Abstract
RUNX1/ETO, the fusion protein resulting from the chromosomal translocation t(8;21), is one of the most frequent translocation products in acute myeloid leukemia. Several in vitro and in vivo studies have shown that the homo-tetramerization domain of ETO, the nervy homology region 2 (NHR2), is essential for RUNX1/ETO oncogenic activity. We analyzed the energetic contribution of individual amino acids within the NHR2 to RUNX1/ETO dimer-tetramer transition and found a clustered area of 5 distinct amino acids with strong contribution to the stability of tetramers. Substitution of these amino acids abolishes tetramer formation without affecting dimer formation. Similar to RUNX1/ETO monomers, dimers failed to bind efficiently to DNA and to alter expression of RUNX1-dependent genes. RUNX1/ETO dimers do not block myeloid differentiation, are unable to enhance the self-renewal capacity of hematopoietic progenitors, and fail to induce leukemia in a murine transplantation model. Our data reveal the existence of an essential structural motif (hot spot) at the NHR2 dimer-tetramer interface, suitable for a molecular intervention in t(8;21) leukemias.
Introduction
Chromosomal translocations are frequent events during malignant cell transformation, particularly during leukemogenesis.1 The translocation t(8;21), one of the most frequent chromosomal anomalies in acute myeloid leukemia (AML), involves the RUNX1 gene (also known as AML1, CBFα2, or PEBP2αB) on chromosome 21 and the ETO gene (also known as MTG8 or RUNX1T1) on chromosome 8. The ubiquitously expressed RUNX1 is a transcription factor and belongs to the key regulators of hematopoietic cell differentiation.2 The fusion protein RUNX1/ETO contains the DNA-binding domain (Runt, RHD) of the RUNX1 transcription factor but lacks the C-terminal transactivation sequence that is replaced by almost the entire ETO protein. 2 forms of RUNX1/ETO coexist in AML-leukemia samples: the originally discovered full-length RUNX1/ETO and a splice variant called RUNX1/ETO9a, which lacks 178 amino acids at the C-terminus. Only RUNX1/ETO9a does not require cooperative events for inducing leukemia development in mice.3,4
We and others have shown that RUNX1/ETO has a modular structure. Besides the Runt domain, RUNX1/ETO contains 4 functional domains, which are generally referred to as nervy homology region (NHR1 to NHR4). The NHR domains serve as docking interface for a variety of different proteins, including the E-protein HEB,5,6 the apoptosis-related protein SON,7 and nuclear corepressor proteins, such as N-CoR, SMRT, mSIN3A, and MTGR1, as well as histone deacetylases (HDACs).8-11
In addition, the NHR2 domain mediates tetramer formation through hydrophobic and ionic/polar interactions. Two α-helices align in a head-to-tail fashion to form an antiparallel dimer. Two dimers subsequently are positioned on top of each other in a sandwich-like fashion. The total interaction area composing all contact points of the 4 α-helices is approximately 10 000 Å.2 Substitution of 7 leucines within the NHR2 sequence by arginine or glutamate was reported to disrupt the tetramer into 4 functionally inactive RUNX1/ETO monomers.12 Similarly, destabilization of the RUNX1/ETO complex by intercalating polypeptides or deletion of the NHR2 domain fully abrogates the oncogenic properties of RUNX1/ETO, arguing for a central role of NHR2-mediated oligomerization in RUNX1/ETO-driven leukemogenesis.12-14 These studies, however, did not study the leukemogenic properties of RUNX1/ETO dimers.
As several reports have shown that RUNX1/ETO activity is required for both the onset and the maintenance of the leukemic phenotype,15,16 interference with RUNX1/ETO tetramer formation represents an attractive strategy for a molecular intervention. However, the large interaction area ascribed to the oligomerization domain makes this approach rather demanding. Increasing evidence suggests that the stability of protein-protein interfaces can depend on critical amino acids (hot spots), which contribute to a large fraction of the binding energy at a particular interface and are often surrounded by energetically less important residues.17-19 Consequently, disruptors or inhibitors of protein-protein interaction do not necessarily need to target the entire interacting surface but rather could be designed to address only those residues located at the hot spots.
The intention of our study was to identify amino acids that disrupt RUNX1/ETO tetramers into 2 dimers to analyze whether this would also abrogate RUNX1/ETO function. Therefore, we used computer-based molecular modeling to evaluate the distribution of the binding free energy along the surface of the NHR2 dimer. Five amino acids were found to form a hot spot at the NHR2 dimer-tetramer interface, which contributed largely to NHR2 tetramer formation and stability. Mutation of these critical amino acids blocks dimer-tetramer transition and abrogates RUNX1/ETO function, including induction of leukemia in vivo.
Methods
MM-GB/SA calculations and free energy decomposition
Molecular dynamics simulations to generate snapshots required for the Molecular Mechanics Generalized Born Surface Area (MM-GB/SA) calculations were performed with the AMBER 8 suite of programs20 together with the force-field as described by Cornell et al21 using modifications suggested by Simmerling et al.22 Further details are provided in the supplemental data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Expression constructs and site-directed mutagenesis
For transient and stable expression RUNX1/ETO, mutants and deletion constructs thereof were cloned into the retroviral vector MSCV. Amino acid substitutions were generated by standard methodologies. The expression plasmid pMigR1-REtr was kindly provided by Dong-Er Zhang (Scripps Research Institute, La Jolla, CA). The composition of all constructs was confirmed by DNA sequencing.
Size exclusion chromatography and CD spectroscopy
Cellular extracts from 293T cells transfected with RUNX1/ETO expression plasmids were fractionated by size-exclusion chromatography and analyzed by Western blotting as described before.10 Circular dichroism (CD) spectroscopy of the 95 amino acids long NHR2 recombinant proteins was performed using a Jasco CD spectrometer. Further details are given in the supplemental data.
Sedimentation velocity measurements
Sedimentation velocity experiments were conducted with 300-μL samples at protein concentrations of 15 or 30μM on an Optima XL-A centrifuge (Beckman Coulter). Data were collected at a wavelength of 280 nm for the wild-type (WT) NHR2 or at 230 nm for the NHR2 mutant m5 and analyzed with SEDFIT and SEDPHAT.23,24 Further details are given in the supplemental data.
ABCD assay and EMSA
5′-Biotinylated oligonucleotides corresponding to the RUNX1 binding sequences within RUNX3 and PU.1 were used for the avidin-biotin complex DNA-binding (ABCD) assay. For electrophoretic mobility shift assay (EMSA) assays, 293T cells were transfected with vectors for RUNX1/ETO or RUNX1/ETO-m5. Further details are given in the supplemental data.
Cells, cell culture, and retroviral transduction
The 293T cells were maintained in DMEM (Invitrogen). The cell lines U937 and K562 were cultured in RPMI 1640 containing 10% fetal calf serum (Invitrogen). Murine lin-depleted progenitor cells were maintained in RPMI supplemented with 10% fetal calf serum, 10 ng/μL mIL-3, 50 ng/μL mIL-6, and 50 ng/μL murine stem cell factor. Retroviral transduction was performed as described before.25 Stable transduced cell lines were purified by cell sorting (FACSAria, BD Biosciences PharMingen).
Apoptosis, differentiation assay, and cell-cycle analysis
Four days after transduction, the percentage of apoptotic U937 cells was determined using annexin V (BD Biosciences PharMingen) by fluorescence-activated cell sorter (FACS) analysis. In cellular differentiation assays, U937 cells were treated with vitamin D3 (10−6M) and transforming growth factor-β (TGF-β; 5nM) for 24 hours or all-trans retinoic acid (ATRA; 10−6M) for 4 days. For flow cytometry, phycoerythrin-conjugated antihuman CD11b and CD14 and mouse monoclonal IgG1 or mouse phycoerythrin-IgG1 isotype control antibodies were used (BD Biosciences PharMingen). For cell-cycle analysis, cells were incubated for 15 minutes with 2μM DRAQ5 (Alexis Biochemicals) at 37°C. Nuclear incorporation of DRAQ5 was measured by FACS (BD Biosciences, FACSCalibur). Multiplets of cells were excluded from analysis using the doublet discrimination module. Data analysis was performed with BD Biosciences Cell Quest Pro Version 4.0.2 software.
Serial replating, transplantation, and immunophenotyping
Serial replating and bone marrow transplantation studies were performed as described recently.3,26 Animal experiments were performed in compliance with the local animal experimentation guidelines and were approved by the regional council (Regierungspräsidium). Further details are given in the supplemental data.
Results
Molecular dynamics simulations and free energy decomposition reveal a spatially clustered arrangement of amino acid hot spots at the interface of the NHR2 tetramer
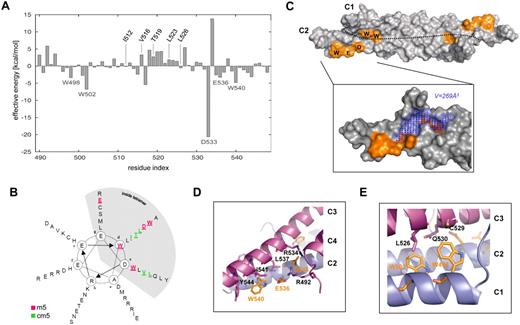
To identify critical amino acids at the interface of the NHR2 tetramer that contribute most to tetramer formation, we performed molecular dynamics simulations and MM-GB/SA calculations combined with a free energy decomposition on a per-residue level using the coordinates of the crystal structure of the NHR2 tetramer.12 Calculations of this type have previously been used successfully by us27 and others28,29 to characterize the origin of binding in terms of contributions from structural subunits of the binding partners. The MM-GB/SA calculation disclosed 5 residues (W498, W502, D533, E536, and W540) whose energy pattern suggested a strong contribution to the stabilization of the NHR2 tetramer structure (Figure 1A; supplemental Figure 1). These residues are located within the inner hydrophobic side of each α-helical stretch of the tetramer at positions “a,” “d,” and “g” in the heptad helical wheel (Figure 1B) and are conserved among all ETO homologs (supplemental Figure 2). The antiparallel orientation of helices C1 and C2 in the NHR2 dimer places W498 and W502 in close proximity to residues D533, E536, and W540, resulting in a spatially compact arrangement of hot spot residues. These residues are not located in the center of the interface, which is rather flat, but at its edges. Here, the protein topography is rugged, and the hot spot arrangement embraces the largest indentation found in the binding epitope with a volume of approximately 269 Å3 (Figure 1C). On tetramer formation with helices C3 and C4, this conformation yields a tight-fitting complex structure that is complementary in shape. One of residues D533, E536, and W540, located at the C-terminal helix end (eg, of C2) involved in interactions with 2 residues mainly located at the C-terminal helix end of C3 (Figure 1D). D533 forms buried salt bridges with R492 of C4 and R534 of C3, in which favorable attractive electrostatic energies prevail over unfavorable desolvation costs. This interaction occurs in place of interactions between hydrophobic amino acids that usually contribute to oligomerization in leucine zippers at these positions.12 E536 forms a solvent-exposed salt bridge with R492 of C4, and this interaction is stabilized by L537 packing to the side chain carbon atoms of E536. W540 forms a hydrophobic contact with I541 and an edge-to-face aromatic interaction with Y544 of C3. Overall, this results in a dense, zipper-like network of nonpolar and polar interactions. Similarly, W498 and W502, located at the N-terminal helix end (eg, of C1), form an indentation into which L526 of C3 inserts (Figure 1E). Conversely, W498 packs into an indentation formed by the hydrophobic parts of the side chains of L526, C529, and Q530 located on C3. As C3 and C4 are related to C1 and C2 by an approximate 2-fold rotational symmetry, the aforementioned interaction patterns occur 4 times in the tetramer interface. In summary, these results suggest the existence of amino acid clusters that form functional epitopes within the structural interface of the NHR2 tetramer.
Molecular dynamics simulation and free energy decomposition of the NHR2 tetramer. (A) MM-GB/SA calculations combined with free energy decomposition were used to determine the contribution of each amino acid within NHR2 to tetramer stabilization. Residues are numbered according to the RUNX1/ETO amino acid sequence. (B) Helical wheel representation of the NHR2 domain showing the positions of the m5 and cm5 residues. (C) NHR2 dimer showing the hot spot generated by residues W498, W502, D533, E536, and W540. The dotted line represents the border between the 2 antiparallel α-helices C1 and C2. The blue dots mark the location of the largest indentation found in the binding epitope. (D) Intermolecular contacts involving W540, D533, and E536. (E) Intermolecular contacts involving W502 and W498.
Molecular dynamics simulation and free energy decomposition of the NHR2 tetramer. (A) MM-GB/SA calculations combined with free energy decomposition were used to determine the contribution of each amino acid within NHR2 to tetramer stabilization. Residues are numbered according to the RUNX1/ETO amino acid sequence. (B) Helical wheel representation of the NHR2 domain showing the positions of the m5 and cm5 residues. (C) NHR2 dimer showing the hot spot generated by residues W498, W502, D533, E536, and W540. The dotted line represents the border between the 2 antiparallel α-helices C1 and C2. The blue dots mark the location of the largest indentation found in the binding epitope. (D) Intermolecular contacts involving W540, D533, and E536. (E) Intermolecular contacts involving W502 and W498.
Substitution of amino acids within the tetramerization hot spot disrupts NHR2 tetramer formation
To study the biochemical properties and functional consequences of amino acid substitution within the NHR2 hot spot domain, we introduced alanine substitutions at amino acid positions 498 (W498A), 502 (W502A), 533 (D533A), 536 (E536A), and 540 (W540A) of RUNX1/ETO to generate a mutant termed m5. As a control, alanine mutations were introduced at amino acid positions with low energy contribution to tetramer stability, namely, positions 512 (I512A), 516 (V516A), 519 (T519A), 523 (L523A), and 526 (L526A) (cm5 mutant, Figure 1B). We also introduced these mutations within a C-terminal truncated version of RUNX1/ETO (REtr) lacking the NHR3 and NHR4 domains because REtr has been shown previously to block differentiation of myeloid cells in vitro and to induce leukemia in mice.3
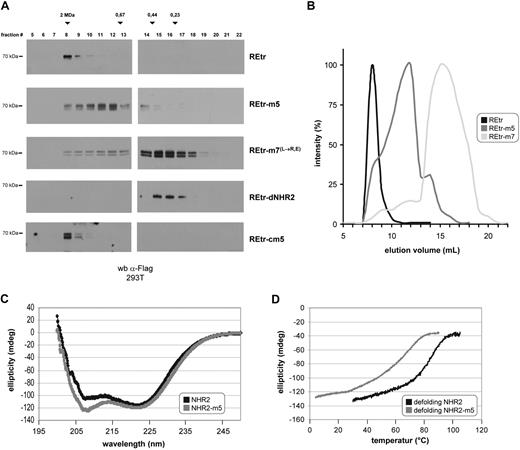
First, we determined the molecular size of REtr-m5 by size exclusion chromatography after expression of Flag-tagged versions of REtr-m5 and WT REtr in 293T cells. Whereas the tetrameric WT REtr migrated with a molecular weight of approximately 2 MDa, the REtr-m5 mutant shifted the migration of REtr to regions of lower-molecular weight species (Figure 2A). In contrast, the migration profile of REtr-cm5 was similar to that of WT REtr, suggesting that REtr-cm5 exists as a tetramer. We also used a mutant, in which the α-helical structure was partially abrogated by leucine to arginine or glutamic acid mutations (REtr-m7).12 As predicted, the elution profile of REtr-m7 was similar to that of REtr monomers lacking the NHR2 domain (REtr-dNHR2) and eluted in fractions of low molecular weight as RUNX1/ETO monomers.12 From the elution profiles of the different RUNX1/ETO variants, it becomes apparent that the elution profile of REtr-m5 peaks between the WT tetrameric (REtr) and the monomeric (REtr-m7) forms (Figure 2B), most probably reflecting a dimeric conformation. Furthermore, the finding that 2 differentially tagged REtr-m5 molecules retain the ability to coimmunoprecipitate implies that the dimeric conformation is stable in vivo (supplemental Figure 3). To investigate the biophysical properties of the m5 mutant in greater detail, a WT NHR2-only domain and the corresponding NHR2-m5 mutant were expressed in bacteria as His-fusion proteins and purified by Ni-affinity chromatography. With these proteins, we performed sedimentation velocity measurements to clarify the oligomerization state of the NHR2-m5 mutant in solution. We found that the NHR2-m5 mutant migrates at a sedimentation coefficient of 2.8 S as a single peak corresponding to 77% to 86% of the material loaded (Table 1; supplemental Figure 4). Based on this observation, the molecular mass for the NHR2-m5 dimer was calculated to be approximately 24 kDa, which is close to the theoretical mass of 22.2 kDa estimated for a dimer of 2 × 95 amino acids. A similar analysis for WT NHR2 confirmed the multimeric structure of this protein (data not shown).
Substitution of critical amino acids disrupts RUNX1/ETO tetramers into dimers without affecting its α-helical structure. (A) Size-exclusion chromatography of RUNX1/ETOtr (REtr) and the indicated mutant forms. Proteins were expressed in 293T cells and fractionated on a superose-6 column. Fractions were analyzed by Western blotting against the Flag epitope. (B) Signal intensity quantification of REtr, REtr-m5, and REtr-m7 elution profiles. (C) CD spectroscopy of the WT NHR2 and m5 mutant protein domains. (D) Ellipticity profile of NHR2 and NHR2-m5 at increasing temperature.
Substitution of critical amino acids disrupts RUNX1/ETO tetramers into dimers without affecting its α-helical structure. (A) Size-exclusion chromatography of RUNX1/ETOtr (REtr) and the indicated mutant forms. Proteins were expressed in 293T cells and fractionated on a superose-6 column. Fractions were analyzed by Western blotting against the Flag epitope. (B) Signal intensity quantification of REtr, REtr-m5, and REtr-m7 elution profiles. (C) CD spectroscopy of the WT NHR2 and m5 mutant protein domains. (D) Ellipticity profile of NHR2 and NHR2-m5 at increasing temperature.
Analysis of NHR2-m5 oligomerization by sedimentation velocity analysis
| Concentration . | S, Svedbergs . | Mass,* kDa . | f/f0† . | r.m.s.‡ . | Percentage contribution§ . |
|---|---|---|---|---|---|
| 15μM | 2.80 | 25.0 | 1.07 | 4.7 × 10−3 | 77 |
| 30μM | 2.75 | 23.0 | 1.09 | 8.0 × 10−3 | 86 |
| Concentration . | S, Svedbergs . | Mass,* kDa . | f/f0† . | r.m.s.‡ . | Percentage contribution§ . |
|---|---|---|---|---|---|
| 15μM | 2.80 | 25.0 | 1.07 | 4.7 × 10−3 | 77 |
| 30μM | 2.75 | 23.0 | 1.09 | 8.0 × 10−3 | 86 |
Sedimentation (S) profile for the NHR2-m5 protein at 2 different loading concentrations. Data were analyzed using the c(s) continuous distribution of Lamm equation solutions with SEDFIT and SEDPHAT.
The molecular mass estimation was obtained using the optimal frictional ratio (f/f0) with SEDFIT.
The ratio f/f0 is the frictional coefficient of a particle of a given axial ratio divided by the frictional coefficient of a sphere of the same volume.
Root-mean-square (r.m.s.) deviation of the global fit in units of absorbance (OD).
The weight percentage contribution of the majority species was obtained by integration of a continuous sedimentation coefficient distribution from approximately 1.5 to approximately 3.5 S with SEDFIT.
Furthermore, we characterized the secondary structure of the NHR2-m5 mutant by CD spectroscopy. The recombinant protein exhibited typical features of an α-helical domain with ellipticity minima at 208 and 222 nm, similar to the WT NHR2 polypeptide (Figure 2C). The content of α-helical secondary structure was identical for NHR2 and NHR2-m5 at room temperature (64% helical content for NHR2 vs 63% for NHR2-m5), thus indicating that the amino acid exchanges in NHR2-m5 do not disturb the α-helical structure of the NHR2 domain. Furthermore, we characterized the thermal stability of NHR2-m5 by thermal denaturation and renaturation experiments and subsequently monitored the changes in ellipticity by CD spectroscopy. In agreement with the values determined previously,12 we found that the melting point for the NHR2-only protein was 85°C, with a sharp transition between the folded and unfolded state, which is indicative of a strong cooperativity between the α-helices in tetramer stabilization (Figure 2D). In comparison, the melting point of the NHR2-m5 mutant protein was 60°C, with a broader transition between the folded and the unfolded state of the helix, suggesting a less tight association between the NHR2-m5 α-helical domains.
RUNX1/ETO-m5 exhibits reduced DNA-binding capacity and fails to activate endogenous RUNX1/ETO target genes
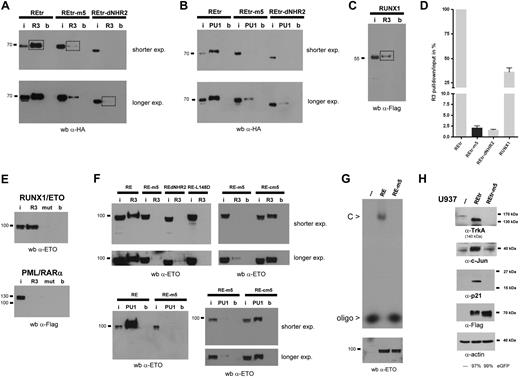
RUNX1/ETO has been shown to down-regulate PU.1 and RUNX3 (R3) gene expression by direct binding to RUNX1 consensus sequences in the promoter region of both genes.30,31 Because the leukemogenic properties of REtr critically depend on its DNA-binding capacity,13 we analyzed the binding properties of REtr-m5 to double-stranded DNA oligonucleotides containing either the PU.1 or the RUNX3 RUNX1 binding site using the ABCD assay. A strong DNA-binding activity to the R3 and PU.1-binding sites was found in cellular extracts of transfected 293T containing REtr (Figure 3A-B). The binding was specific because no binding was found to the empty magnetic beads used to pull down the biotin-labeled oligonucleotides nor to oligos containing mutated RUNX1-binding sites (Figure 3E). In contrast, we found only a weak binding to both oligonucleotides when cell extracts containing REtr-m5 were analyzed in this binding assay. In addition, monomeric forms of REtr (REtr-dNHR2) did not bind to the oligos (Figure 3A-B), confirming the results of others who have shown that tetramerization of RUNX1/ETO is required for specific DNA binding.32 Compared with the binding strength of the REtr tetrameric form, the dimeric and monomeric forms of REtr displayed a 40-fold reduction in binding affinity to the DNA oligos (Figure 3C-D).
RUNX1/ETO dimers lack DNA-binding capacity. (A) ABCD assay for the tetrameric, dimeric, and monomeric forms of REtr. Constructs were tested for binding to biotinylated oligonucleotides containing the RUNX1-binding sites of either RUNX3 (R3) or PU.1 (PU1) promoter sequences. Protein binding was detected by immunoblotting with an anti-HA antibody. (B) REtr and mutants were analyzed for PU.1-oligo binding as in panel A. (C) DNA binding of WT RUNX1 protein to the RUNX3-oligo. The RUNX1-R3-oligo interaction was detected with an anti-Flag antibody. (D) Comparison of the binding strength defined as the signal intensity ratio of R3-pulldown/R3-input to the R3-oligo. Values were obtained from the marked bands in panels A and C. DNA-binding activity of REtr was set to 100%. (E) Binding capacity of full-length RUNX1/ETO (RE) and RE-m5 to the R3 oligonucleotide (upper panel). The binding specificity was confirmed using cell extracts containing PML/RAR-α. (F) Analysis of DNA-binding capacity of RE, RE-m5, RE-cm5, RE-dNHR2, and RE-L148 mutants to the R3 and PU.1 oligonucleotides. (G) DNA-binding capacity of RE and RE-m5 to a radioactively labeled RUNX1 high affinity oligos by EMSA. (Bottom) Western blotting of lysates indicates equal loading for RE and RE-m5. (H) Target gene analysis of REtr and REtr-m5 transduced U937 cells. FACS-sorted (eGFP-positive) cell populations were analyzed for the expression of Trk-A, c-Jun, and p21. Expression of the REtr forms was verified by immunoblotting with an anti-Flag antibody. Actin serves as loading control. I indicates input; R3, RUNX3-oligo; PU1, PU.1-oligo; b, magnetic beads only; and C, RUNX1/ETO-DNA-oligo complex.
RUNX1/ETO dimers lack DNA-binding capacity. (A) ABCD assay for the tetrameric, dimeric, and monomeric forms of REtr. Constructs were tested for binding to biotinylated oligonucleotides containing the RUNX1-binding sites of either RUNX3 (R3) or PU.1 (PU1) promoter sequences. Protein binding was detected by immunoblotting with an anti-HA antibody. (B) REtr and mutants were analyzed for PU.1-oligo binding as in panel A. (C) DNA binding of WT RUNX1 protein to the RUNX3-oligo. The RUNX1-R3-oligo interaction was detected with an anti-Flag antibody. (D) Comparison of the binding strength defined as the signal intensity ratio of R3-pulldown/R3-input to the R3-oligo. Values were obtained from the marked bands in panels A and C. DNA-binding activity of REtr was set to 100%. (E) Binding capacity of full-length RUNX1/ETO (RE) and RE-m5 to the R3 oligonucleotide (upper panel). The binding specificity was confirmed using cell extracts containing PML/RAR-α. (F) Analysis of DNA-binding capacity of RE, RE-m5, RE-cm5, RE-dNHR2, and RE-L148 mutants to the R3 and PU.1 oligonucleotides. (G) DNA-binding capacity of RE and RE-m5 to a radioactively labeled RUNX1 high affinity oligos by EMSA. (Bottom) Western blotting of lysates indicates equal loading for RE and RE-m5. (H) Target gene analysis of REtr and REtr-m5 transduced U937 cells. FACS-sorted (eGFP-positive) cell populations were analyzed for the expression of Trk-A, c-Jun, and p21. Expression of the REtr forms was verified by immunoblotting with an anti-Flag antibody. Actin serves as loading control. I indicates input; R3, RUNX3-oligo; PU1, PU.1-oligo; b, magnetic beads only; and C, RUNX1/ETO-DNA-oligo complex.
We also introduced the m5 mutations into full-length RUNX1/ETO and tested its DNA-binding capability as mentioned in the previous paragraph. Full-length RUNX1/ETO did bind specifically to the R3 oligonucleotide, whereas a mutant of R3 lacking the RUNX1-binding sites was not bound by RUNX1/ETO and DNA binding was undetectable in extracts containing the PML/RAR-α protein (Figure 3E). Similar to the findings with REtr-m5, binding of RE-m5 to the R3 and PU.1 oligonucleotides was barely detectable and could be visualized only after extended exposure times. In contrast to the DNA-binding defect observed with the dimeric RE-m5 molecule, the RE-cm5 control mutant did bind to the R3 and PU.1 oligonucleotides, supporting our previous conclusions that REtr-cm5 forms a tetramer (Figure 3F). In addition, coexpression of the cofactor CBFβ, which enhances DNA-binding of RUNX1,33 did not improve REtr-m5 binding to DNA (data not shown). Similar to the observations of others,32 the monomeric form of RUNX1/ETO (RE-dNHR2) and a DNA-binding defective mutant of RUNX1/ETO (RE-L148D) failed to bind to the R3 oligonucleotide. Loss of RE-m5 DNA binding was also observed by EMSA analysis using a radioactive labeled RUNX1 high-affinity oligonucleotide (Figure 3G). Subsequently, we examined the ability of REtr-m5 to interfere with RUNX1/ETO target gene expression because this property of RUNX1/ETO depends on tetramerization.12 We transduced U937 cells with either WT REtr or REtr-m5. After transduction, cells were sorted for eGFP and analyzed for expression of RUNX1/ETO target genes by Western blotting. The RUNX1/ETO target genes TrkA, c-jun, and p-21 were up-regulated in the presence of the tetrameric form of REtr, but their expression was unaffected in cells expressing the dimeric REtr-m5 protein (Figure 3H). Thus, our cumulative evidence suggests that REtr-m5 lacks substantial DNA-binding activity and does not alter the expression pattern of RUNX1/ETO target genes.
RUNX1/ETO tetramers, but not dimers, trigger apoptosis and block cytokine-induced myeloid differentiation
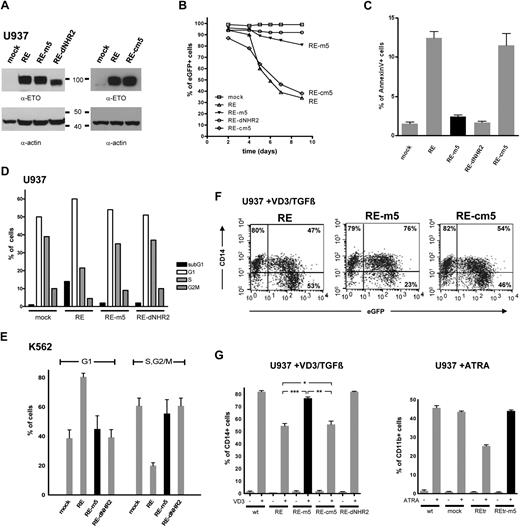
Inhibition of granulocytic and monocytic differentiation as well as cell-cycle arrest and induction of apoptosis are hallmarks of RUNX1/ETO activity.34 To investigate the dependence of these biologic effects on the structural state of RUNX1/ETO, the monomeric (RE-dNHR2), dimeric (RE-m5), and tetrameric forms (RE, RE-cm5) of RUNX1/ETO were retrovirally expressed in U937 cells. Expression of the RUNX1/ETO constructs was verified by Western blotting of cellular lysates (Figure 4A). Cell viability was monitored based on the percentage of eGFP-positive cells in the growing cultures. Five days after transduction, a strong decrease in eGFP-positive cells expressing WT RE or RE-cm5 was observed, whereas the percentage of eGFP-positive cells within the cell population expressing the RE-m5 dimeric form of RUNX1/ETO was only slightly reduced (Figure 4B). No reduction of eGFP-positive cells was observed in the population expressing the monomeric form of RE (RE-dNHR2).
RUNX1/ETO tetramers, but not dimmers, trigger apoptosis and block cytokine-induced myeloid differentiation. (A) Expression of RUNX1/ETO (RE) and mutants thereof in stably transduced U937 cells using anti-ETO and antiactin antibodies. (B) Survival kinetics of RE/eGFP-expressing U937 cells. (C) Apoptosis induction in U937 cells, expressing the indicated forms of RE. (D) Cell-cycle distribution of U937 cells at day 4 after transduction as measured by Draq5 staining. (E) Cell-cycle distribution of stably transduced K562 cells. Draq5 staining was performed 4 days after transduction. (F) Differentiation pattern of U937 cells expressing the indicated RE forms. Two days after transduction, U937 cells were stimulated with vitamin D3/TGF-β for 24 hours and subsequently analyzed for CD14 surface marker expression. (G). Vitamin D3/TGF-β– or ATRA-induced differentiation of U937 cells expressing the indicated RE forms. Statistically significant P values of RE, RE-m5, and RE-cm5 compared with each other are indicated for the vitamin D3 (VD3)/TGF-β–stimulated cells (t test): ***P < .001, **P = .001, *P = .73. Data represent averages of 3 independent experiments.
RUNX1/ETO tetramers, but not dimmers, trigger apoptosis and block cytokine-induced myeloid differentiation. (A) Expression of RUNX1/ETO (RE) and mutants thereof in stably transduced U937 cells using anti-ETO and antiactin antibodies. (B) Survival kinetics of RE/eGFP-expressing U937 cells. (C) Apoptosis induction in U937 cells, expressing the indicated forms of RE. (D) Cell-cycle distribution of U937 cells at day 4 after transduction as measured by Draq5 staining. (E) Cell-cycle distribution of stably transduced K562 cells. Draq5 staining was performed 4 days after transduction. (F) Differentiation pattern of U937 cells expressing the indicated RE forms. Two days after transduction, U937 cells were stimulated with vitamin D3/TGF-β for 24 hours and subsequently analyzed for CD14 surface marker expression. (G). Vitamin D3/TGF-β– or ATRA-induced differentiation of U937 cells expressing the indicated RE forms. Statistically significant P values of RE, RE-m5, and RE-cm5 compared with each other are indicated for the vitamin D3 (VD3)/TGF-β–stimulated cells (t test): ***P < .001, **P = .001, *P = .73. Data represent averages of 3 independent experiments.
Transduced cells were analyzed in parallel for signs of apoptosis by annexin V staining (Figure 4C). Compared with RE or RE-cm5-expressing cells, which showed a 7- to 8-fold increased rate of apoptosis, the apoptosis rate in RE-m5-expressing cells differed only slightly from that of RE-dNHR2 and mock-transduced cells. Further analysis of the cell-cycle distribution revealed an increase of the sub-G1 fraction and a cell-cycle arrest in G1 exclusively in U937 cells expressing RE, but not in RE-m5- or RE-dNR2-expressing cells (Figure 4D).
To investigate the relevance of tetramer formation for cell-cycle arrest in more detail, K562 cells expressing the tetrameric, dimeric, or monomeric forms of RUNX1/ETO were generated. Expression of full-length RUNX1/ETO in these cells leads to a profound arrest in the G1 phase of the cell cycle.3 In agreement with our previous observations, only the tetrameric form of RUNX1/ETO induced a robust cell-cycle arrest in G1, whereas the cell-cycle arrest in RE-m5 or RE-dNHR2 cells was similar to that of mock-transduced cells (Figure 4E). Taken together, expression of the tetrameric, but not the dimeric, form of RUNX1/ETO accounts for a perturbed cell-cycle distribution and apoptosis.
RUNX1/ETO is known to block vitamin D3/TGF-β and ATRA triggered monocytic and granulocytic differentiation. To test the effect of the various RUNX1/ETO forms on myeloid differentiation, transduced U937 cells were stimulated with vitamin D3/TGF-β or ATRA 2 days after transduction (Figure 4F-G). We monitored monocytic differentiation by FACS based on CD14 expression 24 hours after vitamin D3/TGF-β administration. As expected, approximately half of the eGFP-positive RUNX1/ETO-expressing cells remained CD14 negative, whereas the population of nontransduced cells in the same culture was almost completely positive for CD14 (Figure 4F). The tetrameric forms, RE and RE-cm5, both showed similar properties, whereas RE-m5 and RE-dNHR2 failed to block monocytic differentiation, as determined by equal amounts of CD14+ cells in the eGFP-positive and -negative fractions (Figure 4F-G). Similar results were obtained on ATRA-induced myeloid differentiation. In this case, cell differentiation was measured by CD11b staining 3 days after ATRA administration. Again, only cells expressing the tetrameric form of RUNX1/ETO (in this case, REtr) showed a strong reduction in cellular differentiation levels, whereas mock-transduced and REtr-m5-transduced cells did not show any block in myeloid differentiation (Figure 4G). Taken together, the dimeric form of RUNX1/ETO (m5) behaved similarly to the monomeric form (dNHR2) in that both have lost the capacity to block myeloid cell differentiation.
RUNX1/ETO-m5 fails to trigger self-renewal of HSCs
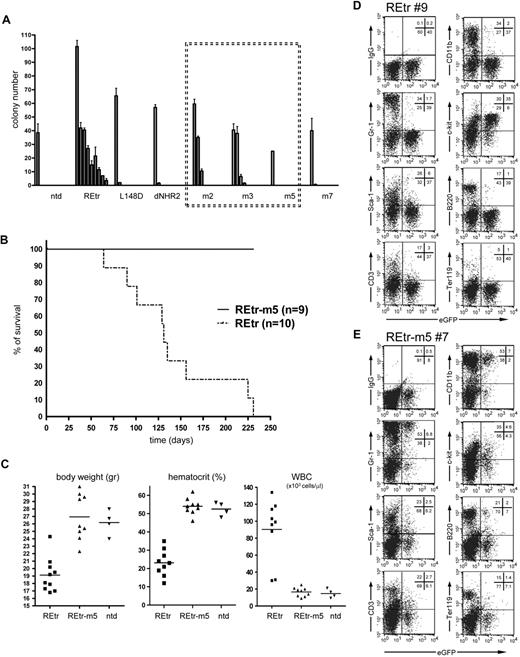
RUNX1/ETO is known to induce self-renewal of immature hematopoietic cells without affecting their lymphoid and myeloid differentiation potential.35-37 We used this property of RUNX1/ETO to test the effect of the NHR2 mutations in primary hematopoietic cells. As a readout system for REtr function in vitro, we made use of a clonogenic assay, which critically depends on RUNX1/ETO tetramer formation.12,26 As expected, expression of REtr in murine progenitor cells induced a strong clonogenic activity as measured by serial replating of REtr-transduced progenitor cells (Figure 5A). In contrast, constructs deficient in DNA-binding activity (REtr-L148D) or tetramer formation (REtr-deltaNHR2 and REtr-m7) completely failed to enhance the clonogenic activity of hematopoietic progenitors. We found that the alanine substitutions in REtr-m5 completely abolished the clonogenic properties of REtr, which thus behaves like the clonogenic inactive forms of REtr, whereas mutants with partial amino acid substitutions in the hot spot (m2, m3; supplemental Figure 2) retained some residual replating capacity (Figure 5A).
RUNX1/ETOtr dimers lost replating capacity and in vivo leukemogenic function. (A) Replating potential of murine lin-depleted bone marrow cells expressing the indicated RUNX1/ETOtr (REtr) forms. Serial replating of bone marrow cells was performed in the presence of IL-3, IL-6, and stem cell factor. (B) Kaplan-Meier survival curves of C57/BL6 mice transplanted with lin-depleted bone marrow cells transduced with either REtr or REtr-m5. (C) Body weight, hematocrit, and white blood cell counts of mice transplanted with nontransduced or lin-depleted cells expressing either REtr or REtr. (D) FACS analysis of bone marrow cells obtained from a mouse transplanted with REtr-transduced cells. (E) Similar to panel D, but transplanted with REtr-m5 expressing cells. The percentage of cells in each quadrant is indicated.
RUNX1/ETOtr dimers lost replating capacity and in vivo leukemogenic function. (A) Replating potential of murine lin-depleted bone marrow cells expressing the indicated RUNX1/ETOtr (REtr) forms. Serial replating of bone marrow cells was performed in the presence of IL-3, IL-6, and stem cell factor. (B) Kaplan-Meier survival curves of C57/BL6 mice transplanted with lin-depleted bone marrow cells transduced with either REtr or REtr-m5. (C) Body weight, hematocrit, and white blood cell counts of mice transplanted with nontransduced or lin-depleted cells expressing either REtr or REtr. (D) FACS analysis of bone marrow cells obtained from a mouse transplanted with REtr-transduced cells. (E) Similar to panel D, but transplanted with REtr-m5 expressing cells. The percentage of cells in each quadrant is indicated.
RUNX1/ETOtr dimers fail to induce leukemia in mice
In contrast to full-length RUNX1/ETO, the truncated form of the oncoprotein (REtr) has been shown to induce leukemia in mice.3,4,13 To evaluate the relevance of tetramer versus dimer formation for the transformation capability of RUNX1/ETO, we transduced lineage-depleted C57/BL6/Ly5.2 murine bone marrow cells with REtr and REtr-m5 along with eGFP. The transduction rate for both constructs ranged between 40% and 60% (not shown). After transduction, the progenitor cells were transplanted into lethally irradiated C57/BL6/Ly5.1 recipient mice. Four weeks after transplantation, engraftment levels ranged between 65% and 85% (not shown). Similar to the results of others,3,4 all mice transplanted with REtr-expressing cells developed leukemia within 6 months (Figure 5B). Remarkably, none of the REtr-m5 transplanted mice developed leukemia, nor did they show any sign of disease. The first indications of REtr-induced leukemia were the reduction in total body weight (Figure 5C) and clinical symptoms, including labored breathing, akinesia, and a hunched posture, none of which was evident in REtr-m5-transplanted animals (not shown). Further analysis of the organs showed greatly enlarged spleens exclusively in the leukemic mice expressing tetrameric REtr. In addition, the percentage of eGFP-positive blood cells was dramatically increased in REtr-transplanted mice compared with stable levels in REtr-m5-transplanted mice (supplemental Table 1). In addition, mice transplanted with REtr-expressing cells showed increased numbers of white blood cells and reduced hematocrit. In contrast, the body weight, white blood cells, and hematocrit values of animals transplanted with REtr-m5 were similar to those transplanted with nontransduced cells (Figure 5C).
Similar to previous reports,3 the bone marrow of REtr mice contained mainly immature cells as estimated by the lack of the differentiation markers Gr-1, CD11b, B220, and CD3 in the eGFP-positive cell fraction (Figure 5D). In contrast, the majority of eGFP-positive cells did express the progenitor cell marker CD117/c-kit. In the same animals, the eGFP-negative fraction included fully differentiated cells, comparable with bone marrow cells obtained from WT animals (data not shown). Most importantly, the fraction of eGFP-positive cells obtained from the bone marrow of REtr-m5 animals showed no signs of differentiation block as estimated by equal proportion of differentiated versus nondifferentiated cells in the eGFP-positive and -negative fractions (Figure 5E). Cytospin preparations of bone marrow cells obtained from REtr-m5 mice revealed a mixture of immature and mature cells, whereas the majority of bone marrow cells of mice harboring REtr were immature myelocytes (supplemental Figure 5). REtr-m5 transplantation mice were healthy and lacked any signs of disease, even 12 months after transplantation. Furthermore, REtr-m5-transduced cells neither accumulated in the bone marrow of transplanted animals nor did they display a block in myeloid and lymphoid differentiation in vivo. Taken together, our results indicate that the REtr-m5 mutant completely failed to induce leukemia in vivo.
Discussion
We have used binding free energy calculations together with a free energy decomposition to identify essential amino acids involved in RUNX1/ETO dimer-tetramer transition. These methods have been previously used among others to define amino acid hot spots at the interaction surfaces of H-Ras/C-Raf1 and H-Ras/RalGDS27 and to define amino acid recognition by aspartyl-tRNA synthetase.28 The computational analysis of NHR2 suggested an uneven distribution of effective binding energy along the tetramer interface and identified 5 critical amino acids that provide more than 50% of the binding energy to dimer-tetramer transition. According to the crystal structure of the NHR2 homo-tetrameric α-helical bundle, we have identified a hot spot for RUNX1/ETO tetramerization at both edges of each dimer. Notably, not all amino acids contributing to the hot spot were clustered on one α-helix. Rather, the hot spot is shaped by the close spatial proximity of W498 and W502 to D533, E536, and W540 once 2 α-helical monomers associate in an antiparallel orientation to form a dimer. Because our computational analysis was focused on the dimer-tetramer transition, leucine residues, which largely contribute to the stability of each NHR2 dimer, were not scored as essential amino acids for tetramer formation. Among the 5 amino acids involved, we found tryptophan and aspartate, 2 amino acids that are capable of forming multiple types of favorable interactions, such as π-π-, hydrogen bonding, and hydrophobic interactions, and are frequently found in hot spots of various protein-protein interactions.18 The relevance of these amino acids in tetramer stabilization is strengthened by the fact that salt bridge networks often stabilize complexes of α-helical stretches by reducing their thermal motion, which results in increased thermostability.38 The amino acid residues at the NHR2 hot spot are located at the “a,” “d,” and “g” positions of the α-helix at the inner side of the tetramer and become deeply buried in the interface. Moreover, these amino acids cluster around a cleft on the dimer surface, identified as the largest indentation on the dimer surface. Alanine substitutions at these 5 positions destroyed tetramer formation, resulting in RUNX1/ETO dimers as demonstrated by biophysical and biochemical experiments (Figure 2; supplemental Figures 3-4; Table 1). It has to be considered, however, that in most experiments data were acquired after overexpression of RUNX1/ETO and its mutants. Therefore, we cannot exclude the existence of additional configurations at physiologic concentrations.
RUNX1/ETO dimers display a reduced ability to block myeloid differentiation, do not enhance self-renewal of hematopoietic progenitor cells in vitro, and have no leukemogenic potential in vivo. Similar observations were recently reported for the transcription factor STAT5. Here, the leukemogenic potential of a constitutively activated STAT5 molecule was entirely dependent on the ability to form tetramers, as mutants engineered to exclusively form dimers failed to induce leukemia in mice.39
Because nuclear localization and intranuclear mobility of the dimeric RUNX1/ETO were not altered compared with the tetrameric counterpart (data not shown), the lack of leukemogenic properties of RUNX1/ETO dimers may result from impaired binding to cofactors or to a defect in DNA-binding activity essential for cellular transformation. RUNX1/ETO tetramers are known to associate with a wide variety of factors, including the ETO family members ETO, MTG16, and MTGR1 and with proteins involved in induction of repressive chromatin, although their contribution to the leukemogenic potential of RUNX1/ETO is still not clear. The lack of leukemic potential of RUNX1/ETO dimers is doubtful because of defective cofactor binding because monomers of RUNX1/ETO (RE-m7) have been shown to bind to HDAC1, HDAC2, HDAC3, N-CoR, SMRT, PKA-RIIα, PLZF, mSin3A, and HEB with similar affinities as to the RUNX1/ETO tetramer. Likewise, the binding affinity to ETO and its homologs MTG16 and MTGR1 was reduced but not eliminated in RUNX1/ETO monomers, which partially retained NHR2 α-helicity.12 Furthermore, an FKBP homo-oligomerization domain (4 × FKBP) can functionally replace the NHR2 domain,26 suggesting that, at least in vitro, the NHR2-binding proteins are dispensable for RUNX1/ETO activity. Our experimental evidence suggests that the DNA-binding affinity of RUNX1/ETO dimers is severely reduced compared with that of WT RUNX1 and RUNX1/ETO. Indeed, the DNA-binding strength of REtr-m5 to the RUNX3 oligonucleotide was reduced by 40-fold compared with WT REtr, whereas a 10-fold decrease in binding affinity was observed compared with RUNX1. Although RUNX1 binds DNA, monomers of RUNX1/ETO lacking the oligomerization domain or RUNX1/ETO dimers bind DNA with severely reduced affinity. RUNX1 is known to bind DNA as a heterodimer and to undergo a conformational shift on binding, which is further stabilized after heterodimerization with CBFβ.40 CBFβ is known to enhance the DNA-binding affinity of the Runt domain and binds more efficiently to the DNA-bound Runt domain than to the Runt domain alone.41,42 In our study, however, coexpression of CBFβ did not increase the DNA-binding affinity of the RUNX1/ETO dimer, probably reflecting the poor association of the dimer with DNA.
RUNX1/ETO tetramers have been shown to bind preferentially to DNA stretches containing 2 RUNX1-binding sites.32 Although the RUNX3 and PU.1 oligonucleotides used in our study did contain 2 RUNX1 consensus-binding sites, RE-m5 or REtr-m5 failed to bind to these oligonucleotides. One can anticipate that the antiparallel orientation of both α-helices in the RUNX1/ETO-m5 dimer does not favor the conformational change observed on DNA binding for RUNX1. Thus, only the close proximity of 2 Runt domains, as it occurs in the RUNX1/ETO tetramer, may generate a high-affinity DNA-binding site. This hypothesis is reinforced by the observations made with RUNX1 dimers formed by 2 parallel oriented RUNX1 helices, as it exists in the fusion protein TEL/RUNX1.43 In addition, the replacement of the NHR2 domain in RUNX1/ETO with 4 tandem repeats of a self-oligomerizing mutant of FKBP results in homo-oligomers with the ability to bind DNA with similar affinity as RUNX1/ETO (data not shown) and to enhance the clonogenic potential of primary murine bone marrow cells.26 The tetrameric configuration of RUNX1/ETO is reminiscent of that described for the BCR/ABL oncoprotein. In BCR/ABL, 2 antiparallel-oriented monomers associate to form a coiled coil dimer and 2 dimers assemble into a tetramer. In the tetrameric configuration, the functional domains of 2 opposite dimers are placed in close proximity to each other (∼ 24 Å), whereas in the dimer, the antiparallel orientation of the monomers places the functional domains of each monomer approximately 60 Å apart from each other.44 In analogy, only the tetrameric configuration in RUNX1/ETO may provide the spatial configuration required for tight DNA binding.
We propose the NHR2 tetramerization domain of RUNX1/ETO as a challenging but reasonable and promising target for a molecular intervention in t(8;21) leukemias. Although protein-protein interfaces were originally thought to be difficult targets for drug development, recent advances in the understanding of structural constraints at interacting surfaces in combination with structural and mutational analysis has allowed for the development of small-molecule mimics of hot spots that have been found to inhibit protein-protein interactions.45-48 In addition, effective modulation of protein-protein interactions has been described for small-molecule binding to clefts or grooves at interface regions.49,50 It appears promising that the NHR2 hot spot is located around the largest indentation on the dimer surface. Furthermore, the 2-fold rotational symmetry of the RUNX1/ETO dimer creates 2 hot spot regions per dimer, which are expected to act as 2 independent ligand-binding sites for appropriate modulators, thus enhancing their specific effect. As oligomerization events represent an important subset of protein-protein interactions in leukemia,51-53 the rational approach adopted in our work could be further expanded to other oligomerization domains for target evaluation and the development of novel therapeutic approaches.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank C. Gerum and H. Kunkel for expert technical assistance, O. Heidenreich and R. Piekorz for critical comments on the manuscript, and A. Metz for fruitful discussions.
This work was supported by the Jose Carreras Leukämie Stiftung (grant DJCLS R 05/08; M.G.), the NGFNplus Cancer Network (grant 01GS0879), Graduate School Biologicals (GK1172), the Goethe-University of Frankfurt (Targeting aberrant PPI in leukemia), and the Deutsche Krebshilfe (grant 102362, TP7).
Authorship
Contribution: C.W. conceived the study, performed experiments, analyzed the data, supervised the project, and wrote the manuscript; Y.B., L.C.-W., A.V., J.H., and S.M. performed experiments; V.V. and W.M. performed the sedimentation velocity analysis; J.K. and J.L. contributed to the design of the experiments and to the writing of the manuscript; H.G. performed the computations and analyzed the data; and H.G. and M.G. conceived the study and analyzed the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christian Wichmann, Institute for Biomedical Research, Georg-Speyer-Haus, Paul-Ehrlich-Strasse 42-44, 60596 Frankfurt, Germany; e-mail: c.wichmann@em.uni-frankfurt.de; and Manuel Grez, Institute for Biomedical Research, Georg-Speyer-Haus, Paul-Ehrlich-Strasse 42-44, 60596 Frankfurt, Germany; e-mail: grez@em.uni-frankfurt.de.
References
Author notes
H.G. and M.G. share senior authorship.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal