.0. (H) Representative flow cytometric profiles of the BM cells isolated from a recipient of Evi1- or mock-transduced BM cells. The surface marker profiles of Evi1-induced leukemic cells were almost the same. These cells expressed c-kit and Mac-1. Lymphoid markers such as CD3 and B220 were negative except for some deviations in the intensity of B220. In contrast, mock-transduced cells were hardly detected, which suggested that the transplanted cells did not engraft. The numbers in the figure show the percentage of cells gated in each quadrant. (I) Survival of Evi1-induced leukemic mice (top; n = 20 in total, 3 clones were transplanted), or that of TEL/PDGFβR-AML1/ETO-induced leukemic mice and AML1_S291fsX300-induced leukemic mice (bottom; n = 20 in total for each leukemia, 2 clones were transplanted for each) treated with vehicle or rapamycin. DMSO indicates dimethyl sulfoxide; ERK1/2, extracellular signal-regulated kinase 1/2; STAT3, signal transducer and activator of transcription 3; STAT5, signal transducer and activator of transcription 5.
Evi1 represses PTEN protein level and activates downstream AKT/mTOR signaling. (A-B) Analysis of indicated protein levels in Evi1 or mock-transduced BM cells. BM cells (1-2 × 106) were used in these assays. Experiments were repeated for > 2 times, and the representative figures are presented. (C) Comparison of PTEN mRNA expression between various oncogene-transduced BM cells. Mock, AML1/ETO, E2A/HLF, PML/RARα, or Evi1-transduced BM cells were prepared and were analyzed after 1 week of G418 selection. Error bars indicate SD (n = 4; *P < .01). Results were represented as the averages of 4 independent experiments performed in duplicate. (D) Comparison of PTEN/AKT/mTOR signaling of the indicated cells analyzed by Western blotting. BM cells (1-2 × 106) were used in these assays, and 4 independent experiments were performed and representative figures are shown (n = 2 for each). (E) Rapamycin was added to each oncogene- or mock-transduced BM cells with the indicated concentrations in semisolid medium. The average colony counts were converted to percentages, defining the colony number without rapamycin as the cell viability of 100%. P values were calculated by comparing percentages of viable cells at 0.4nM rapamycin. Error bars indicate SD (n = 8 from 4 independent experiments; *P < .00001). (F) Cell cycle analysis of Evi1- or mock-transduced BM cells with/without addition of rapamycin. Representative fluorescence-activated cell sorting data (left), and average percentage of cells in G2/M phase (right) were shown. Error bars indicate SD (n = 3; *P < .01). (G) A BM smear of Evi1-induced AML, stained with Wright-Giemsa, showed an increase of myeloblasts. Slides were examined by Olympus BH-2 microscope with 40×/0.75 NA oil objective. Picture was taken with Olympus DP20-E camera and analyzed with Adobe Photoshop 7.0. (H) Representative flow cytometric profiles of the BM cells isolated from a recipient of Evi1- or mock-transduced BM cells. The surface marker profiles of Evi1-induced leukemic cells were almost the same. These cells expressed c-kit and Mac-1. Lymphoid markers such as CD3 and B220 were negative except for some deviations in the intensity of B220. In contrast, mock-transduced cells were hardly detected, which suggested that the transplanted cells did not engraft. The numbers in the figure show the percentage of cells gated in each quadrant. (I) Survival of Evi1-induced leukemic mice (top; n = 20 in total, 3 clones were transplanted), or that of TEL/PDGFβR-AML1/ETO-induced leukemic mice and AML1_S291fsX300-induced leukemic mice (bottom; n = 20 in total for each leukemia, 2 clones were transplanted for each) treated with vehicle or rapamycin. DMSO indicates dimethyl sulfoxide; ERK1/2, extracellular signal-regulated kinase 1/2; STAT3, signal transducer and activator of transcription 3; STAT5, signal transducer and activator of transcription 5.
We next assessed the effect of rapamycin on colony-forming activity of these oncogene- or mock-transduced BM cells. Evi1-transduced BM cells showed increased sensitivity to rapamycin (half maximal inhibitory concentration < 0.2nM) compared with control cells (half maximal inhibitory concentration > 0.6nM) (Figure 3E). Rapamycin also slightly reduced the colony-forming activity of AML1/ETO-, E2A/HLF-, or PML/RARα-transduced cells, probably through a nonspecific cytotoxic effect. We also treated these cells with phosphatidylinositol 3-kinase (PI3K) inhibitor LY294002 and obtained similar results (supplemental Figure 3A). We performed similar experiments with inhibitors of the nuclear factor-κB pathway (BMS345541) and the Notch pathway (DAPT), but neither of them has significant effects on Evi1-expressing cells compared with AML1/ETO-, E2A/HLF-, or PML/RARα-transduced BM cells (supplemental Figure 3B-C). We also found that Evi1 overexpression promoted cell cycling progression compared with control cells, and the effect was cancelled by adding the mTOR inhibitor rapamycin (Figure 3F) or overexpression of PTEN (supplemental Figure 4).
To evaluate the effect of rapamycin on Evi1-induced leukemia in vivo, we generated Evi1-expressing leukemic mice by BM transplantation. Wild-type BM cells were transduced with the pMYs-Evi1-IRES-GFP (n = 8; ID: 1-8 shown in supplemental Table 3) or an empty vector (n = 11; ID: 9-19), and were injected into sublethally irradiated mice. All of the recipient mice received a transplant with Evi1-expressing cells died with AML within 6-11 months after transplantation, whereas development of leukemia was not observed in control mice. AMLs were characterized by large numbers of blasts in the BM smear (Figure 3G; supplemental Table 3), positivity for myeloid markers of leukemic cells (Figure 3H), and marked splenomegaly (supplemental Table 3). Because some of the clones were positive for B220, we checked IgH gene rearrangements of leukemic cells, and no clonal JH rearrangements were detected in both B220+ and B220− leukemic cells (supplemental Figure 5A). Expression of Evi1 protein was confirmed in these leukemic cells (supplemental Figure 5B). Then isolated leukemic cells (3 clones) were transplanted into sublethally irradiated secondary recipient mice. These mice were treated with daily injections of vehicle (n = 10) or rapamycin (0.4 mg/kg per day; n = 10). Although all of the secondary recipient mice developed AML, rapamycin significantly prolonged the survival of recipient mice compared with vehicle-treated mice (Figure 3I). We also developed murine AML models with the use of TEL/PDGFβR-AML1/ETO and AML1 mutant (AML1_S291fsX300)35,41 (supplemental Figure 5B-C), but rapamycin did not show any effects on the survival of these mice (Figure 3I). These data suggest that the AKT/mTOR pathway has a role in the proliferation and survival of Evi1-expressing leukemic cells both in vitro and in vivo.
PTEN inversely correlates with Evi1 in human leukemia
To address the potential relevance of the findings to human disease, we analyzed gene expression data of leukemic BM cells of 57 cases with human AML. All samples contained 80%-99% blast cells (Table 1 for the patient characteristics). We evaluated Evi1 and PTEN levels by real-time PCR and found an inverse correlation between Evi1 and PTEN levels with statistical significance (Figure 4A). We further examined Evi1 and PTEN expression in BM cells of CML because high Evi1 expression is observed in patients with CML4 (n = 44; Table 2). We found that Evi1 seemed to be activated in CML during a blastic phase (Figure 4B), and we identified a statistically significant inverse relationship between Evi1 and PTEN expression levels again, suggesting that PTEN down-regulation by Evi1 may play a role in the progression of the disease from the chronic to the acute phase.
Clinical and molecular characteristics of the 57 patients with AML
| Characteristics . | Value . |
|---|---|
| Sex, n (%) | |
| Male | 37 (65) |
| Female | 20 (35) |
| Age group, y, n (%) | |
| < 35 | 8 (14) |
| 35-60 | 22 (32) |
| > 60 | 25 (54) |
| Age, y | |
| Median | 57 |
| Range | 16-85 |
| Bone marrow blasts count, % | |
| Median | 87.1 |
| Range | 80-99 |
| Disease status, n (%) | |
| Diagnosis | 40 (70) |
| Relapse 1 | 8 (14) |
| Relapse > 2 or refractory | 9 (16) |
| French-American-British classification, n (%) | |
| M0 | 4 (7) |
| M1 | 12 (21) |
| M2 | 19 (39) |
| M3 | 4 (7) |
| M4 | 5 (9) |
| M5 | 7 (12) |
| M6 | 1 (4) |
| M7 | 0 (0) |
| Not determined | 5 (9) |
| Cytogenetic abnormalities, n (%) | |
| t(8;21) | 8 (14) |
| t(15;17) | 3 (5) |
| inv(16)/t(16;16) | 2 (4) |
| t(11q23) | 3 (5) |
| Complex karyotype (> 3 chromosomal abnormalities) | 3 (5) |
| Other abnormal karyotypes* | 2 (4) |
| Normal karyotype | 23 (40) |
| Not determined/no data | 13 (23) |
| Characteristics . | Value . |
|---|---|
| Sex, n (%) | |
| Male | 37 (65) |
| Female | 20 (35) |
| Age group, y, n (%) | |
| < 35 | 8 (14) |
| 35-60 | 22 (32) |
| > 60 | 25 (54) |
| Age, y | |
| Median | 57 |
| Range | 16-85 |
| Bone marrow blasts count, % | |
| Median | 87.1 |
| Range | 80-99 |
| Disease status, n (%) | |
| Diagnosis | 40 (70) |
| Relapse 1 | 8 (14) |
| Relapse > 2 or refractory | 9 (16) |
| French-American-British classification, n (%) | |
| M0 | 4 (7) |
| M1 | 12 (21) |
| M2 | 19 (39) |
| M3 | 4 (7) |
| M4 | 5 (9) |
| M5 | 7 (12) |
| M6 | 1 (4) |
| M7 | 0 (0) |
| Not determined | 5 (9) |
| Cytogenetic abnormalities, n (%) | |
| t(8;21) | 8 (14) |
| t(15;17) | 3 (5) |
| inv(16)/t(16;16) | 2 (4) |
| t(11q23) | 3 (5) |
| Complex karyotype (> 3 chromosomal abnormalities) | 3 (5) |
| Other abnormal karyotypes* | 2 (4) |
| Normal karyotype | 23 (40) |
| Not determined/no data | 13 (23) |
Patients with 3q26 abnormalities are not included in this study.
PTEN expression is inversely correlated with Evi1 expression in human leukemia. (A) Correlation between Evi1 and PTEN mRNA expressions in AML (n = 57; Pearson coefficient = −0.339 [P = .0097], Spearman coefficient = −0.298 [P = .024]). The same samples of patients 1-5 were subjected to ChIP analysis (Figure 5G). (B) Correlation between Evi1 and PTEN mRNA expressions in CML with disease status of each patient are shown (n = 44; Pearson coefficient = −0.347 [P = .020], Spearman coefficient = −0.368 [P = .013]).
PTEN expression is inversely correlated with Evi1 expression in human leukemia. (A) Correlation between Evi1 and PTEN mRNA expressions in AML (n = 57; Pearson coefficient = −0.339 [P = .0097], Spearman coefficient = −0.298 [P = .024]). The same samples of patients 1-5 were subjected to ChIP analysis (Figure 5G). (B) Correlation between Evi1 and PTEN mRNA expressions in CML with disease status of each patient are shown (n = 44; Pearson coefficient = −0.347 [P = .020], Spearman coefficient = −0.368 [P = .013]).
Clinical and molecular characteristics of the 44 patients with CML
| Characteristics . | Value . |
|---|---|
| Sex, n (%) | |
| Male | 34 (77) |
| Female | 10 (23) |
| Age group, y, n (%) | |
| < 35 | 9 (21) |
| 35-60 | 27 (61) |
| > 60 | 8 (18) |
| Age, y | |
| Median | 47 |
| Range | 17-78 |
| Bone marrow blasts count, % | |
| Median | 3.6 |
| Range | 0.5-96 |
| Disease status, n (%) | |
| Chronic phase | 14 (32) |
| Accelerated phase | 4 (9) |
| Blastic crisis | 26 (59) |
| Characteristics . | Value . |
|---|---|
| Sex, n (%) | |
| Male | 34 (77) |
| Female | 10 (23) |
| Age group, y, n (%) | |
| < 35 | 9 (21) |
| 35-60 | 27 (61) |
| > 60 | 8 (18) |
| Age, y | |
| Median | 47 |
| Range | 17-78 |
| Bone marrow blasts count, % | |
| Median | 3.6 |
| Range | 0.5-96 |
| Disease status, n (%) | |
| Chronic phase | 14 (32) |
| Accelerated phase | 4 (9) |
| Blastic crisis | 26 (59) |
Patients with 3q26 abnormalities are not included in this study.
These expression analyses indicate that the inverse correlation between Evi1 and PTEN observed in murine models is recapitulated in human AML and CML.
Evi1 interacts with PcG proteins to repress PTEN
It has been shown that Evi1 recruits several histone methyltransferases (HMTs) for regulation of gene transcription, such as SUV39H1 and G9a.20-22 EZH2 is another HMT which is a core component of PRC2/3/4 and imparts methyltransferase activity to the complexes. We therefore investigated whether HMTs are actively involved in Evi1-mediated PTEN repression. By the reporter assays that used Jurkat cells, we evaluated the effect of Evi1 on PTEN4_wt promoter in the presence of SUV39H1, G9a, EZH2, and their catalytically inactive mutants. Although SUV39H1 and G9a showed little to no effect on the PTEN repression (Figure 5A-B), Evi1 and EZH2 repressed PTEN promoter activity in a synergistic manner (Figure 5C). Moreover, EZH2-H689A, a construct carrying an inactivating point mutation within the HMT domain of EZH2, completely abolished the transcriptional repression mediated by Evi1. Similar results were obtained with THP-1 and COS7 cells (supplemental Figure 1E and F, respectively). These data suggest that Evi1 requires EZH2 for PTEN down-regulation.
Evi1 recruits polycomb complexes to repress PTEN. (A) Reporter assays that used Jurkat cells and PTEN4_wt promoter in the presence of Evi1, G9a, or dominant-negative (DN) G9a (n = 4). Error bars indicate SD; *P < .05. (B) Reporter assays that used Jurkat cells and PTEN4_wt promoter in the presence of EVi1, SUV39H1, or DN-SUV39H1 (n = 4). Error bars indicate SD; *P < .05. (C) Reporter assays that used Jurkat cells and PTEN4_wt promoter in the presence of Evi1, EZH2, or EZH2-H689 (n = 6). Error bars indicate SD; *P < .05, **P < .01, and ***P < .001. (D-E) shRNAs (shEZH2-A, -B, -C, -D or control) were retrovirally delivered to Evi1-transduced BM cells, and the efficacy of these constructs and the effects on the PTEN/AKT/mTOR signaling were examined by quantitative real-time PCR (D; n = 3) and immunoblotting (E; experiments were performed twice and the representative figures are presented). BM cells (1-2 × 106) were used for Western blotting. Error bars indicate SD; *P < .05. (F) ChIP assays for PTEN promoter region as shown in Figure 2F using primers 3 (Figure 1D) (n = 3). Error bars indicate SD; *P < .05, **P < .01, ***P < .001, and ****P < .0001. (G) ChIP assays for PTEN promoter region that used human AML blasts (n = 5). Primers used in these assays amplify sequences, including a putative Evi1 binding site (5′-AGAAGATAA-3′ fragment at position 1875/1884 downstream of the initiation codon ATG [+1] of human PTEN). P value was calculated by comparing variables of patients with low-intermediate Evi1 expression (n = 3) and patients with high Evi1 expression (n = 2).
Evi1 recruits polycomb complexes to repress PTEN. (A) Reporter assays that used Jurkat cells and PTEN4_wt promoter in the presence of Evi1, G9a, or dominant-negative (DN) G9a (n = 4). Error bars indicate SD; *P < .05. (B) Reporter assays that used Jurkat cells and PTEN4_wt promoter in the presence of EVi1, SUV39H1, or DN-SUV39H1 (n = 4). Error bars indicate SD; *P < .05. (C) Reporter assays that used Jurkat cells and PTEN4_wt promoter in the presence of Evi1, EZH2, or EZH2-H689 (n = 6). Error bars indicate SD; *P < .05, **P < .01, and ***P < .001. (D-E) shRNAs (shEZH2-A, -B, -C, -D or control) were retrovirally delivered to Evi1-transduced BM cells, and the efficacy of these constructs and the effects on the PTEN/AKT/mTOR signaling were examined by quantitative real-time PCR (D; n = 3) and immunoblotting (E; experiments were performed twice and the representative figures are presented). BM cells (1-2 × 106) were used for Western blotting. Error bars indicate SD; *P < .05. (F) ChIP assays for PTEN promoter region as shown in Figure 2F using primers 3 (Figure 1D) (n = 3). Error bars indicate SD; *P < .05, **P < .01, ***P < .001, and ****P < .0001. (G) ChIP assays for PTEN promoter region that used human AML blasts (n = 5). Primers used in these assays amplify sequences, including a putative Evi1 binding site (5′-AGAAGATAA-3′ fragment at position 1875/1884 downstream of the initiation codon ATG [+1] of human PTEN). P value was calculated by comparing variables of patients with low-intermediate Evi1 expression (n = 3) and patients with high Evi1 expression (n = 2).
To assess the genetic requirement of EZH2 for Evi1-mediated PTEN regulation in BM cells, we designed 4 independent shRNAs targeting murine EZH2 (shEZH2-A, -B, -C, and -D) and transduced them into Evi1-expressing BM cells. shEZH2-A, -B, and -C strongly reduced EZH2 expression compared with control shRNA, whereas shEZH2-D could not (Figure 5E). PTEN expression was significantly higher in EZH2-knockdown cells than in control cells at both mRNA and protein levels (Figure 5D-E). Furthermore, EZH2 knockdown resulted in decreased AKT/mTOR phosphorylation (Figure 5E). Thus, EZH2 is genetically required for PTEN down-regulation and subsequent AKT/mTOR activation by Evi1. In contrast, PTEN expression was not changed when these shRNAs were transduced into whole mononuclear BM cells, in which Evi1 expression is low (supplemental Figure 6A-B). Furthermore, overexpression of EZH2 in fluorouracil-primed BM cells did not repress Evi1 mRNA and protein levels (supplemental Figure 6C-D). These results support the idea that EZH2 or polycomb complex is recruited to the PTEN promoter and epigenetically induces repressive chromatin modifications only in cells with high Evi1 expression.
We then performed ChIP assays with the use of murine BM cells transduced with Flag-Evi1, Flag-Evi1_ΔZF1-7, or mock. Evi1 and EZH2 are both enriched in the PTEN promoter only in Evi1-expressing cells (Figure 5F). In addition, strong enrichment of SUZ12, another component of PRC2/3/4, was observed in the same genomic region. Consistent with the binding of PRC2/3/4, H3K27me3 was significantly enriched. Interestingly, BMI1, a component of PRC1, was also detected. Furthermore, we found a decrease in trimethylation of histone H3 lysine 4 and H3 acetylation marks, indicating the repressive epigenetic modification in this genomic region in Evi1-expressing cells. In contrast, we observed no epigenetic modification in other genomic regions, or when we used an unrelated antibody (anti-HA) (supplemental Figure 7A-D). In Evi1_ΔZF1-7–expressing cells, we observed no enrichment of EZH2, SUZ12, BMI1, or H3K27me3 in the PTEN promoter, indicating that ZF1-7 is a central domain for Evi1-mediated PcG recruitment.
To further address the clinical relevance of PcG proteins to PTEN down-regulation, we performed ChIP assays with additionally available samples derived from patients with AML (patients 1-5 indicated in Figure 4A). Primers for human PTEN promoter were designed to amplify fragments that correspond to the murine PTEN promoter regions depicted in Figure 1D. In leukemic cells with high Evi1 expression (patients 4 and 5), Evi1, EZH2, SUZ12, BMI1, and H3K27me3 tended to be enriched in the PTEN promoter (Figure 5G). However, H3K4me3 and H3 acetylation marks had a tendency to be reduced in these cells. Anti-Flag antibody was used as a negative control. These results indicate that Evi1 recruits both PRC2/3/4 and PRC1 to the PTEN promoter region and induces histone modification to repress PTEN transcription in leukemic cells with highly expressed Evi1.
These data prompted us to test whether Evi1 physically interacts with PcG proteins. We introduced HA-tagged Evi1 and components of polycomb complex (Flag-tagged EZH2, Myc-tagged SUZ12, or Flag-tagged EED) into 293T cells. Immunoblot analysis of anti–HA-Evi1 immunoprecipitates showed the existence of Flag-EZH2, Myc-SUZ12, and Flag-EED in a complex with Evi1 protein, under stringent washing condition (500mM NaCl) (Figure 6A, B, and C, respectively). Identical results were obtained by the reciprocal coimmunoprecipitation experiments (supplemental Figure 8A-C). To investigate the existence of the interactions between Evi1 and PcG proteins in leukemic cells, we performed coimmunoprecipitation assays with the use of leukemic cells derived from Evi1-induced leukemia mice and found that Evi1 interacts with PRC2/3/4 proteins in these leukemic cells (Figure 6D). Importantly, similar results were obtained with the use of human leukemia cells derived from the patient with AML with high Evi1 expression (Figure 6E). We next assessed the interaction between Evi1 and PRC1 complexes and found that Evi1 interacts with BMI1, RING1, RING2, and HPH2 (Figure 6F-I; supplemental Figure 9A-D), but immunoglobulin does not interact with HPH1 or HPC proteins (CBX2, CBX4, CBX6, CBX7, and CBX8) (data not shown). In addition, domain-mapping experiments showed that ZF1-7 of Evi1 is responsible for the interaction with EZH2, EED, and BMI1 (Figure 6J; supplemental Figure 10A and B, respectively), although the interaction between Evi1 and SUZ12 is mediated through multiple regions (supplemental Figure 10C). By more detailed domain-mapping experiments, we found that Evi1 binds to EZH2 via its ZF1 domain (supplemental Figure 10D). Evi1_ΔZF1 can bind to DNA but fails to interact with EZH2, whereas Evi1_R205N can interact with EZH2 but does not bind to DNA (supplemental Figure 10D). These results suggest that complex formation of Evi1 and EZH2 is primarily mediated by protein-protein interaction, rather than by DNA, although we cannot exclude potential participation of DNA. Considering that neither of these mutants efficiently represses PTEN transcription, both binding to DNA and interaction with EZH2 are required for Evi1 to repress PTEN expression.
Evi1 interacts with PcG proteins. (A-C) Immunoprecipitation of HA-Evi1 identified EZH2 (A), SUZ12 (B), and EED (C) as interacting proteins in 293T cells. Vertical lines have been inserted to indicate a repositioned gel lane in panel A. (D) Interaction between Evi1 and endogenous PRC2/3/4 proteins in Evi1-induced murine leukemic cells. (E) Interactions between endogenous Evi1 and PRC2/3/4 in human leukemia cells derived from the patient with AML patient. (F-I) Immunoprecipitation of HA-Evi1 identified BMI1 (F), RING1 (G), RING2 (H), and HPH2 (I) as interacting proteins in 293T cells. A vertical line has been inserted to indicate a repositioned gel lane in panel I. (J) ZF1-7 domain of Evi1 is responsible for the physical interaction with EZH2. IgG indicates immunoglobulin G; WB, Western blotting.
Evi1 interacts with PcG proteins. (A-C) Immunoprecipitation of HA-Evi1 identified EZH2 (A), SUZ12 (B), and EED (C) as interacting proteins in 293T cells. Vertical lines have been inserted to indicate a repositioned gel lane in panel A. (D) Interaction between Evi1 and endogenous PRC2/3/4 proteins in Evi1-induced murine leukemic cells. (E) Interactions between endogenous Evi1 and PRC2/3/4 in human leukemia cells derived from the patient with AML patient. (F-I) Immunoprecipitation of HA-Evi1 identified BMI1 (F), RING1 (G), RING2 (H), and HPH2 (I) as interacting proteins in 293T cells. A vertical line has been inserted to indicate a repositioned gel lane in panel I. (J) ZF1-7 domain of Evi1 is responsible for the physical interaction with EZH2. IgG indicates immunoglobulin G; WB, Western blotting.
We further examined whether Evi1 and PcG proteins could colocalize in cells with the use of immunofluorescence analysis, and we found that Evi1 and each PcG protein formed speckles that partially overlap (yellow) in nuclei, confirming the association between the 2 proteins in vivo (supplemental Figure 11A-G).
Myeloid-transforming activity of Evi1 depends on PcG proteins
Finally, we evaluated a role for polycomb complexes in Evi-1–induced myeloid transformation. Evi1- or E2A/HLF-mediated transformed BM cells from the third to fourth round of in vitro replating were infected with retrovirus encoding shRNAs and replated in each dish. Efficient knockdown of EZH2 mediated by shEZH2-A, -B, and -C in Evi1–transduced BM cells significantly reduced their colony-forming activity (Figure 7A). In contrast, knockdown of EZH2 did not impair BM transformation by E2A/HLF. We used E2A/HLF as a control because the contribution of Evi1 to colony-forming activity is relatively small in E2A/HLF-transformed cells.1 We also designed 4 independent shRNAs for murine SUZ12 and EED, respectively. As shown in Figure 7B, shSUZ12-B, -C, -D and shEED-B, -C, -D strongly reduced the corresponding proteins, whereas other shRNAs showed little effect. Again, we found a significant decrease in colony-forming activity of Evi1-expressing BM cells when we used effective shRNAs for SUZ12 or EED (Figure 7C). We also confirmed that down-regulation of SUZ12 or EED did not affect the colony formation of E2A/HLF-transduced cells. Thus, major components of PRC2/3/4 are specifically required for maintenance of transformation mediated by Evi1.
PcG proteins are required for the serial replating capacity of Evi1-transduced BM cells. (A,C) Serial replating capacity of Evi1-expressing cells was markedly reduced with EZH2 (A), SUZ12, or EED (C) shRNAs (n = 8 from 4 independent experiments. Error bars indicate SD; *P < .01 and **P < .001; P values were calculated by comparisons with the colony numbers of control cells at the second replating. (B) RNAi of SUZ12 and EED. (D) A scheme showing that Evi1 recruits polycomb complexes to the PTEN promoter and represses PTEN transcription through H3K27me3-mediated chromatin remodeling, which, in turn, activates downstream AKT/mTOR pathway.
PcG proteins are required for the serial replating capacity of Evi1-transduced BM cells. (A,C) Serial replating capacity of Evi1-expressing cells was markedly reduced with EZH2 (A), SUZ12, or EED (C) shRNAs (n = 8 from 4 independent experiments. Error bars indicate SD; *P < .01 and **P < .001; P values were calculated by comparisons with the colony numbers of control cells at the second replating. (B) RNAi of SUZ12 and EED. (D) A scheme showing that Evi1 recruits polycomb complexes to the PTEN promoter and represses PTEN transcription through H3K27me3-mediated chromatin remodeling, which, in turn, activates downstream AKT/mTOR pathway.
Discussion
It is well known that many of the critical regulators of leukemic transformation are transcription factors. Among them is Evi1, and intense attention has been focused on the molecular mechanisms underlying Evi1-mediated transcriptional regulation. The present study showed several important aspects of Evi1-related leukemia.
First, the dependency of Evi1-expressing leukemic cells on AKT/mTOR signaling provides a potential therapeutic target in a genetically distinctive subset of poor-prognosis leukemia defined by high Evi1 expression. Our study clearly showed that inhibition of the AKT/mTOR pathway antagonizes the leukemogenic properties of Evi1-expressing leukemic cells in vitro and in vivo. Thus, rapamycin or other inhibitors of the PI3K/AKT/mTOR signaling will have a therapeutic effect on Evi1-related leukemia. Importantly, it is the first example of targeted therapeutic modalities that suppress the leukemogenic activity of Evi1. Evi1 has a variety of oncogenic potentials, which is probably one of the reasons that rapamycin did not cure the diseased mice in our experiments. However, this will be the first step to overcome Evi1-related leukemia, and the possibility of combination therapy that targets several functions of Evi1 should be tested in the future.
Second, our results strongly suggest that activated Evi1 induces epigenetic regulation on PTEN transcription. Although several studies have shown that the PI3K/AKT pathway is often deregulated in AML,42 underlying mechanisms of such deregulation are unclear. Some RAS mutations, PTEN mutations, or PTEN phosphorylation can result in AKT activation, but the importance of PTEN in causing AKT activation in AML has remained undetermined.42 Because histone modifiers or, in particular, PcG proteins that epigenetically regulate PTEN transcription have not been reported in hematologic malignancies, our model will shed light on a new mechanisms of AKT activation in a genetically defined AML subgroup, which provides a molecular basis for specific treatment (Figure 7D).
Third, note that Evi1 interacts with both PRC2/3/4 and PRC1. The ability of Evi1 to interact with various PcG proteins suggests that Evi1 acts as an anchor for polycomb complexes to DNA to organize gene transcription (Figure 7D). Epigenetic changes mediated by PRC2/3/4 make attractive therapeutic targets because they are potentially reversible processes. In addition, Evi1 also interacts with PRC1 and seems to recruit PRC1 to the PTEN promoter. Recently, Boukarabila et al43 reported that PLZF/RARα fusion protein interacts with both PRC2/3/4 and PRC1, and the recruitment of PRC1 leads to a deep influence on the maintenance of target gene repression. Therefore, PTEN repression by Evi1 may be firmly maintained by the recruitment of PRC1, and the interactions between Evi1 and PRC1 will be another therapeutic target.
Fourth, PTEN is the first described repressive target of Evi1. Because Evi1 rarely changes transcription levels of genes by ≥ 1.4-fold as shown in our previous gene expression analysis that used Evi1 conditional knockout mice,1 it is essential to detect subtle changes of gene expression. Several studies have shown the crucial function of PTEN in multiple cellular processes and its involvement in human diseases, which suggests that PTEN needs to be deliberately regulated, and subtle changes in PTEN expression levels have profound effects on tumorigenesis.44-48 Therefore, we performed an extensive promoter analysis of PTEN. Furthermore, primary BM cells seem appropriate for detecting Evi1 targets in the hematopoietic system because transcriptional regulation by Evi1 is highly context dependent. In fact, we did not find the effect of Evi1 on PTEN in other cell lines, such as 32D, HEK293T, or NIH3T3 cells (data not shown). Recently, Song et al49 have reported that BMI1 induces epithelial-mesenchymal transition partially through transcriptional repression of PTEN in nasopharyngeal epithelial cells, although it seems yet to be determined whether PTEN is a universal target for BMI1.49 Our observation in hematopoietic system clearly indicates that BMI1 and other PcG proteins are recruited to the PTEN promoter only in cells with high expression of Evi1 but not in cells with low Evi1 expression. These results show a critical role for Evi1 in anchoring PcG proteins to the PTEN promoter (Figure 7D). Meanwhile, no anchor protein like Evi1 has been identified in BMI1-mediated PTEN regulation in nasopharyngeal epithelial cells or other cells. Because the activated PI3K/PTEN/AKT pathway is well documented for many types of human malignancies and is also associated with an aggressive phenotype,50 further investigations are warranted for elucidating the epigenetic regulation of PTEN/AKT signaling in various types of cancer cells.
Fifth, we established a novel murine model of Evi1-induced leukemia, which will be a valuable tool for analyzing a mechanistic basis or drug sensitivity of human leukemia with elevated Evi1 expression. Previous studies reported that Evi1 overexpression induced myelodysplastic syndrome in murine BM transplantation assays,14,36 and most of the mice died of severe anemia. In contrast, all of the mice that received a transplant in our study died of AML. This discrepancy may derive from the high efficiency of gene transfer into BM cells in our experiments (Figure 1A) or from the difference in virus vectors used in these assays, although other possibilities may exist. Furthermore, the relatively long latency of leukemia development in this mouse model suggests that other genetic events are required for the onset of full-blown leukemia.
We previously showed that Evi1 is essential for proliferation of HSCs and myeloid leukemia cells. It was also shown that inactivation of PTEN in HSCs causes their short-term expansion, but long-term decline, primarily because of an enhanced level of HSC activation.27,28 Moreover, we observed that PTEN expression was higher in Evi1-deficient HSCs than in wild-type HSCs. Therefore, it is tempting to speculate that the modest down-regulation of PTEN by Evi1 results in HSC expansion without inducing its exhaustion. In addition, given that PTEN is also known as a key regulator in leukemic stem cells, activation of Evi1 may contribute to leukemic stem cell generation through PTEN down-regulation. Thus, the role of Evi1-PTEN pathway in normal and malignant stem cells should be clarified in the future.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank D. Allis, D. Reinberg, Y. Shi, R. Shiekhattar, P. Freemont, M. Bollen, A. Eynde, H. Wang, D. Wotton, P. Farnham, M. Tomasson, K. Yamamoto, T. Kim, D. Rotin, X. Wang, T. Nakamura, T. Inaba, I. Kitabayashi, F. Fuks, and W. Cavenee for providing essential plasmids; S. Ogawa for technical supports of the microarray experiment; Y. Shimamura for expert technical assistance; Kyowa Hakko Kirin Co Ltd for cytokines.
This study was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology (KAKENHI 19679004).
Authorship
Contribution: A.Y., S.G., and M.K. designed the study; A.Y. performed most of the experiments and wrote the paper with S.G. and M.K.; A.Y. and N.W.-O. performed animal studies; A.Y., Y.N., and M.N. performed microarray analyses; S.G., Y.Y., E.N., S.A., T.S., and M.S. participated in plasmid/protein preparation; M.N. and Y.I. provided technical advice and support; T.K. supervised the animal studies and assisted with manuscript preparation; and M.K. supervised all of the experiments and data interpretation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mineo Kurokawa, Department of Hematology and Oncology, Graduate School of Medicine, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8655, Japan; e-mail: kurokawa-tky@umin.ac.jp.

![Figure 1. Evi1 down-regulates PTEN expression in BM cells. (A) Schematic representation of gene expression analysis. Evi1-GFP– or GFP-transduced BM cells (n = 4 for each) were sorted and subjected to gene expression analysis. Representative fluorescence-activated cell sorting data show the gene transfer efficiencies of 78% and 90% for Evi1-GFP and GFP-expressing retrovirus, respectively. (B) Real-time PCR for PTEN expression. Error bars indicate SD (n = 6; *P < .0001). (C) PTEN mRNA expression in lineage−, c-Kit+, Sca-1+ cells derived from Mx-Cre Evi1flox/flox mice. Error bars indicate SD (n = 3; *P < .05). Evi1 was retrovirally added back. The Cre-mediated Evi1 deletion and the expression of wild-type Evi1 were evaluated by PCR with the use of genome DNA and cDNA, respectively, and representative figures are shown. Detailed experimental methods are shown in supplemental Data. (D) Schematic representation of mouse PTEN promoter region, possible Evi1 binding site predicted by rVISTA 2.0 (http://rvista.dcode.org/), constructs cloned into pGL4.10[Luc2], mutagenesis strategy, probe sets used for EMSAs, and primers for ChIP assays. (E) Relative luciferase activity of Evi1 on each PTEN promoter. Error bars indicate SD (n = 6; *P < .01). Jurkat cells were used. Error bars indicate SD. (F) Protein expression of Flag-tagged wild-type (WT) Evi1 and Evi1_R205N used for EMSAs. Western blot analysis was done with anti-Flag antibody. (G) Results of EMSAs are shown. Probe sets 3 or 3_mut are indicated in Figure 1D. Corresponding cold-specific competitors or nonspecific competitors were added as indicated. KO indicates knockout.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/13/10.1182_blood-2009-12-261602/4/m_zh89991168490001.jpeg?Expires=1767709576&Signature=DVaLodaicqf8dIdb3XdplR20KO~ihbBX5GIeE3VbRwy~bQOcdwTsVth5zb2Q5C1i8ofI-ZbxwI09NKi91dPJdgNofKov9qFXx3rUCMoE3-2lcDkk-Npk3p6kQJIxqYZFfezleVxGhr6jzt9Rg3ExOofbP8LduKEy3Tj6TmXjcgpM9lZYlHELc3jncmwETFlTzBFRn5AOZn7fJZ1NHRJzYybhuGjj2JJUyzCFxyr3cEMO~lFm-rn7omdKfDBSJxfk8B6FiFCQoJnj-y-wAxdBZmjFuJQxaQHo6w6FhLPU2YFzaA-0I5O8t-fUdTLJlc55ueEncPn3wcXC9QEhpJYZTA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 4. PTEN expression is inversely correlated with Evi1 expression in human leukemia. (A) Correlation between Evi1 and PTEN mRNA expressions in AML (n = 57; Pearson coefficient = −0.339 [P = .0097], Spearman coefficient = −0.298 [P = .024]). The same samples of patients 1-5 were subjected to ChIP analysis (Figure 5G). (B) Correlation between Evi1 and PTEN mRNA expressions in CML with disease status of each patient are shown (n = 44; Pearson coefficient = −0.347 [P = .020], Spearman coefficient = −0.368 [P = .013]).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/13/10.1182_blood-2009-12-261602/4/m_zh89991168490004.jpeg?Expires=1767709576&Signature=Y86eY7ap3CNvnNjv~mFkVJtiWLePAxYdvy9xsOCVZnHSKOdQRRQm9FbdjTkq2uFHmXZRmgy~1K7lhDSqtFrPOPsNlyeLbxxzcJ9hlE9YPjCJhFZvwLaqfSuIqkixvRCihoiOzAAydMbwWbDdVYF7-YholEmgknRtKSd8m4cue-J6OCo~sZw5rthm2BLbW7ynFNh3um5f0bpZxK5RhC5VDSklgsA8hdrEnL1JPtyal1GoW7VeECWHPGL~oueh856IgwwP00MwfhErmEMPF8qyp-ykSG-o1UBkmYPZAIcwvrCDobGCjgnkzN3Lrpy4n10pxrCbtBlij8GycdeFw~BqVQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Evi1 recruits polycomb complexes to repress PTEN. (A) Reporter assays that used Jurkat cells and PTEN4_wt promoter in the presence of Evi1, G9a, or dominant-negative (DN) G9a (n = 4). Error bars indicate SD; *P < .05. (B) Reporter assays that used Jurkat cells and PTEN4_wt promoter in the presence of EVi1, SUV39H1, or DN-SUV39H1 (n = 4). Error bars indicate SD; *P < .05. (C) Reporter assays that used Jurkat cells and PTEN4_wt promoter in the presence of Evi1, EZH2, or EZH2-H689 (n = 6). Error bars indicate SD; *P < .05, **P < .01, and ***P < .001. (D-E) shRNAs (shEZH2-A, -B, -C, -D or control) were retrovirally delivered to Evi1-transduced BM cells, and the efficacy of these constructs and the effects on the PTEN/AKT/mTOR signaling were examined by quantitative real-time PCR (D; n = 3) and immunoblotting (E; experiments were performed twice and the representative figures are presented). BM cells (1-2 × 106) were used for Western blotting. Error bars indicate SD; *P < .05. (F) ChIP assays for PTEN promoter region as shown in Figure 2F using primers 3 (Figure 1D) (n = 3). Error bars indicate SD; *P < .05, **P < .01, ***P < .001, and ****P < .0001. (G) ChIP assays for PTEN promoter region that used human AML blasts (n = 5). Primers used in these assays amplify sequences, including a putative Evi1 binding site (5′-AGAAGATAA-3′ fragment at position 1875/1884 downstream of the initiation codon ATG [+1] of human PTEN). P value was calculated by comparing variables of patients with low-intermediate Evi1 expression (n = 3) and patients with high Evi1 expression (n = 2).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/13/10.1182_blood-2009-12-261602/4/m_zh89991168490005.jpeg?Expires=1767709576&Signature=PrZVpuQQrSoMjrxM4bFlD0MjTRFPwM8w34irPXE-kb~WYqINU5tr2LWMbqBdRpp~e5VUCRXFhTsrq-4YfcQzcLy1A0lCxOLKmxhPUTp8hCAdURJROxLDQZZK1B-YVC1YycLX05jucs9FJE-3FCyPT1I9dndVjdlD1Gv90aohVZVzQUeP6POSMkYCgTup7u2TN8zsbTxIYjEDR~KXMnz-ppWwwGk0EaupZUMPuHtK~EiZ4wyR2O-rjgnv8OeDGDgEjHjcKTT7mWh7WnSmrvsBY~yBL0rmev2OJX0k-nAE97FcF1708Z9FKev8~NLVqWgyWUkMa1VQmH9c2j-D1pPgGw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal