Abstract
To resolve the genetic heterogeneity within pediatric high-risk B-precursor acute lymphoblastic leukemia (ALL), a clinically defined poor-risk group with few known recurring cytogenetic abnormalities, we performed gene expression profiling in a cohort of 207 uniformly treated children with high-risk ALL. Expression profiles were correlated with genome-wide DNA copy number abnormalities and clinical and outcome features. Unsupervised clustering of gene expression profiling data revealed 8 unique cluster groups within these high-risk ALL patients, 2 of which were associated with known chromosomal translocations (t(1;19)(TCF3-PBX1) or MLL), and 6 of which lacked any previously known cytogenetic lesion. One unique cluster was characterized by high expression of distinct outlier genes AGAP1, CCNJ, CHST2/7, CLEC12A/B, and PTPRM; ERG DNA deletions; and 4-year relapse-free survival of 94.7% ± 5.1%, compared with 63.5% ± 3.7% for the cohort (P = .01). A second cluster, characterized by high expression of BMPR1B, CRLF2, GPR110, and MUC4; frequent deletion of EBF1, IKZF1, RAG1-2, and IL3RA-CSF2RA; JAK mutations and CRLF2 rearrangements (P < .0001); and Hispanic ethnicity (P < .001) had a very poor 4-year relapse-free survival (21.0% ± 9.5%; P < .001). These studies reveal striking clinical and genetic heterogeneity in high-risk ALL and point to novel genes that may serve as new targets for diagnosis, risk classification, and therapy.
Introduction
Overall survival in pediatric B-precursor acute lymphoblastic leukemia (ALL) now exceeds 80% on contemporary treatment regimens. These therapeutic advances have been achieved through the progressive intensification of chemotherapy and the development of risk classification schemes that target children to more intensive therapies based on their relative relapse risk.1,2 Current risk classification schemes incorporate pretreatment clinical characteristics (white blood cell count [WBC], age, and the presence of extramedullary disease), the presence or absence of recurring cytogenetic abnormalities, and measures of minimal residual disease (MRD) at the end of induction therapy to classify children with B-precursor ALL into “low,” “standard/intermediate,” “high,” or “very high” risk categories.2 Yet, despite these advances, more than 20% of children still relapse, and the majority of these relapses occur in children who are initially classified as “standard/intermediate” or “high” risk. Thus, although overall outcomes in pediatric ALL have significantly improved, children classified with “high” or “very high” risk ALL, those who have relapsed, or those of Hispanic or Native American race or ethnicity3 continue to have relatively poor survival and require the development of novel therapies for cure.
Shuster et al previously demonstrated that the prospective identification of children with “high-risk” B-precursor ALL using the National Cancer Institute (NCI)/Rome criteria (age ≥ 10 years and/or presenting WBC ≥ 50 000/μL) could be refined using age, sex, and WBC to identify a subgroup of approximately 12% of B-precursor ALL patients with a very poor outcome, with less than 50% relapse-free survival (RFS).4 In contrast to children with favorable “low-risk” ALL (associated with t(12;21)/ETV6-RUNX1 or trisomies of chromosomes 4, 10, and 17) or those with unfavorable “very-high” risk disease (associated with t(9;22)/BCR-ABL1 or hypodiploidy), the recurring genetic abnormalities uniquely associated with “high-risk” B-precursor ALL are only now just beginning to be described.5-11 To identify novel biologic and genetically defined subgroups within high-risk ALL and genes that might serve as new diagnostic or therapeutic targets, we performed gene expression profiling in a cohort of 207 uniformly treated high-risk B-precursor ALL patients who were enrolled in the Children's Oncology Group (COG) P9906 trial using the Shuster et al criteria.4,12 Under the auspices of a National Cancer Institute TARGET Project (Therapeutically Applicable Research to Generate Effective Treatments; www.target.cancer.gov), we have also assessed genome-wide DNA copy number abnormalities (CNAs) in leukemic DNA in this same cohort of patients,5 and we have performed selective gene resequencing to identify mutated genes in leukemic cells.6,8,10,11 Herein we report the discovery of 8 distinct gene expression-based patient cluster groups, defined by shared patterns of gene expression, within clinically defined “high-risk” B-precursor ALL. Although 2 clusters were associated with known recurring cytogenetic abnormalities (either t(1;19)/TCF3-PBX1 or MLL translocations), the remaining 6 cluster groups had no known sentinel cytogenetic lesion. Each of the 8 gene expression-based cluster groups was characterized by distinct patterns of genome-wide DNA CNAs and with expression of unique sets of “outlier” genes. Such outlier genes are of great interest as their aberrant expression, significantly above or below the mean, may arise as a result of their involvement in underlying recurring genetic abnormalities.13-15 Two of the unique clusters were also associated with strikingly different preclinical characteristics and treatment outcomes. These studies reveal the striking biologic and genetic heterogeneity within high-risk ALL and identify genes that may serve as new targets for discovery of novel recurrent genetic abnormalities and improved diagnosis, risk classification, and therapy.
Methods
Patient selection and characteristics
COG Trial P9906 enrolled 272 eligible children and adolescents with high-risk B-precursor ALL between March 15, 2000 and April 25, 2003 (http://www.acor.org/ped-onc/diseases/ALLtrials/9906.html).12 This trial targeted a subset of patients with high-risk features (older age and higher WBC), as defined by Shuster et al,4 that had experienced poor outcomes (< 50% 4-year RFS) in prior trials. Patients were first enrolled in the COG P9000 classification study and received a 4-drug induction regimen. Patients in complete remission with less than 5% bone marrow blasts after either 4 or 6 weeks of induction were then eligible to participate in COG P9906 if they met the age and WBC criteria described4 or had overt central nervous system or testicular involvement at diagnosis. Patients who met these criteria but had favorable (t(12;21)/ETV6-RUNX1 or trisomy of 4 and 10) or unfavorable genetic features (t(9;22)/BCR-ABL1 or hypodiploidy) were excluded.12 Patients enrolled in COG P9906 were uniformly treated with a modified augmented BFM regimen.16,17 The majority of patients had MRD assessed by flow cytometric analysis at day 29 at the end of induction therapy12,18 ; cases were defined as MRD-positive or MRD-negative using a threshold of 0.01%.
For this study, cryopreserved pretreatment leukemia specimens were available on a representative cohort of 207 of the 272 (76%) patients. As previously described,9 these 207 patients did not differ significantly from the full 272 patients accrued to the trial (supplemental Table 1 and supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Treatment protocols were approved by the National Cancer Institute (NCI) and participating institutions through their Institutional Review Boards. Informed consent for participation in these research studies was obtained from all patients or their guardians in accordance with the Declaration of Helsinki. Outcome data for all patients were frozen as of October 2006; the median time to event or censoring was 3.7 years. An independent cohort of 99 patients with high-risk B-precursor ALL (defined as high-risk using NCI/Rome criteria), previously selected as a case (failure)/control (continuous complete remission) study, was used as a validation cohort.19 This cohort was derived from COG CCG Trial 1961, and gene expression profiles were derived using the same Affymetrix microarray platform as for this study (Supplemental data).
Gene expression profiling
As previously described,9 RNA was isolated from pretreatment diagnostic ALL samples in the 207 patients (131 bone marrow, 76 peripheral blood) using TRIzol (Invitrogen); all samples had more than 80% leukemic blasts. cDNA labeling, hybridization, and scanning were performed as previously described.9 A mask to remove uninformative probe pairs and Affymetrix controls was applied to all the arrays (detailed in Supplemental data), and the default Affymetrix MAS 5.0 normalization was used. Array experimental quality was assessed using the following parameters, and all arrays met these criteria for inclusion: GAPDH more than 5000, more than 20% expressed genes, GAPDH 3′/5′ ratios less than 4; and linear regression r2 values of spiked poly(A) controls more than 0.90. This gene expression dataset may be accessed via the NCI caArray site (https://array.nci.nih.gov/caarray) or at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE11877.
Unsupervised clustering methods and selection of outlier genes
Microarray gene expression profiling data were available from an initial 54 504 probe sets after masking and filtering of minimal probe sets and controls (Supplemental data). Three different unsupervised, unbiased methods were used to select genes for standard hierarchical clustering: High Coefficient of Variation (HC) as originally described by Eisen et al,20 Cancer Outlier Profile Analysis (COPA),13-15 and Recognition of Outliers by Sampling Ends (ROSE), a novel method similar to COPA developed in our laboratory (Supplemental data). In HC, the 54 504 probe sets were ordered by their coefficients of variation and the highest 254 probe sets were used for clustering; this method identifies probe sets having an overall high variance relative to mean intensities. COPA13-15 selects “outlier” probe sets, also in an unsupervised fashion, on the basis of their absolute deviation from median at a fixed point (typically the 95th percentile). ROSE was developed by our group as an alternative to COPA, and selects probe sets both on the basis of the size of the outlier group they identify as well as the magnitude of the deviation from expected intensity (Supplemental data; ROSE and COPA). For all 3 probe selection methods, the top 254 probe sets (supplemental Table 7A) were clustered using EPCLUST (http://www.bioinf.ebc.ee/EP/EP/ EPCLUST, Version 0.9.23 beta, Euclidean distance, average linkage UPGMA). A threshold branch distance was applied, and the largest distinct branches above this threshold containing more than 8 patients were retained and labeled. The HC method was used as the basis of cluster definition and nomenclature, with each of the 8 predominant clusters first identified through HC being assigned a number (H1-H8). All clusters are prefixed by the method of their probe set selection (H indicates HC; C, COPA; and R, ROSE), with COPA and ROSE numbers being assigned based on the similarity of a specific cluster group's membership (patient membership) to that seen in the original H clusters. The top 100 median rank order probe sets for each ROSE cluster are provided in Supplemental data. In the validation cohort (COG CCG 1961), the same initial masking criteria were applied to the raw data, yielding 54 504 probe sets for analysis. Applying ROSE with the same parameters used for the COG P9906 ALL cohort (Supplemental data), 167 probe sets were identified for clustering. The selection criteria used for COG P9906 was also used for COPA and HC, and the top 167 probe sets derived from these methods were used for hierarchical clustering (supplemental Table 7A).
Assessment of genome-wide DNA CNAs
Copy number alterations, analyzed in 198 of the 207 patients in the COG P9906 cohort who had paired leukemic and germline DNA available for analysis, were detected as previously described and reported by Mullighan et al5 Briefly, DNA from the diagnostic leukemic cells and from a sample obtained after remission induction therapy (germline) was extracted and genotyped using either the 250K Sty and Nsp single nucleotide polymorphism arrays (Affymetrix). Single nucleotide polymorphism array data preprocessing and inference of DNA CNA and loss of heterozygosity were performed as previously described.5,21
Statistical analyses
Log-rank analysis was used to evaluate RFS.22 Kaplan-Meier survival analyses and hazard ratios were also calculated for comparisons of group RFS.23,24 Kruskal-Wallis rank-sum tests were used to analyze age and WBC counts; Fisher exact test was used to evaluate the binary variables.22 All statistical analyses were performed using R25 (http://www.R-project.org, Version 2.10.0, with basic and survival packages).
Results
Reflective of their initial classification as “high-risk” B-precursor ALL, the 207 uniformly treated children and adolescents studied herein had a median age of 13.1 years (range, 1-20 years), a median WBC at disease presentation of 62 300/μL, a male predominance (66%), and high rates of MRD (35%) at the end of induction therapy (supplemental Table 2). Nearly 25% were of self-reported Hispanic ethnicity. Whereas 10% (21 of 207) had translocations involving MLL on chromosome 11q23 and 11% (23 of 207) had t(1;19)/TCF3-PBX1, the remaining 79% (163 of 207) of cases lacked any previously known recurring chromosomal abnormality (supplemental Table 2). RFS was 66.3% plus or minus 3.5% and overall survival was 83% at 4 years.
Unsupervised hierarchical clustering defines 8 gene expression cluster groups
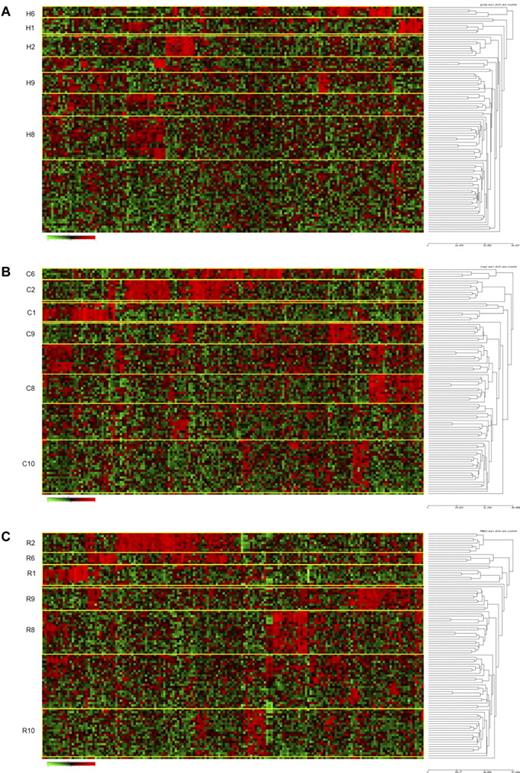
We hypothesized that the most statistically robust patient cluster groups, defined by shared patterns of gene expression, would be repeatedly identified using more than one clustering method. Thus, several unbiased methods for probe selection for unsupervised hierarchical clustering were applied to the gene expression profiles. First, using the top 254 genes (full list, supplemental Table 7A) selected by the standard approach of high coefficient of variation20 followed by hierarchical clustering, we identified 8 unique gene expression-based patient cluster groups that were labeled H1 through H8 (Figure 1A). Interestingly, whereas cluster H1 contained 20 of 21 cases with an MLL translocation and cluster H2 contained all 23 cases with a t(1;19)/TCF3-PBX1, the remaining 6 clusters (H3-H8) were unique and lacked association with any known recurring cytogenetic abnormality (Table 1; Figure 1A). Alternatively, using probe sets selected by 2 unsupervised methods designed to first find “outlier” genes (COPA13-15 and ROSE; probe lists/genes provided in supplemental Table 7A) followed by hierarchical clustering, all of the same patient cluster groups were identified using ROSE (R1-R8), whereas COPA (C1-C3, C5-C8) identified all patient cluster groups with the sole exception of cluster 4 (Figure 1B-C; Table 1). The degree of overlap across these 3 unsupervised clustering methods was highly significant (Table 2). The membership of the patient cluster groups defined by HC and ROSE was the most similar (93.2% identical); however, all pairwise comparisons were approximately 90% identical (Table 2). Even with no cluster 4 identified by COPA, the consensus overlap of all 3 methods was 86.5%. This is particularly noteworthy because only 37% of the clustering probe sets were shared by all 3 methods (supplemental Table 7B).
Hierarchical clustering identifies 8 cluster groups in high-risk ALL. Hierarchical clustering using 254 genes (provided in supplemental Table 7A) was used to identify clusters of patients with shared patterns of gene expression. Rows indicate 207 high-risk ALL patients from COG P9906; and columns, 254 probe sets. Shades of red represent expression levels higher than the median; and green, levels lower than the median. The cluster groups are numbered and prefixed by their method of probe set selection: H indicates high CV; C, COPA; and R, ROSE. (A) HC method for selection of probe sets. (B) COPA selection of probe sets. (C) ROSE selection of probe sets.
Hierarchical clustering identifies 8 cluster groups in high-risk ALL. Hierarchical clustering using 254 genes (provided in supplemental Table 7A) was used to identify clusters of patients with shared patterns of gene expression. Rows indicate 207 high-risk ALL patients from COG P9906; and columns, 254 probe sets. Shades of red represent expression levels higher than the median; and green, levels lower than the median. The cluster groups are numbered and prefixed by their method of probe set selection: H indicates high CV; C, COPA; and R, ROSE. (A) HC method for selection of probe sets. (B) COPA selection of probe sets. (C) ROSE selection of probe sets.
Association of clinical and outcome features with gene expression cluster groups
| . | Method* . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | Sum . | P† . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of cases per cluster group | H | 20 | 23 | 8 | 11 | 9 | 19 | 95 | 22 | 207 | — |
| C | 20 | 23 | 10 | — | 11 | 21 | 102 | 20 | 207 | — | |
| R | 21 | 23 | 12 | 14 | 10 | 21 | 82 | 24 | 207 | — | |
| Median age, years | H | 6.9 | 13.1 | 13.8 | 14.2 | 14. 7 | 14.5 | 11.4 | 13.8 | 13.1 | .002 |
| C | 6.9 | 13.1 | 15.2 | — | 14.7 | 14.5 | 11.7 | 14.3 | 13.1 | <.001 | |
| R | 4.7 | 13.1 | 15.2 | 14.3 | 14.5 | 14.5 | 7.8 | 14.1 | 13.1 | <.001 | |
| Sex (male) | H | 11/20 | 11/23 | 4/8 | 10/11 | 7/9 | 15/19 | 64/95 | 15/22 | 137/207 | .165 |
| C | 11/20 | 11/23 | 5/10 | — | 8/11 | 17/21 | 71/102 | 14/20 | 137/207 | .196 | |
| R | 11/21 | 11/23 | 6/12 | 13/14 | 8/10 | 17/21 | 54/82 | 17/24 | 137/207 | .043 | |
| Hispanic ethnicity | H | 3/20 | 6/23 | 2/8 | 2/11 | 0/8 | 3/18 | 22/95 | 13/22 | 51/205 | .018 |
| C | 3/20 | 6/23 | 2/10 | — | 0/10 | 3/20 | 25/102 | 12/20 | 51/205 | .008 | |
| R | 4/21 | 6/23 | 2/12 | 3/14 | 0/9 | 3/20 | 18/82 | 15/24 | 51/205 | .004 | |
| MLL rearrangement | H | 20/20 | 0/23 | 0/8 | 0/11 | 0/9 | 0/19 | 1/95 | 0/22 | 21/207 | < .001 |
| C | 20/20 | 0/23 | 0/10 | — | 0/11 | 0/21 | 1/102 | 0/20 | 21/207 | <.001 | |
| R | 21/21 | 0/23 | 0/12 | 0/14 | 0/10 | 0/21 | 0/82 | 0/24 | 21/207 | <.001 | |
| TCF3-PBX1 | H | 0/20 | 23/23 | 0/8 | 0/11 | 0/9 | 0/19 | 0/95 | 0/22 | 23/207 | < .001 |
| C | 0/20 | 23/23 | 0/10 | — | 0/11 | 0/21 | 0/102 | 0/20 | 23/207 | <.001 | |
| R | 0/21 | 23/23 | 0/12 | 0/14 | 0/10 | 0/21 | 0/82 | 0/24 | 23/207 | <.001 | |
| Positive MRD (day 29) | H | 8/16 | 0/20 | 0/7 | 2/11 | 7/9 | 6/19 | 27/88 | 17/21 | 67/191 | < .001 |
| C | 9/17 | 0/20 | 1/9 | — | 8/11 | 6/21 | 26/94 | 17/19 | 67/191 | <.001 | |
| R | 9/17 | 0/20 | 1/11 | 3/14 | 8/10 | 6/21 | 21/75 | 19/23 | 67/191 | <.001 | |
| WBC, × 103/μL, median | H | 129.4 | 67.2 | 139.0 | 13.3 | 32.6 | 31.4 | 59.9 | 197.5 | 62.3 | <.001 |
| C | 129.4 | 67.2 | 33.5 | — | 32.6 | 26.0 | 52.5 | 158.3 | 62.3 | .028 | |
| R | 125.8 | 67.2 | 49.6 | 9.2 | 31.5 | 26.0 | 68.8 | 153.8 | 62.3 | <.001 | |
| 4-year RFS, ± SE | H | 65.0 ± 10.7 | 73.9 ± 9.2 | 75.0 ± 15.3 | 58.2 ± 16.9 | 88.9 ± 10.5 | 94.1 ± 5.7 | 67.4 ± 5.1 | 23.0 ± 10.3 | 66.3 ± 3.5 | — |
| C | 70.0 ± 10.3 | 73.9 ± 9.2 | 70.0 ± 14.5 | — | 78.7 ± 13.4 | 94.7 ± 5.1 | 66.4 ± 5.0 | 15.1 ± 9.3 | 66.3 ± 3.5 | — | |
| R | 66.7 ± 10.3 | 73.9 ± 9.2 | 72.7 ± 13.4 | 75.0 ± 12.9 | 78.7 ± 13.4 | 94.7 ± 5.1 | 66.2 ± 5.5 | 21.0 ± 9.5 | 66.3 ± 3.5 | — | |
| Log-rank P for RFS‡ | H | .722 | .409 | .582 | .930 | .185 | .018 | .993 | <.001 | — | — |
| C | .808 | .409 | .788 | — | .364 | .010 | .944 | <.001 | — | — | |
| R | .881 | .409 | .615 | .259 | .366 | .010 | .680 | <.001 | — | — | |
| Hazard ratio for RFS‡ | H | 1.152 | 0.704 | 0.675 | 1.046 | 0.286 | 0.133 | 0.998 | 3.491 | — | — |
| C | 0.901 | 0.704 | 0.853 | — | 0.527 | 0.117 | 1.017 | 4.382 | — | — | |
| R | 1.060 | 0.704 | 0.744 | 0.520 | 0.528 | 0.117 | 1.110 | 3.878 | — | — |
| . | Method* . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | Sum . | P† . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of cases per cluster group | H | 20 | 23 | 8 | 11 | 9 | 19 | 95 | 22 | 207 | — |
| C | 20 | 23 | 10 | — | 11 | 21 | 102 | 20 | 207 | — | |
| R | 21 | 23 | 12 | 14 | 10 | 21 | 82 | 24 | 207 | — | |
| Median age, years | H | 6.9 | 13.1 | 13.8 | 14.2 | 14. 7 | 14.5 | 11.4 | 13.8 | 13.1 | .002 |
| C | 6.9 | 13.1 | 15.2 | — | 14.7 | 14.5 | 11.7 | 14.3 | 13.1 | <.001 | |
| R | 4.7 | 13.1 | 15.2 | 14.3 | 14.5 | 14.5 | 7.8 | 14.1 | 13.1 | <.001 | |
| Sex (male) | H | 11/20 | 11/23 | 4/8 | 10/11 | 7/9 | 15/19 | 64/95 | 15/22 | 137/207 | .165 |
| C | 11/20 | 11/23 | 5/10 | — | 8/11 | 17/21 | 71/102 | 14/20 | 137/207 | .196 | |
| R | 11/21 | 11/23 | 6/12 | 13/14 | 8/10 | 17/21 | 54/82 | 17/24 | 137/207 | .043 | |
| Hispanic ethnicity | H | 3/20 | 6/23 | 2/8 | 2/11 | 0/8 | 3/18 | 22/95 | 13/22 | 51/205 | .018 |
| C | 3/20 | 6/23 | 2/10 | — | 0/10 | 3/20 | 25/102 | 12/20 | 51/205 | .008 | |
| R | 4/21 | 6/23 | 2/12 | 3/14 | 0/9 | 3/20 | 18/82 | 15/24 | 51/205 | .004 | |
| MLL rearrangement | H | 20/20 | 0/23 | 0/8 | 0/11 | 0/9 | 0/19 | 1/95 | 0/22 | 21/207 | < .001 |
| C | 20/20 | 0/23 | 0/10 | — | 0/11 | 0/21 | 1/102 | 0/20 | 21/207 | <.001 | |
| R | 21/21 | 0/23 | 0/12 | 0/14 | 0/10 | 0/21 | 0/82 | 0/24 | 21/207 | <.001 | |
| TCF3-PBX1 | H | 0/20 | 23/23 | 0/8 | 0/11 | 0/9 | 0/19 | 0/95 | 0/22 | 23/207 | < .001 |
| C | 0/20 | 23/23 | 0/10 | — | 0/11 | 0/21 | 0/102 | 0/20 | 23/207 | <.001 | |
| R | 0/21 | 23/23 | 0/12 | 0/14 | 0/10 | 0/21 | 0/82 | 0/24 | 23/207 | <.001 | |
| Positive MRD (day 29) | H | 8/16 | 0/20 | 0/7 | 2/11 | 7/9 | 6/19 | 27/88 | 17/21 | 67/191 | < .001 |
| C | 9/17 | 0/20 | 1/9 | — | 8/11 | 6/21 | 26/94 | 17/19 | 67/191 | <.001 | |
| R | 9/17 | 0/20 | 1/11 | 3/14 | 8/10 | 6/21 | 21/75 | 19/23 | 67/191 | <.001 | |
| WBC, × 103/μL, median | H | 129.4 | 67.2 | 139.0 | 13.3 | 32.6 | 31.4 | 59.9 | 197.5 | 62.3 | <.001 |
| C | 129.4 | 67.2 | 33.5 | — | 32.6 | 26.0 | 52.5 | 158.3 | 62.3 | .028 | |
| R | 125.8 | 67.2 | 49.6 | 9.2 | 31.5 | 26.0 | 68.8 | 153.8 | 62.3 | <.001 | |
| 4-year RFS, ± SE | H | 65.0 ± 10.7 | 73.9 ± 9.2 | 75.0 ± 15.3 | 58.2 ± 16.9 | 88.9 ± 10.5 | 94.1 ± 5.7 | 67.4 ± 5.1 | 23.0 ± 10.3 | 66.3 ± 3.5 | — |
| C | 70.0 ± 10.3 | 73.9 ± 9.2 | 70.0 ± 14.5 | — | 78.7 ± 13.4 | 94.7 ± 5.1 | 66.4 ± 5.0 | 15.1 ± 9.3 | 66.3 ± 3.5 | — | |
| R | 66.7 ± 10.3 | 73.9 ± 9.2 | 72.7 ± 13.4 | 75.0 ± 12.9 | 78.7 ± 13.4 | 94.7 ± 5.1 | 66.2 ± 5.5 | 21.0 ± 9.5 | 66.3 ± 3.5 | — | |
| Log-rank P for RFS‡ | H | .722 | .409 | .582 | .930 | .185 | .018 | .993 | <.001 | — | — |
| C | .808 | .409 | .788 | — | .364 | .010 | .944 | <.001 | — | — | |
| R | .881 | .409 | .615 | .259 | .366 | .010 | .680 | <.001 | — | — | |
| Hazard ratio for RFS‡ | H | 1.152 | 0.704 | 0.675 | 1.046 | 0.286 | 0.133 | 0.998 | 3.491 | — | — |
| C | 0.901 | 0.704 | 0.853 | — | 0.527 | 0.117 | 1.017 | 4.382 | — | — | |
| R | 1.060 | 0.704 | 0.744 | 0.520 | 0.528 | 0.117 | 1.110 | 3.878 | — | — |
— indicates not applicable.
H indicates gene expression cluster groups determined by selection of genes using high CV and standard hierarchical clustering (HC); C, COPA; and R, ROSE.
All P values are calculated for Fisher exact test (all variables except age and WBC) or Kruskal-Wallis rank-sum test (age and WBC) using R (Version 2.9.1, survival and stats packages).
Log-rank P values and HRs calculated separately for each cluster using R (Version 2.9.1, stats package).
Comparison of cluster group membership
| . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | Overall identity, % . |
|---|---|---|---|---|---|---|---|---|---|
| HC vs COPA | 19 | 23 | 8 | NA | 9 | 19 | 88 | 19 | 89.4 |
| HC vs ROSE | 20 | 23 | 8 | 10 | 9 | 19 | 82 | 22 | 93.2 |
| COPA vs ROSE | 20 | 23 | 10 | NA | 10 | 21 | 82 | 20 | 89.9 |
| HC vs COPA vs ROSE | 19 | 23 | 8 | NA | 9 | 19 | 82 | 19 | 86.5 |
| . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | Overall identity, % . |
|---|---|---|---|---|---|---|---|---|---|
| HC vs COPA | 19 | 23 | 8 | NA | 9 | 19 | 88 | 19 | 89.4 |
| HC vs ROSE | 20 | 23 | 8 | 10 | 9 | 19 | 82 | 22 | 93.2 |
| COPA vs ROSE | 20 | 23 | 10 | NA | 10 | 21 | 82 | 20 | 89.9 |
| HC vs COPA vs ROSE | 19 | 23 | 8 | NA | 9 | 19 | 82 | 19 | 86.5 |
NA indicates not applicable; and HC, gene expression cluster groups determined by selection of genes using high CV and standard hierarchical clustering.
In addition to the significant association (P < .001) observed between clusters 1 and 2 and MLL translocations or t(1;19)/TCF3-PBX1, respectively, significant associations were seen between several clinical and outcome features and the other unique cluster groups, including age (P < .001-.002), Hispanic ethnicity (P = .004-.018), end-induction MRD (P < .001), and RFS (Table 1; Figure 2). Of particular note was the significant variation in RFS among the clusters, with 2 of the unique clusters (clusters 6 and 8) having statistically different survivals compared with the overall cohort by independent log-rank analysis using all 3 clustering methods (cluster 6: P = .010-.018, hazard ratio [HR] = 0.117-0.133;cluster 8: P < .001, HR = 3.491-4.382) (Table 1; Figure 2). In contrast to an overall 4-year RFS of 66.3% plus or minus 3.5% in the entire cohort of 207 ALL patients, patients who were clustered in cluster 6 by each method had a significantly superior outcome, with 4-year RFS ranging from 94.1% plus or minus 5.7% to 94.7% plus or minus 5.1% (Table 1; Figure 2). COPA and ROSE identified the largest patient clusters (21 members) for this cluster group with the best RFS. In contrast to patients in cluster 6, patients who were in cluster 8 had a 4-year RFS that ranged from 15.1% plus or minus 9.3% using COPA to 23.0% plus or minus 10.3% for HC (Table 1; Figure 2). ROSE cluster R8 was the largest, containing 24 members, with a 4-year RFS of 21.0% plus or minus 9.5%. The time to relapse also varied among the cluster groups. Although all relapses in clusters 1, 2, and 6 occurred within the first 3 years, patients in the remaining clusters, particularly in cluster 8, continued to experience relapses in years 3 to 5. Among all cluster groups, patients in cluster 8 were also distinguished by the highest frequency of MRD positivity at the end of induction therapy (81.0%-89.5% of cases) and self-reported Hispanic/Latino ethnicity (59.1%-62.5%).
RFS in gene expression cluster groups. RFS is shown for each of the high CV clusters (A), COPA clusters (B), and ROSE clusters (C). Only the H6, C6, and R6 clusters (curves shown in blue) have a significantly better outcome compared with the entire cohort (dense line), whereas the H8, C8, and R8 clusters (curves shown in red) have a significantly poorer RFS. Hazard ratios and P values are shown in the bottom left of each panel.
RFS in gene expression cluster groups. RFS is shown for each of the high CV clusters (A), COPA clusters (B), and ROSE clusters (C). Only the H6, C6, and R6 clusters (curves shown in blue) have a significantly better outcome compared with the entire cohort (dense line), whereas the H8, C8, and R8 clusters (curves shown in red) have a significantly poorer RFS. Hazard ratios and P values are shown in the bottom left of each panel.
Given the high degree of concordance between the clustering methods, ROSE was selected as the reference method for the remaining analyses. Provided in Table 3 are the 113 “outlier” probe sets that overlapped between the 254 probe sets used for ROSE clustering (full list provided supplemental Table 7A) and those probe sets that were among the top 100 rank-ordered probe sets that defined each ROSE cluster group (the full rank-ordered lists are provided in Supplemental data). The majority of the outlier probe sets/genes that defined cluster R1, which contained all of the patients with MLL translocations, included MEIS1, PROM1, RUNX2, and members of the HOX gene family, all of which have been frequently reported as characteristic of ALL cases with MLL translocations.26 Several other interesting outlier genes were also found associated with cluster R1/MLL translocations (Table 3; supplemental Table 9), such as CTGF, which was previously reported to be associated with a poor outcome in adult ALL27 ; the correlation between CTGF expression and MLL translocations was not previously reported. Outlier genes distinguishing cluster R2, containing all 23 cases with t(1;19)/TCF3-PBX1, included PBX1 itself, which is directly involved in the underlying t(1;19) translocation. Because several of the outlier genes uniquely associated with clusters R1 and R2 are involved in the underlying recurrent cytogenetic abnormalities associated with these cluster groups, we postulated that the outlier genes associated with the other ROSE clusters were also interesting candidates for genes, which may be involved in novel underlying genetic abnormalities, or, genes whose expression might be perturbed by novel genetic abnormalities. Consistent with this hypothesis was the presence of several notable outlier genes that defined cluster R8 (including GAB1, MUC4, PON2, GPR110, SEMA6, and SERPINB9; supplemental Tables 15, 17, and 18). High expression of these genes was previously reported to be predictive of a poor outcome in t(9;22)/BCR-ABL1 ALL,28 yet the ALL cases in R8 lacked the classic t(9;22)/BCR-ABL1. This “activated kinase” or “BCR-ABL1-like” signature found by our group5 and Den Boer et al7 has been reported to be associated with IKAROS/IKZF1 deletions and poor outcomes in pediatric ALL. As discussed in “Correlation of acquired JAK mutations with ROSE clusters,” this discovery led us to sequence tyrosine kinases in this high-risk ALL cohort, leading to the discovery of JAK family mutations in high-risk ALL.6 Also as discussed in “Correlation of genome-wide DNA copy number changes with ROSE clusters,” the recognition of CRLF2 as an outlier gene in cluster R8, in concert with the observation of DNA copy number variations in the region of CRLF2, led to our discovery of novel genomic rearrangements of CLRF2, leading to marked elevations of CRLF2 expression in high-risk ALL,8,29 a discovery also recently reported by other groups.30,31 These discoveries demonstrate the power of outlier analysis methods for the identification of genes involved in novel recurring genetic abnormalities.
ROSE outlier genes/probe sets used for clustering and definition of unique ROSE cluster groups
| R1 . | R2 . | R3 . | R4 . | R5 . | R6 . | R7 . | R8 . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Probe set . | Gene symbol . | Probe set . | Gene symbol . | Probe set . | Gene symbol . | Probe set . | Gene symbol . | Probe set . | Gene symbol . | Probe set . | Gene symbol . | Probe set . | Gene symbol . | Probe set . | Gene symbol . |
| 220416_at | ATP8B4 | 227441_s_at | ANKS1B | 213808_at | ADAM23* | 203949_at | MPO | 212062_at | ATP9A | 242457_at | — | 219837_s_at | CYTL1 | 229975_at | BMPR1B |
| 219463_at | C20orf103 | 227440_at | ANKS1B | 203865_s_at | ADARB1 | 203948_s_at | MPO | 228297_at | CNN3* | 241535_at | — | 212192_at | KCTD12 | 208303_s_at | CRLF2 |
| 205899_at | CCNA1 | 227439_at | ANKS1B | 230128_at | IGL@ | 202273_at | PDGFRB | 209604_s_at | GATA3 | 204066_s_at | AGAP1 | 238689_at | GPR110 | ||
| 209101_at | CTGF | 243533_x_at | ANKS1B* | 231513_at | KCNJ2* | 203476_at | TPBG | 213362_at | PTPRD | 240758_at | AGAP1* | 235988_at | GPR110 | ||
| 218468_s_at | GREM1 | 234261_at | ANKS1B* | 203726_s_at | LAMA3 | 229661_at | SALL4 | 233225_at | AGAP1* | 236489_at | GPR110 | ||||
| 213150_at | HOXA10 | 202207_at | ARL4C | 232914_s_at | SYTL2 | 213258_at | TFPI | 219470_x_at | CCNJ | 207651_at | GPR171 | ||||
| 235521_at | HOXA3 | 202206_at | ARL4C | 225496_s_at | SYTL2 | 210665_at | TFPI | 203921_at | CHST2 | 212592_at | IGJ | ||||
| 213844_at | HOXA5 | 212077_at | CALD1 | 210664_s_at | TFPI | 206756_at | CHST7 | 213371_at | LDB3 | ||||||
| 214651_s_at | HOXA9 | 223786_at | CHST6 | 1552398_a_at | CLEC12A/B | 217110_s_at | MUC4 | ||||||||
| 209905_at | HOXA9 | 205489_at | CRYM | 231166_at | GPR155 | 217109_at | MUC4 | ||||||||
| 218847_at | IGF2BP2 | 206070_s_at | EPHA3 | 202409_at | IGF2 | 204895_x_at | MUC4 | ||||||||
| 201105_at | LGALS1 | 201579_at | FAT1 | 215177_s_at | ITGA6 | ||||||||||
| 1557534_at | LOC339862 | 231455_at | FLJ42418 | 201656_at | ITGA6 | ||||||||||
| 202890_at | MAP7 | 239657_x_at | FOXO6 | 211340_s_at | MCAM | ||||||||||
| 242172_at | MEIS1 | 235666_at | ITGA8? | 210869_s_at | MCAM | ||||||||||
| 204069_at | MEIS1 | 235911_at | K03200* | 215692_s_at | MPPED2 | ||||||||||
| 1559477_s_at | MEIS1 | 213005_s_at | KANK1 | 205413_at | MPPED2 | ||||||||||
| 204304_s_at | PROM1 | 208567_s_at | KCNJ12 | 202336_s_at | PAM | ||||||||||
| 202976_s_at | RHOBTB3 | 210150_s_at | LAMA5 | 228863_at | PCDH17 | ||||||||||
| 232231_at | RUNX2 | 228262_at | MAP7D2 | 227289_at | PCDH17 | ||||||||||
| 226415_at | VAT1L | 206028_s_at | MERTK | 205656_at | PCDH17 | ||||||||||
| 231899_at | ZC3H12C | 204114_at | NID2 | 230537_at | PCDH17 | ||||||||||
| 212151_at | PBX1 | 203335_at | PHYH | ||||||||||||
| 212148_at | PBX1 | 203329_at | PTPRM | ||||||||||||
| 205253_at | PBX1 | 1555579_s_at | PTPRM | ||||||||||||
| 227949_at | PHACTR3 | 220059_at | STAP1 | ||||||||||||
| 202178_at | PRKCZ | 1554343_a_at | STAP1 | ||||||||||||
| 242385_at | RORB | ||||||||||||||
| 231040_at | RORB? | ||||||||||||||
| 46665_at | SEMA4C | ||||||||||||||
| 206181_at | SLAMF1 | ||||||||||||||
| 225483_at | VPS26B | ||||||||||||||
| R1 . | R2 . | R3 . | R4 . | R5 . | R6 . | R7 . | R8 . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Probe set . | Gene symbol . | Probe set . | Gene symbol . | Probe set . | Gene symbol . | Probe set . | Gene symbol . | Probe set . | Gene symbol . | Probe set . | Gene symbol . | Probe set . | Gene symbol . | Probe set . | Gene symbol . |
| 220416_at | ATP8B4 | 227441_s_at | ANKS1B | 213808_at | ADAM23* | 203949_at | MPO | 212062_at | ATP9A | 242457_at | — | 219837_s_at | CYTL1 | 229975_at | BMPR1B |
| 219463_at | C20orf103 | 227440_at | ANKS1B | 203865_s_at | ADARB1 | 203948_s_at | MPO | 228297_at | CNN3* | 241535_at | — | 212192_at | KCTD12 | 208303_s_at | CRLF2 |
| 205899_at | CCNA1 | 227439_at | ANKS1B | 230128_at | IGL@ | 202273_at | PDGFRB | 209604_s_at | GATA3 | 204066_s_at | AGAP1 | 238689_at | GPR110 | ||
| 209101_at | CTGF | 243533_x_at | ANKS1B* | 231513_at | KCNJ2* | 203476_at | TPBG | 213362_at | PTPRD | 240758_at | AGAP1* | 235988_at | GPR110 | ||
| 218468_s_at | GREM1 | 234261_at | ANKS1B* | 203726_s_at | LAMA3 | 229661_at | SALL4 | 233225_at | AGAP1* | 236489_at | GPR110 | ||||
| 213150_at | HOXA10 | 202207_at | ARL4C | 232914_s_at | SYTL2 | 213258_at | TFPI | 219470_x_at | CCNJ | 207651_at | GPR171 | ||||
| 235521_at | HOXA3 | 202206_at | ARL4C | 225496_s_at | SYTL2 | 210665_at | TFPI | 203921_at | CHST2 | 212592_at | IGJ | ||||
| 213844_at | HOXA5 | 212077_at | CALD1 | 210664_s_at | TFPI | 206756_at | CHST7 | 213371_at | LDB3 | ||||||
| 214651_s_at | HOXA9 | 223786_at | CHST6 | 1552398_a_at | CLEC12A/B | 217110_s_at | MUC4 | ||||||||
| 209905_at | HOXA9 | 205489_at | CRYM | 231166_at | GPR155 | 217109_at | MUC4 | ||||||||
| 218847_at | IGF2BP2 | 206070_s_at | EPHA3 | 202409_at | IGF2 | 204895_x_at | MUC4 | ||||||||
| 201105_at | LGALS1 | 201579_at | FAT1 | 215177_s_at | ITGA6 | ||||||||||
| 1557534_at | LOC339862 | 231455_at | FLJ42418 | 201656_at | ITGA6 | ||||||||||
| 202890_at | MAP7 | 239657_x_at | FOXO6 | 211340_s_at | MCAM | ||||||||||
| 242172_at | MEIS1 | 235666_at | ITGA8? | 210869_s_at | MCAM | ||||||||||
| 204069_at | MEIS1 | 235911_at | K03200* | 215692_s_at | MPPED2 | ||||||||||
| 1559477_s_at | MEIS1 | 213005_s_at | KANK1 | 205413_at | MPPED2 | ||||||||||
| 204304_s_at | PROM1 | 208567_s_at | KCNJ12 | 202336_s_at | PAM | ||||||||||
| 202976_s_at | RHOBTB3 | 210150_s_at | LAMA5 | 228863_at | PCDH17 | ||||||||||
| 232231_at | RUNX2 | 228262_at | MAP7D2 | 227289_at | PCDH17 | ||||||||||
| 226415_at | VAT1L | 206028_s_at | MERTK | 205656_at | PCDH17 | ||||||||||
| 231899_at | ZC3H12C | 204114_at | NID2 | 230537_at | PCDH17 | ||||||||||
| 212151_at | PBX1 | 203335_at | PHYH | ||||||||||||
| 212148_at | PBX1 | 203329_at | PTPRM | ||||||||||||
| 205253_at | PBX1 | 1555579_s_at | PTPRM | ||||||||||||
| 227949_at | PHACTR3 | 220059_at | STAP1 | ||||||||||||
| 202178_at | PRKCZ | 1554343_a_at | STAP1 | ||||||||||||
| 242385_at | RORB | ||||||||||||||
| 231040_at | RORB? | ||||||||||||||
| 46665_at | SEMA4C | ||||||||||||||
| 206181_at | SLAMF1 | ||||||||||||||
| 225483_at | VPS26B | ||||||||||||||
Correlation of genome-wide DNA copy number changes with ROSE clusters
To gain further insights into the genetic heterogeneity in high-risk ALL, we next correlated the gene expression profiles with genome-wide DNA CNA measured using single nucleotide polymorphism arrays. These CNAs were previously reported in 198 of the 207 cases studied herein,5 but we now correlate these CNAs with the novel ROSE gene expression-based cluster groups (Table 4; supplemental Table 20). As shown in Table 4, whereas certain CNAs (such as those in seen in CDKN2A/B and PAX5) were seen in many ROSE clusters, other abnormalities were more uniquely associated with a specific cluster. As expected, 1q gain and TCF3 loss were highly associated with cluster R2 containing TCF3-PBX1 cases, reflecting the unbalanced t(1;19) translocations that lead to duplication of chromosome 1 telomeric to PBX1 and deletion of chromosome 19 telomeric to TCF3. ERG deletions, as previously described by Mullighan et al,32 were seen almost exclusively in cluster R6. EBF1 deletions were seen only in clusters R7 and R8. Although IKAROS/IKZF1 deletions, which were previously reported to be associated with a poor outcome in ALL,5 were found in several cluster groups, even in cluster R6, which had an extremely good outcome (Table 1; Figure 2), they were particularly prevalent and significantly associated with cluster R8 (Table 4), which had an extremely poor outcome (Table 1; Figure 2). Interestingly, however, ALL patients who had IKAROS/IKZF1 deletions and who were in cluster 8 had a poorer RFS than the remaining ALL patients in the cohort who had IKAROS/IKZF1 deletions but were not clustered in R8 (P = .008; supplemental Figure 3), implying that the constellation of genetic abnormalities associated with cluster R8 must contribute to the worse overall outcome in these patients. Other DNA deletions significantly associated with the R8 cluster included RAG1-2, NUP160-PTPRJ, IL3RA-CSF2RA, C20orf94, and ADD3. The findings of CRLF2 as an outlier gene and DNA copy number variations in the pseudoautosomal region (PAR1) of X and Y immediately adjacent to CRLF2 (Table 4; the IL3RA-CSF2RA deletion) led our group8,29 and Russell et al30 to recently discover novel genomic rearrangements (IGH-CRLF2 and P2RY8-CRLF2 translocations), resulting in activated expression of wild-type CRLF2 in ALL, further demonstrating the power of identification of outlier genes in the discovery of novel underlying genetic abnormalities in cancer cells. Of the 30 CRLF2 genomic rearrangements discovered in this cohort of 207 high-risk ALL cases, 18 were in cluster R8, 11 were in R7, and the remaining case was in R4 (Table 4).
Correlation of genome-wide DNA CNAs and acquired mutations or genomic rearrangements with ROSE gene-expression cluster groups
| . | R1 . | R2 . | R3 . | R4 . | R5 . | R6 . | R7 . | R8 . | Total . | FET (P)* . | Comments . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases evaluated | 20 | 22 | 12 | 13 | 10 | 21 | 76 | 24 | 198 | ||
| 1q (gain)† | 0 | 14 | 1 | 1 | 0 | 0 | 1 | 0 | 17 | < .0001 | R2 contains TCF3-PBX1 |
| IKZF1 | 1 | 0 | 0 | 3 | 3 | 6 | 24 | 22 | 59 | < .0001 | |
| CDKN2A/B | 4 | 9 | 11 | 11 | 1 | 5 | 40 | 15 | 96 | < .0001 | |
| TCF3 | 0 | 14 | 0 | 0 | 2 | 2 | 2 | 0 | 20 | < .0001 | R2 contains TCF3-PBX1 |
| ERG | 0 | 0 | 0 | 1 | 0 | 8 | 0 | 0 | 9 | < .0001 | |
| VPREB1 | 0 | 0 | 0 | 5 | 1 | 8 | 23 | 14 | 51 | < .0001 | |
| B-cell pathway | 5 | 17 | 5 | 12 | 4 | 12 | 54 | 23 | 132 | < .0001 | |
| B pathway w/VPREB1 | 5 | 17 | 5 | 12 | 5 | 14 | 56 | 24 | 138 | < .0001 | |
| PAX5 | 1 | 9 | 4 | 11 | 0 | 3 | 28 | 7 | 63 | < .0001 | |
| EBF1 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 9 | 13 | .0001 | |
| TBL1XR1 | 0 | 0 | 3 | 0 | 1 | 1 | 0 | 0 | 5 | .0005 | |
| NUP160-PTPRJ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 4 | .0028 | |
| ETV6 | 1 | 0 | 2 | 2 | 4 | 1 | 14 | 0 | 24 | .0055 | |
| IL3RA-CSF2RA | 0 | 0 | 0 | 0 | 1 | 0 | 6 | 7 | 14 | .0064 | High CRLF2 expression |
| DMD | 0 | 5 | 1 | 0 | 2 | 3 | 3 | 0 | 14 | .0109 | |
| C20orf94 | 0 | 0 | 0 | 1 | 1 | 0 | 7 | 7 | 16 | .0102 | |
| RAG1/2 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 5 | 7 | .0144 | |
| ADD3 | 0 | 1 | 0 | 2 | 0 | 0 | 7 | 7 | 17 | .0156 | |
| NF1 | 1 | 1 | 0 | 0 | 2 | 0 | 0 | 1 | 5 | .0269 | |
| ARMC2-SESN1 | 0 | 2 | 0 | 1 | 2 | 0 | 3 | 5 | 13 | .0297 | |
| JAK1 (mutation) | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 3 | .9448 | High CRLF2 expression |
| JAK2 (mutation) | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 11 | 16 | < .0001 | High CRLF2 expression |
| CRLF2 rearrangement: IGH@-CRLF2 | 0 | 0 | 0 | 1 | 0 | 0 | 7 | 11 | 19 | < .0001 | High CRLF2 expression |
| CRLF2 rearrangement: P2RY8-CRLF2 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 7 | 11 | .0041 | High CRLF2 expression |
| . | R1 . | R2 . | R3 . | R4 . | R5 . | R6 . | R7 . | R8 . | Total . | FET (P)* . | Comments . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases evaluated | 20 | 22 | 12 | 13 | 10 | 21 | 76 | 24 | 198 | ||
| 1q (gain)† | 0 | 14 | 1 | 1 | 0 | 0 | 1 | 0 | 17 | < .0001 | R2 contains TCF3-PBX1 |
| IKZF1 | 1 | 0 | 0 | 3 | 3 | 6 | 24 | 22 | 59 | < .0001 | |
| CDKN2A/B | 4 | 9 | 11 | 11 | 1 | 5 | 40 | 15 | 96 | < .0001 | |
| TCF3 | 0 | 14 | 0 | 0 | 2 | 2 | 2 | 0 | 20 | < .0001 | R2 contains TCF3-PBX1 |
| ERG | 0 | 0 | 0 | 1 | 0 | 8 | 0 | 0 | 9 | < .0001 | |
| VPREB1 | 0 | 0 | 0 | 5 | 1 | 8 | 23 | 14 | 51 | < .0001 | |
| B-cell pathway | 5 | 17 | 5 | 12 | 4 | 12 | 54 | 23 | 132 | < .0001 | |
| B pathway w/VPREB1 | 5 | 17 | 5 | 12 | 5 | 14 | 56 | 24 | 138 | < .0001 | |
| PAX5 | 1 | 9 | 4 | 11 | 0 | 3 | 28 | 7 | 63 | < .0001 | |
| EBF1 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 9 | 13 | .0001 | |
| TBL1XR1 | 0 | 0 | 3 | 0 | 1 | 1 | 0 | 0 | 5 | .0005 | |
| NUP160-PTPRJ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 4 | .0028 | |
| ETV6 | 1 | 0 | 2 | 2 | 4 | 1 | 14 | 0 | 24 | .0055 | |
| IL3RA-CSF2RA | 0 | 0 | 0 | 0 | 1 | 0 | 6 | 7 | 14 | .0064 | High CRLF2 expression |
| DMD | 0 | 5 | 1 | 0 | 2 | 3 | 3 | 0 | 14 | .0109 | |
| C20orf94 | 0 | 0 | 0 | 1 | 1 | 0 | 7 | 7 | 16 | .0102 | |
| RAG1/2 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 5 | 7 | .0144 | |
| ADD3 | 0 | 1 | 0 | 2 | 0 | 0 | 7 | 7 | 17 | .0156 | |
| NF1 | 1 | 1 | 0 | 0 | 2 | 0 | 0 | 1 | 5 | .0269 | |
| ARMC2-SESN1 | 0 | 2 | 0 | 1 | 2 | 0 | 3 | 5 | 13 | .0297 | |
| JAK1 (mutation) | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 3 | .9448 | High CRLF2 expression |
| JAK2 (mutation) | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 11 | 16 | < .0001 | High CRLF2 expression |
| CRLF2 rearrangement: IGH@-CRLF2 | 0 | 0 | 0 | 1 | 0 | 0 | 7 | 11 | 19 | < .0001 | High CRLF2 expression |
| CRLF2 rearrangement: P2RY8-CRLF2 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 7 | 11 | .0041 | High CRLF2 expression |
P values are derived from Fisher exact test.
All abnormalities are deletions or chromosomal losses unless otherwise indicated.
Correlation of acquired JAK mutations with ROSE clusters
The discovery of the activated kinase or BCR-ABL1-like gene expression signature in virtually all cases in cluster R8 and in some cases of cluster R7 led us to sequence tyrosine kinases in the 198 cases with available DNA samples in the P9906 ALL cohort.6 Table 4 provides the correlation of JAK mutation status with each ROSE cluster group. Of these 198 patients, 19 had mutations of either JAK1 (n = 3) or JAK2 (n = 16). There was a highly significant association of JAK1 and JAK2 mutations with cluster R8, with all 19 of the mutations being either in R8 (n = 12) or in the less tightly clustered group R7 (n = 7). As we have recently reported, nearly all of the JAK mutations occurred in patients with CRLF2 genomic rearrangements.8 Thus, patients in the R8 cluster are characterized by a constellation of genomic abnormalities (IKZF1 deletions, CRLF2 rearrangements, and JAK mutations, as well as other DNA deletions) that may contribute to their overall poor outcome.
Validation of the significance of the ROSE clusters in an independent high-risk ALL cohort
We next determined whether the unique cluster groups found in the COG P9906 high-risk ALL cases could be found in a second high-risk ALL cohort. All 3 clustering methods were thus applied to the expression profiles derived from a second independent cohort of 99 children and adolescents with high-risk ALL treated on COG CCG Trial 1961 (“Patient selection and characteristics” and Supplemental data). Although smaller than COG P9906, the COG CCG 1961 cohort was accrued using traditional NCI/Rome rather than Shuster et al criteria4 and contained a more diverse spectrum of sentinel cytogenetic lesions, including cases with t(12;21)/ETV6-AML1, BCR-ABL1, and favorable trisomies.12 As shown in Figure 3, all clustering methods identified the same 4 clusters seen in the P9906: clusters 1, 2, 6, and 8. Similar to the initial cohort, clusters 1 and 2 contained all of cases with MLL or TCF3-PBX1 translocations. Because of the smaller size of the CCG 1961 cohort, it is possible that the other 3 clusters seen in P9906 (clusters 3-5) were not detected because there simply were not enough patients with these gene expression signatures to be detected as a robust cluster. In contrast to the COG P9906 cohort, 2 new cluster groups were detected: clusters 9 and 10 (Figure 3); cluster 9 was determined to contain ALL cases with t(12;21)/ETV6-AML1 translocations, whereas cluster 10, identified using outlier methods with both COPA and ROSE, appeared to be a new unique cluster group (supplemental Table 19). As reported by others,33 ALL cases in this cohort with BCR-ABL1/t(9;22) did not tightly cluster because of their divergent expression profiles.
Hierarchical clustering identifies similar clusters in an independent high-risk ALL cohort. Hierarchical clustering using 167 probe sets (provided in supplemental Table 7A) was used to identify clusters of patients with shared patterns of gene expression in a second cohort of high-risk ALL patients previously accrued to COG Trial CCG 1961. Rows indicate 99 patients from COG CCG 1961; and columns, 167 probe sets. Shades of red represent expression levels higher than the median; and green represents levels lower than the median. The cluster groups are prefixed by their method of probe set selection: H indicates high CV; C, COPA; and R, ROSE. (A) HC method for selection of probe sets. (B) COPA selection of probe sets. (C) ROSE selection of probe sets.
Hierarchical clustering identifies similar clusters in an independent high-risk ALL cohort. Hierarchical clustering using 167 probe sets (provided in supplemental Table 7A) was used to identify clusters of patients with shared patterns of gene expression in a second cohort of high-risk ALL patients previously accrued to COG Trial CCG 1961. Rows indicate 99 patients from COG CCG 1961; and columns, 167 probe sets. Shades of red represent expression levels higher than the median; and green represents levels lower than the median. The cluster groups are prefixed by their method of probe set selection: H indicates high CV; C, COPA; and R, ROSE. (A) HC method for selection of probe sets. (B) COPA selection of probe sets. (C) ROSE selection of probe sets.
The 3 methods used for selecting probe sets yielded more divergent lists (provided in supplemental Table 7B) than the P9906 cohort, with only 25.1% of probe sets common among all 3 methods. This lower similarity was primarily the result of the difference between those probe sets identified by HC and those found by the 2 outlier methods (COPA and ROSE), which were more similar. Although the same cluster groups found in P9906 and CCG 1961 were defined by the same sets of outlier genes, the 167 genes derived for ROSE and COPA clustering (supplemental Table 7C) contained many unique genes compared with P9906, in large part because of the different composition of the CCG 1961 cohort containing ALL cases with BCR-ABL1 and ETV6-AML1 translocations.
Similar to the P9906 high-risk ALL cohort, patients from the COG CCG 1961 cohort who were in cluster 8 had very poor 4-year RFS (HR = 2.36-4.51; P = .001-.028) depending on the clustering method (Figure 4). Although only 5 patients with the features of cluster 6 were present in the CCG 1961 cohort (Figure 3), only one of these patients relapsed. Overall, these results confirm the robust nature of the outlier clustering methods, the genetic and clinical heterogeneity within high-risk ALL, and the very poor outcome consistently associated with cluster 8 gene expression profiles.
RFS in an independent high-risk ALL cohort. RFS for the 99 high-risk ALL patients on COG Trial CCG 1961 who were either clustered in cluster 8 or were in the remaining cohort using each different clustering method: HC (A), COPA (B), and ROSE (C). By each method, ALL patients clustered as H8 (A), C8 (B), or R8 (C) had a significantly worse RFS than the remaining patients in the cohort. Hazard ratios and P values are shown in the bottom left of each panel.
RFS in an independent high-risk ALL cohort. RFS for the 99 high-risk ALL patients on COG Trial CCG 1961 who were either clustered in cluster 8 or were in the remaining cohort using each different clustering method: HC (A), COPA (B), and ROSE (C). By each method, ALL patients clustered as H8 (A), C8 (B), or R8 (C) had a significantly worse RFS than the remaining patients in the cohort. Hazard ratios and P values are shown in the bottom left of each panel.
Discussion
Using 3 different unbiased, unsupervised methods to analyze and cluster gene expression profiles, we have identified 8 unique gene expression-based cluster groups among children and adolescents with high-risk B-precursor ALL in a cohort of 207 uniformly treated children accrued to COG Trial P9906. These 8 cluster groups were distinguished by high levels of expression of unique “outlier” genes, distinct DNA CNAs, variable clinical features, and significantly different rates of RFS. These studies reveal the striking biologic, genetic, and clinical heterogeneity within high-risk ALL and point to novel genes that may serve as new targets for the discovery of unique underlying recurrent genetic abnormalities as well as for improved diagnosis, risk classification, and therapy.
Particularly striking among the unique cluster groups were 2 clusters found by all methods (clusters 6 and 8) with strikingly different rates of RFS. In contrast, a 4-year RFS of 66.3% plus or minus 3.5% in the entire ALL cohort, patients in cluster 6 had a significantly superior 4-year RFS ranging from 94.1% plus or minus 5.7% to 94.7% plus or minus 5.1% depending on the clustering method (P = .010-.018; HR = 0.117-0.133). These patients were characterized by high expression of several unique “outlier” genes that distinguished this cluster (AGAP1, CCNJ, CHST2/7, CLEC12A/B, and PTPRM) and by intragenic ERG DNA deletions. Although the superior outcome in these ALL patients has not been previously reported, the expression profile of cluster group 6 is highly similar to a “novel” ALL cluster first reported by Yeoh et al,33 which has been further characterized by Mullighan et al32 Whereas only 5 patients with the cluster 6 expression signature were found in the independent validation cohort of 99 high-risk ALL patients treated on COG Trial CCG1961, only one of these patients has relapsed, further emphasizing the superior outcome of this group.
In contrast to the patients in cluster 6, the high-risk ALL patients in cluster 8 had an extremely poor survival, with 4-year RFS ranging from 15.1% plus or minus 9.3% to 23.0% plus or minus 10.3% depending on the clustering method (P < .001; HR = 3.491-4.382). A similar poor outcome was seen in the ALL patients clustered in R8 in the independent validation cohort. A particularly interesting feature of cluster 8 was the significant association with Hispanic/Latino ethnicity (P < .001). Hispanic and Native American children with ALL have been reported to have poorer outcomes than non-Hispanic white children when treated with conventional ALL therapy.3,34,35 Rather than relying on self-reported race, we have recently studied large cohorts of pediatric ALL patients from COG and St Jude Children's Research Hospital and determined the genetic ancestry of children with ALL using genome-wide single nucleotide polymorphisms and comparing genomic variation to that of reference populations. These studies have confirmed that children whose ethnicity is self-declared as “Hispanic” have high Native American genetic ancestry. (J.Y., C. Cheng, M.D., X. Cao, Y. Fan, D. Campana, W. Yang, G. Neale, N. Cox, P. Scheet, M.J.B., N. Winick, P.L. Martin, C.L.W., W.P.B., B.C., A.J.C., G.H.R., W.L.C., M. Loh, S.P.H., C.-H. Pui, W.E. Evans, M.V.R., manuscript submitted). Whether outcome disparities result from differences in disease biology, host pharmacogenetic responses to therapy, or social and behavioral factors remain to be explored. Whether children of different genetic ancestries are susceptible to the acquisition of different genetic abnormalities that predispose to ALL is also an important area for future investigation.
The extremely poor outcomes seen in the ALL patients within cluster group 8 must in part result from the unique genetic features and expression signatures that characterize this cluster. These features include expression of high levels of a distinguishing set of “outlier” genes, including BMPR1B, CRLF2, GPR110, GPR171, IGJ, LDB3, and MUC4, and several DNA copy number variations, including deletions in EBF1, NUP160-PTPRJ, IL3RA-CSF2RA (adjacent to CRLF2), C20orf94, and ADD3. Deletions of IKZF1and VPREB1 were also frequent in cluster 8, occurring in 20 of 24 and 14 of 24 R8 cases, respectively, and have been previously associated with poorer outcomes in ALL.5,7 Somewhat surprisingly, deletions in these genes were also found in cluster 6 (IKZF1: 6 of 21 cases, only one of which relapsed; VPREB1: 8 of 21 cases) associated with a superior outcome. The RFS patients with IKAROS/IKZF1 deletions who were clustered within cluster 8 were significantly worse than patients with IKZF1 deletions in the remaining cohort (P = .008), implying that overall outcome in ALL probably results from and is best predicted by a constellation of genetic abnormalities rather than a single lesion. In this regard, assays that measure the expression of genes that distinguish the novel cluster groups or application of gene expression classifiers strongly predictive of outcome discovered using supervised learning methods9 may be most useful in the clinical setting for the prospective identification of patients at very high risk of treatment failure.
The discovery of CRLF2 as an outlier gene associated with cluster 8, combined with the discovery of DNA deletions in the pseudo-autosomal region of Xp/Yp adjacent to the CRLF2 locus (IL3RA-CSF2RA) in cluster 8 patients, led to our recent discovery of novel recurring genomic alterations involving CRLF2 in high-risk ALL patients and in Down syndrome children with ALL,8,29 as also reported by other groups.30,31,36 Another distinguishing feature of cluster 8, which lacked t(9;22)/BCR-ABL1 translocations, was a gene expression signature reflective of activated tyrosine kinases, which has been referred to as the BCR-ABL1-like signature.7 Some of these genes in this signature, such as GAB1, were previously reported to be predictive of outcome and imatinib response in ALL with t(9;22)/BCR-ABL1.28 Supported by a NCI TARGET Initiative, this discovery led us to sequence several tyrosine kinases in the COG P9906 ALL cohort leading to the discovery of JAK family mutations in 12 of 24 patients in cluster 8 and in 7 patients in cluster 7.6 We used next generation sequencing methods to identify the other kinases that may be responsible for the BCR-ABL1-like signature in the remaining cluster 8 cases.11 Thus, ALL patients in cluster 8 are characterized by a constellation of genomic abnormalities (CRLF2 rearrangements, JAK mutations, IKAROS/IKZF1 deletions, BCR-ABL1-like signatures, as well as other DNA deletions) that may cooperate to promote leukemogenesis and contribute to the exceedingly poor outcome in this group. Importantly, the discovery of these new genetic abnormalities in ALL attests to the power of outlier gene expression analysis and comprehensive analysis of DNA copy number variation for the discovery of novel recurring genetic abnormalities in cancer cells. As such, we are focusing on the unique outlier genes and DNA copy number variations associated with the other novel cluster groups in this high-risk ALL cohort to discover additional novel underlying genetic abnormalities. These new genes and genetic abnormalities will not only improve diagnosis and risk classification but also serve as important new targets for therapy in a group of patients who have not adequately responded to today's intensive treatment regimens and require the development of new targeted therapies for cure.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Institutes of Health Department of Health and Human Services, National Cancer Institute Strategic Partnerships to Evaluate Cancer Gene Signatures Program (grant NCI U01 CA11476, principal investigator C.L.W.; and grant NCI U10CA98543 Supporting the Children's Oncology Group and Statistical Center, principal investigator G.H.R.), the American Lebanese Syrian Associated Charities (J.Y.), the National Childhood Cancer Foundation, COG (cell banking grant U24 CA114766) (G.H.R.), and a Leukemia & Lymphoma Society Specialized Center of Research (program grant 7388-06) (principal investigator C.L.W.). Sandia is a multiprogram laboratory operated by Sandia Corporation, a Lockheed Martin Company, for the U.S. Department of Energy's National Nuclear Security Administration (contract DE-AC04-94AL85000). University of New Mexico Cancer Center Shared Facilities (KUGR Genomics, Biostatistics, and Bioinformatics & Computational Biology) are supported in part by the National Cancer Institute (grant NCI P30 CA118100) and were critical for this work. S.P.H. holds the Ergen Family Chair in Pediatric Cancer.
National Institutes of Health
Authorship
Contribution: R.C.H. performed microarray studies and statistical and data analysis and wrote manuscript; C.G.M. performed CNA research and analysis; X.W. performed statistical and data analysis (hierarchical clustering); K.K.D. performed data analysis (COPA); G.S.D. performed data analysis (VxInsight); E.J.B. performed statistical analysis and helped develop the ROSE method; I-M.C. performed COG 9906 microarray studies and analyzed data; S.R.A., H.K., and M.D., performed statistical and data analysis; K.A. performed COG 9906 microarray studies and helped develop the ROSE method; C.S.W., W.W., and M.S. performed data analysis and wrote manuscript; M.M. performed data analysis; A.J.C. performed cytogenetic analysis; M.J.B. performed flow studies and designed research; W.P.B. designed COG studies; J.R.D. and M.R. performed CNA statistical and data analysis; J.Y. performed CNA research and statistical and data analysis; D.B. performed COG CCG 1961 research and analysis; W.L.C. designed COG studies and performed COG CCG 1961 research and analysis; B.C. designed COG studies and wrote manuscript; G.H.R. designed COG and CCG studies; S.P.H. designed COG studies, reviewed and assisted in manuscript writing; and C.L.W. designed COG studies, performed data analysis and wrote manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Cheryl L. Willman, University of New Mexico Cancer Center, MSC08 4630 1, University of New Mexico, Albuquerque, NM 87131; e-mail: cwillman@salud.unm.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal