Adhesion properties of hematopoietic stem cells (HSCs) in the bone marrow (BM) niches control their migration and affect their cell-cycle dynamics. The serum response factor (Srf) regulates growth factor–inducible genes and genes controlling cytoskeleton structures involved in cell spreading, adhesion, and migration. We identified a role for Srf in HSC adhesion and steady-state hematopoiesis. Conditional deletion of Srf in BM cells resulted in a 3-fold expansion of the long- and short-term HSCs and multipotent progenitors (MPPs), which occurs without long-term modification of cell-cycle dynamics. Early differentiation steps to myeloid and lymphoid lineages were normal, but Srf loss results in alterations in mature-cell production and severe thrombocytopenia. Srf-null BM cells also displayed compromised engraftment properties in transplantation assays. Gene expression analysis identified Srf target genes expressed in HSCs, including a network of genes associated with cell migration and adhesion. Srf-null stem cells and MPPs displayed impair expression of the integrin network and decreased adherence in vitro. In addition, Srf-null mice showed increase numbers of circulating stem and progenitor cells, which likely reflect their reduced retention in the BM. Altogether, our results demonstrate that Srf is an essential regulator of stem cells and MPP adhesion, and suggest that Srf acts mainly through cell-matrix interactions and integrin signaling.
Introduction
The serum response factor Srf is required in multiple biologic processes such as inductive development, cell morphology, growth, migration, and adhesion.1,2 Germline loss of Srf in mouse results in a severe gastrulation defect and embryonic death at day 8.5.3 In adult fibroblasts and muscle and neuronal cells, Srf is an important regulator of actin cytoskeleton dynamic and contractile processes, whereas its transcriptional activity appears dispensable for cell proliferation. Its implication in cellular transformation has been only recently suggested in humans, in solid tumors through its contribution to the tumor cell invasiveness4,5 and in acute myeloblastic leukemia through the deregulation of its coactivator MAL.6,7 The ability of Srf to coordinate cytoskeleton dynamics and cell cycles makes it an interesting candidate for regulating hematopoietic stem cell (HSC) homeostasis.
Srf regulates transcription by binding to a sequence of CC(A/T)6GG known as the CArG motif. The transcriptional activity of Srf essentially depends on signal-regulated cofactors. These include the ternary complex factors (Tcf) of the ETS-domain family activated by the classical mitogen-activated protein (MAP) kinase pathways and the myocardin-related transcription factors (Mrtf) composed of 3 members: myocardin, megakaryocytic acute leukemia (MAL)/Mrtf-A (also known as megakaryoblastic leukemia 1, MKL1), and Mrtf-B. The activity of the latter is controlled by guanosine triphosphate hydrolase enzymes (GTPases) from the Rho family through the regulation of actin dynamics.1 Direct actin binding regulates the subcellular localization of MAL/Mrtf-A and Mrtf-B in fibroblasts,8,9 while myocardin is constitutively nuclear. Tcf compete with Mrtf proteins for Srf interaction, and this balance was proposed to control the switch between differentiation and proliferative state in smooth muscle cells.10
Comparatively less is known about the functions of Srf and its Srf accessory protein 1 coactivators in the hematopoietic system. The Tcf family members Sap-1, Ets-like gene 1 (Elk-1), and Net act downstream of the extracellular signal-related kinase (ERK) pathway to regulate the expression of immediate early genes (IEGs). Gene knockout studies demonstrated the involvement of Sap-1 for positive selection and T-cell receptor–dependent IEG activation in double positive thymocytes,11 while loss of Elk-1 and Net did not affect the hematopoietic development.12,13 The embryonic lethality associated with constitutive loss-of-function of myocardin and Mrtf-B has precluded biologic analysis in the hematopoietic system.14,15 Mice with Mrtf-A germline deficiency did not show prominent defects in stem cells and progenitors development,16 possibly due to functional redundancy with other gene family members.17 Chromosomal rearrangements affecting the MAL gene are specific for pediatric acute megakaryoblastic leukemia (AMKL).6,7 The resulting OTT-MAL/RBM15-MKL1 fusion protein affects Srf target genes transcription.18,19
To gain insight into the role of Srf in hematopoiesis, we inactivated Srf20 in mouse hematopoietic cells using the Mx1-Cre–inducible system.21 We found that Srf acts as a key regulator of stem cells and progenitors adhesion through regulation of membrane-associated cytoskeleton genes and appears dispensable for their cell-cycle progression. Our results demonstrate an essential role of Srf in HSC homeostasis.
Methods
Mice, peripheral blood hematology, and hematopoietic reconstitutions
Srfflx/flx mice20 were crossed with the polyinosine-polycytidine (pIpC)–inducible Mx1-Cre-recombinase mouse line.21 Six-week-old mice were injected with 200 μg of pIpC (Cayla-InvivoGen) every 2 days and analyzed between days 4 and 35 after the last injection. Blood samples were analyzed with an MS-9 hematology analyzer (Melet Schloesing Technologies). Bone marrow (BM) transplantations were performed by retro-orbital injection of donor BM cells into 6-week-old lethally irradiated (1200 rad) recipients. Outlines of transplantation experiments are given in the figures legends. This study received Inserm review board approval for the use of mice.
Cell cultures
Five hundred Lin−Sca-1+c-Kit+ cells (LSKs) were cultured in StemSpan serum-free expansion medium (SFEM; StemCell Technologies) supplemented with 5% fetal bovine serum (StemCell Technologies), interleukin-3 (IL-3; 10 ng/mL), IL-11 (25 ng/mL), stem cell factor (SCF; 25 ng/mL), fms-like tyrosine kinase 3 ligand (Flt-3; 25 ng/mL), erythropoietin (EPO; 25 ng/mL), and thrombopoietin (TPO; 25 ng/mL; PeproTech).
Flow cytometry and cell sorting
Peripheral blood (PB) was collected from anesthetized mice by intracardiac perfusion of 50 mL of phosphate-buffered saline (PBS)–heparin.
PB, spleen, and BM cells were stained with antibodies raised against specific markers of hematopoietic lineages as reported.22,–24 For in vivo bromodeoxyuridine (BrdU) incorporation studies, mice received 4 intraperitoneal injections of 400 μg of BrdU (Sigma-Aldrich) every 2 hours and were killed 24 hours later. BrdU staining was performed using the BrdU Flow Kit according to manufacturer's instructions (Becton Dickinson). Flow cytometric data were acquired using a CYAN ADP (Beckman Coulter) or a FACSAria (Becton Dickinson) and analyzed using the FlowJo Version 7.2.4 software (TreeStar).
In vivo homing assay
Fluorescence activated cell sorting (FACS)–sorted LSKs were labeled with the cytoplasmic dye carboxyfluorescein diacetate succinimidyl diester (CFSE) according to manufacturer's instructions (Invitrogen). Lethally irradiated (1200 rads) wild-type mice were injected retro-orbitally with 5 × 104 CFSE-stained LSKs. The percentage of homing was calculated as described.25
In vitro differentiation assays
BM (2 × 104), spleen (2 × 105), and PB (5 × 105) cells were cultured in methylcellulose-based medium (M3434; StemCell Technologies) and scored for CFUs (colony-forming units) using combined scoring for burst-forming unit erythroids (BFU-Es), CFU-granulocyte macrophages (CFU-GMs), and CFU-granulocyte erythroid macrophage megakaryocytes (CFU-GEMMs) after 7 days. All live colonies were counted for each of the 2 35-mm dishes plated per sample.
Mobilization assay
SrfΔ/Δ;Mx1-Cre and Srf+/+;Mx1-Cre mice were injected with 200 μg/kg/d during 5 days with granulocyte colony-stimulating factor (G-CSF; Neupogen; Amgen) or NaCl 0.9%. One hundred microliters of PB were plated in methylcellulose medium (M3434; StemCell Technologies) before the first injection and after the last injection and scored for colonies after 7 days of culture.
Gene expression profiling
Total RNAs were collected from 105 LSKs invalidated or not for Srf, using the RNeasy micro kit (QIAGEN). RNA was amplified and labeled using the NuGen Ovation RNA Amplification System V2 and the FL-Ovation cDNA Biotin Module V2 (NuGEN Technologies Inc). In addition, 3.75 μg of amplified cDNA were hybridized to Affymetrix GeneChip Mouse Genome 430 arrays (PartnerChip). Data were normalized using the GCRMA (GC content adjusted-robust multiarray) algorithm and GeneSpring MS Version GX 7.3 software (Agilent Technologies). Lists of differentially expressed genes were obtained with Focus Version 5.5 software. Expression levels were verified by quantitative reverse-transcription–polymerase chain reaction (qRT-PCR) for a selection of genes. The biologic significance of differential representation of standard functional gene categories was obtained using Ingenuity Pathway analysis. All microarray data are now referenced as GSE23556 on the Gene Expression Omnibus (GEO) Web site.
qRT-PCR
cDNA was synthesized from total RNA purified from LSK, common myeloid progenitor (CMP), megakaryocyte-erythroid progenitor (MEP), and granulocyte-macrophage progenitor (GMP) populations using the Superscript II reverse transcriptase (Invitrogen). TaqMan probes were purchased from Applied Biosystems. The expression level of each gene was assessed by qRT-PCR with an ABI PRISM 7000; each sample was analyzed in triplicate and normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh).
Ex vivo adhesion experiments
Ninety-six–well plates were coated with 25 μg/mL Matrigel, 25 μg/mL fibronectin, 25 μg/mL laminin, 50 μg/mL collagen IV (from Becton Dickinson), 200 μg/mL poly-L-lysine, or 20 μg/mL fibrinogen (Sigma-Aldrich). One thousand LSKs were plated for 2 hours at 37°C in serum-free StemSpan SFEM medium (StemCell Technologies) supplemented with IL-3 (10 ng/mL), IL-6 (10 ng/mL), and stem cell factor (10 ng/mL; PeproTech), and adherent cells were counted as reported.26
Statistical analysis
Results are expressed as means values ± SEM. Data were analyzed using the Mann-Whitney test. All statistical analyses were performed using GraphPad Prism Version 5.03 software (GraphPad Software).
Results
Srf is expressed in all hematopoietic progenitor subsets
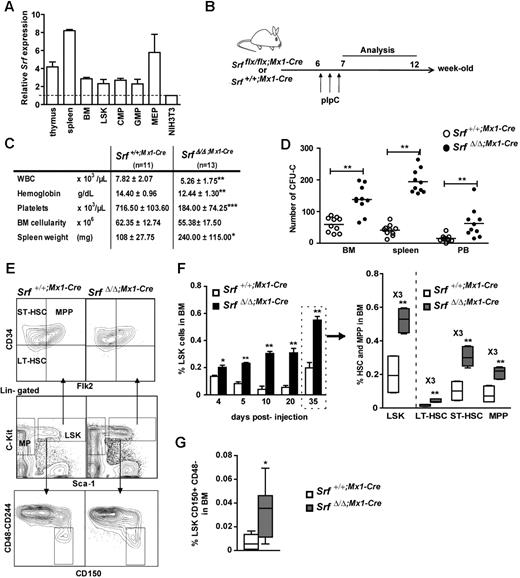
We first determined the relative expression level of Srf in wild-type hematopoietic cells using qRT-PCR. All hematopoietic tissues displayed higher Srf expression levels than the NIH3T3 murine fibroblastic cell line, with increases of 3-fold in the BM, 4-fold in the thymus, and 8-fold in the spleen (Figure 1A). We next analyzed Srf expression in sorted hematopoietic stem and progenitor populations including the multipotent LSKs and the myeloid-committed GMPs, CMPs, and MEPs.22 While LSKs, GMPs, and CMPs displayed similar levels of Srf transcription, MEPs showed a 2-fold increase (Figure 1A). Altogether, these results show a significant expression level of Srf across the hematopoietic tree.
Loss of SRF induces the expansion of stem cells and their multipotent progenitors. (A) Relative expression of Srf in hematopoietic organs and sorted hematopoietic populations from control mice (n = 2) analyzed by qRT-PCR. Expression levels are normalized to Gapdh and results are expressed relative to the level of Srf expression in NIH3T3 fibroblasts (set to 1). (B) Experimental outline of Srf deletion. (C) Table of the hematologic parameters, BM cellularity, and spleen weight for each genotype 5 weeks post-pIpC treatment. Values shown are mean ± SEM (n = 11-13 per group, *P < .05, **P < .01, ***P < .001 by Mann-Whitney test). (D) In vitro colony assays. BM (2 × 104), spleen (2 × 105), and peripheral blood (PB,5 × 105) cells were plated in methylcellulose medium and scored for CFU-Cs (combined scoring for BFU-Es, CFU-GMs, and CFU-GEMMs) after 7 days. Values shown are mean ± SEM (n = 10 per genotype, **P < .01). (E) Representative FACS profile of LSKs and their subpopulations (LT-HSCs, ST-HSCs, MPPs) in the BM of Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre mice 5 weeks after pIpC treatment. (F) Time course analyses of dynamic changes in LSKs (left) and their subpopulations (right) after pIpC injection. Values shown are mean ± SEM (n = 4 for each genotype at 4, 5, 10, and 20 days, and n = 10 at 35 days, *P < .05 and **P < .01 by Mann-Whitney test). (G) Quantification of the percentage of CD150+CD48-CD244- cells in the LSK compartment of Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre mice 5 weeks after pIpC treatment. Values shown are mean ± SEM (n = 4 per genotype, *P < .05 by Mann-Whitney test). For box-and-whisker plots, midline indicates the median value; upper and lower edges of the box plot are the 75th and 25th percentiles.
Loss of SRF induces the expansion of stem cells and their multipotent progenitors. (A) Relative expression of Srf in hematopoietic organs and sorted hematopoietic populations from control mice (n = 2) analyzed by qRT-PCR. Expression levels are normalized to Gapdh and results are expressed relative to the level of Srf expression in NIH3T3 fibroblasts (set to 1). (B) Experimental outline of Srf deletion. (C) Table of the hematologic parameters, BM cellularity, and spleen weight for each genotype 5 weeks post-pIpC treatment. Values shown are mean ± SEM (n = 11-13 per group, *P < .05, **P < .01, ***P < .001 by Mann-Whitney test). (D) In vitro colony assays. BM (2 × 104), spleen (2 × 105), and peripheral blood (PB,5 × 105) cells were plated in methylcellulose medium and scored for CFU-Cs (combined scoring for BFU-Es, CFU-GMs, and CFU-GEMMs) after 7 days. Values shown are mean ± SEM (n = 10 per genotype, **P < .01). (E) Representative FACS profile of LSKs and their subpopulations (LT-HSCs, ST-HSCs, MPPs) in the BM of Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre mice 5 weeks after pIpC treatment. (F) Time course analyses of dynamic changes in LSKs (left) and their subpopulations (right) after pIpC injection. Values shown are mean ± SEM (n = 4 for each genotype at 4, 5, 10, and 20 days, and n = 10 at 35 days, *P < .05 and **P < .01 by Mann-Whitney test). (G) Quantification of the percentage of CD150+CD48-CD244- cells in the LSK compartment of Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre mice 5 weeks after pIpC treatment. Values shown are mean ± SEM (n = 4 per genotype, *P < .05 by Mann-Whitney test). For box-and-whisker plots, midline indicates the median value; upper and lower edges of the box plot are the 75th and 25th percentiles.
Srf loss alters homeostasis of the mature hematopoietic lineages
To evaluate the contribution of Srf to hematopoietic differentiation, Srfflx/flx and Srf+/+ mice were bred with Mx1-Cre animals21 and Cre induction was achieved in 6-week-old animals by injection of pIpC (Figure 1B). Srf deletion was analyzed 5 weeks post-pIpC injection by PCR analyses of genomic DNA, which confirmed the presence of the inactivated allele (SrfΔ) in all the hematopoietic tissues of the injected animals (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Analysis of the PB of pIpC-treated Srfflx/flx;Mx1-Cre mice (called thereafter SrfΔ/Δ;Mx1-Cre) 5 weeks after induction revealed a statistically significant decrease in hemoglobin and platelet levels compared with pIpC-treated Srf+/+;Mx1-Cre control littermates (Figure 1C). A significant decline in white blood cell (WBC) counts was also observed in SrfΔ/Δ;Mx1-Cre mice, which was primarily due to an approximately 40% decrease in the numbers of circulating B and T lymphocytes. Srf-null mice showed a decrease in the frequency of B220+IgM+ B-lymphocytes in both BM and spleen (supplemental Figure 2A) and a skewed ratio of double negative and single positive CD4+ and CD8+ cells in the thymus (not shown). These results agree with a previous report showing that Srf inactivation impacts on the development and homeostasis of the B- and T-lymphoid lineages.27 In vitro CFU assays using Srf-null BM, spleen, and PB samples showed an increased number of colony-forming cells (CFCs) in Srf-null animals (Figure 1D), which were a majority of CFU-GMs (not shown) and confirmed to be SrfΔ/Δ by PCR genotyping (not shown). In addition, Srf-null mice showed a statistically significant increase in the frequency and absolute numbers of CD11b/macrophage-1 antigen (Mac1+) and Gr-1+ cells in the BM and spleen indicating a skewed myelomonocytic differentiation in these animals (supplemental Figure 3A-B; supplemental Table 1).
Growth and maturation of megakaryocytes and platelet production were overly affected in SrfΔ/Δ;Mx1-Cremice.16 In particular, Srf-null mice suffered from a severe thrombocytopenia, with circulating platelets counts lower than 200 000/μL (700 000 to 1 000 000/μL in control mice; Figure 1C) and showed the presence of numerous megakaryocytes in BM and spleen.16 Such megakaryocytic hyperplasia, in addition to granulocytosis, may account for the splenomegaly that developed over time in all the mutant mice (Figure 1C). Limited abnormalities were observed in the erythroid differentiation pathway in the mutant BM and spleen, with an erythroid differentiation slightly increased in the spleen most likely because of an extramedullary hematopoiesis (not shown). Finally, Srf loss did not affect the BM cellularity nor the overall viability of BM cells, as estimated by 7-amino-actinomycin D (7-AAD)/annexin V staining (supplemental Figure 4).
Srf loss induces the expansion of stem and MPP cells
Because the hematopoietic phenotypes of SrfΔ/Δ;Mx1-Cre mice associate multiple differentiation defects, we analyzed the LSK compartment in the BM since it contains the most immature stem cells and multipotent progenitors (MPPs; Figure 1E). The overall frequency of LSK was measured at several time points after pIpC injection and was found to be significantly increased in SrfΔ/Δ;Mx1-Cre BM compared with control BM (Figure 1F). This approximately 3-fold LSK overrepresentation was observed 5 days after inactivation and persisted over time. To determine whether specific subpopulations were amplified, we used a combination of antibodies to CD34 and Flt3/Flk2/CD135 that distinguish between long-term HSCs, (LT-HSCs: CD34−Flt3−), short-term HSCs (ST-HSCs: CD34+Flt3−) and MPPs (CD34+Flt3+) within the LSK compartment.23 These investigations were performed 5 weeks (35 days) after induction (Figure 1E). In fact, all 3 subpopulations were similarly increased by a 3-fold factor in SrfΔ/Δ;Mx1-Cre BM compared with control BM (∼ 0.043% vs ∼ 0.016% for LT-HSCs, ∼ 0.30% vs ∼ 0.10% for ST-HSCs, ∼ 0.21% vs ∼ 0.08% for MPPs; Figure 1F right panel). We confirmed the amplification of both LT-HSCs and ST-HSCs in mutant BM using the signaling lymphocyte activation molecule (SLAM)–family of cell surface markers CD48, CD244, and CD150/SlamF1 (Figures 1E-G).24 Finally, Hoechst 33 342 dye exclusion assay,28 confirmed an increase in the most immature side population (SP) cells in Srf-null animals (not shown). Altogether, these complementary approaches demonstrate that Srf loss lead to a marked expansion of the HSC and early multipotent progenitor compartments.
Srf loss affects lineage-committed progenitor functions without changing their numbers in the BM
We next investigated the lineage-committed progenitors downstream of the HSC compartment. In contrast to the LSK fraction, the myeloid progenitors (MPs: Lin−c-Kit+Sca-1−) compartment remained unchanged 5 weeks after pIpC-induction and Srf deletion (supplemental Figure 5A-B). Similar percentages and absolute numbers of myeloid lineage progenitors CMP, GMP, and MEP were observed in the BM of control and mutant mice (supplemental Figure 5B right; supplemental Table 1). In addition, mutant animals showed normal BM cellularity (Figure 1C) and unchanged rate of apoptosis in both LSK and MP compartments (supplemental Figure 4). Furthermore, the frequency and absolute numbers of common lymphoid progenitors (CLP: Lin−IL-7Rα+c-kitintSca-1int) were similar in both control and mutant mice supplemental Figure 2B; supplemental Table 1). These results contrast with both the LSK expansion and the defective production of mature lineages observed in Srf-null mice. Lymphocyte defects and thrombocytopenia can be explained by the specific function of Srf on essential developmental programs regulating these lineages, hence resulting in impaired differentiation in Srf-null cells.27 Enhanced granulopoiesis can be explained by the amplification of more differentiated granulocytic precursors and/or by a skewing toward granulocytic differentiation from Srf-null CMPs and GMPs. In support of these hypotheses, we found a 1.8-fold increase in the numbers of CFU-GM colonies generated from mutant LSKs and CMPs in methylcellulose assays (supplemental Figure 5C). The observation that HSCs and MPPs are increased in numbers in the BM of Srf-deficient mice, while the lineage-committed progenitors are not, suggests either faster differentiation rates toward these lineages or increased migration to the periphery.
Srf loss affects the stem and progenitor cells functions in a cell-autonomous manner
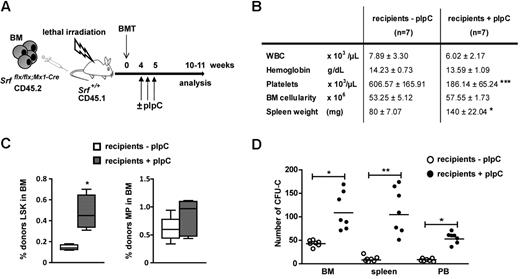
The Mx1-Cre transgene is also active in the BM microenvironment. To ensure the cell autonomous nature of the phenotype, CD45.2 Srfflx/flx;Mx1-Cre BM cells (4 × 106) were transplanted into lethally irradiated CD45.1 wild-type recipients, and the engrafted mice were treated 5 weeks later with pIpC to induce Cre expression and Srf inactivation exclusively in SrfΔ/Δ;Mx1-Cre hematopoietic cells (Figure 2A). Flow cytometric analyses of BM cells harvested 5 weeks after Cre induction revealed a similar LSK amplification (Figure 2C) and abnormalities in platelet levels (Figure 2B) compared with primary Srfflx/flx;Mx1-Cre mice. Notably, recipient mice exhibited identical defects in myeloid differentiation, that is, increased in vitro activity of MPs (Figure 2D) and elevated numbers of megakayocytes and myelo-monocytic cells in the BM and spleen (supplemental Figure 6B). As conditional inactivation of Srf in the microenvironment of recipient mice did not result in similar expansion of the LSK subset (supplemental Figure 6E), these findings demonstrate that Srf regulates LSK amplification and MPP development in a cell autonomous fashion.
Analysis of the cell-intrinsic effects of Srf deletion. (A) Experimental outline of transplantation experiments. Srfflx/flx;Mx1-Cre whole BM cells (CD45.2, 4 × 106 totals) were transplanted into 6-week-old lethally irradiated wild-type littermate recipients (CD45.1). The recipients were treated or not with pIpC 4 weeks postengraftment to induce Srf-deletion exclusively in hematopoietic cells and not in BM stromal cells. Mice were analyzed 5 weeks after Cre induction. (B) Table of the hematologic parameters, BM cellularity, and spleen weight. Values shown are mean ± SEM (n = 7 per group, ***P < .001 by Mann-Whitney test). (C) Cell autonomous expansion of the LSK and MP compartments in recipient mice treated or not with pIpC. (n = 7 per group, *P < .05). (D) BM (2 × 104), spleen (2 × 105) and PB (5 × 105) cells of recipient mice treated or not with pIpC were plated in methylcellulose medium and scored for CFU-Cs after 7 days (n = 7 per group, *P < .05 and **P < .01).
Analysis of the cell-intrinsic effects of Srf deletion. (A) Experimental outline of transplantation experiments. Srfflx/flx;Mx1-Cre whole BM cells (CD45.2, 4 × 106 totals) were transplanted into 6-week-old lethally irradiated wild-type littermate recipients (CD45.1). The recipients were treated or not with pIpC 4 weeks postengraftment to induce Srf-deletion exclusively in hematopoietic cells and not in BM stromal cells. Mice were analyzed 5 weeks after Cre induction. (B) Table of the hematologic parameters, BM cellularity, and spleen weight. Values shown are mean ± SEM (n = 7 per group, ***P < .001 by Mann-Whitney test). (C) Cell autonomous expansion of the LSK and MP compartments in recipient mice treated or not with pIpC. (n = 7 per group, *P < .05). (D) BM (2 × 104), spleen (2 × 105) and PB (5 × 105) cells of recipient mice treated or not with pIpC were plated in methylcellulose medium and scored for CFU-Cs after 7 days (n = 7 per group, *P < .05 and **P < .01).
Srf loss results in a rapid expansion of HSCs and MPPs without long-term modification of cell-cycle dynamics
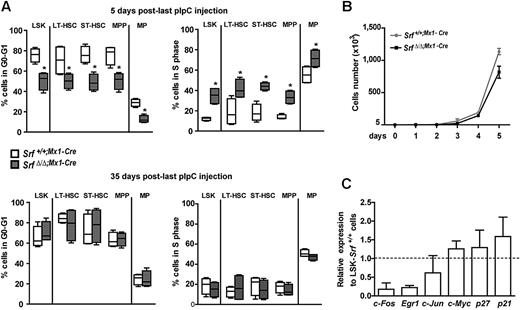
To investigate the cell-cycle parameters of Srf-null HSCs and MPPs, we measured the number and frequency of cells transiting through the cell cycle by in vivo BrdU incorporation assays after pIpC treatment (Figure 3A). A statistically significant increase in proliferation rates was observed in SrfΔ/Δ;Mx1-Cre LSKs, LT-HSCs, ST-HSCs, MPPs, and myeloid-committed MPs 5 days after pIpC induction, compared with populations isolated from similarly treated control littermates. In contrast, by 35 days after pIpC induction, equivalent percentages of G0/G1 and S phase cells were observed in all the BM compartments of control and mutant mice (Figure 3A). Labeling of the LSK fraction with the proliferation marker Ki67 antigen also showed similar proportions of proliferative cells between Srf-null and control mice (not shown), and in vitro culture of LSKs demonstrated similar proliferation rates (Figure 3B). Taken together, these results indicate that Srf loss results in a rapid and transient cell-cycle acceleration occurring after pIpC, in contrast to the expansion of the LSK compartment that persists over time. We also examined the relative transcription level of several cell-cycle regulators in mutant cells. Expression levels of c-Myc, p21Cip1, and p27Kip1 were unchanged in Srf-null LSKs, confirming that Srf loss does not affect the overall proliferation status of HSCs and MPPs. In contrast, Srf-null LSKs showed a marked decrease in mRNA levels of the IEG c-fos (∼ 80% decrease) and early growth response factor 1 (Egr-1; ∼ 77%; Figure 3C), which are direct Srf-target genes29 involved in cell growth regulation. Consistently, Srf−/− murine embryonic stem cells also show defective expression of these IEG but continue to proliferate normally.30 Taken together, these results demonstrate that Srf loss affects the biology of the HSC compartment without long-term effects on the proliferation rates of stem cells and MPPs.
Loss of Srf does not induce persistent changes in stem and progenitor cell cycle. (A) BrdU incorporation following a 24 hours in vivo pulse in Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre mice. Cell cycle indices (G0-G1 and S) of BM LSKs, LT-HSCs, ST-HSCs, MPPs, and MPs are shown for each genotype at 5 (top) and 35 (bottom) days after pIpC injection (n = 4 mice per genotype *P < .05 by Mann-Whitney test). Box-and-whisker plots show medians, quartiles, highest and lowest values. (B) In vitro growth of LSKs isolated from Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre mice 35 days after pIpC treatment. Values shown are mean ± SEM (n = 3 per genotype). (C) qRT-PCR analysis of cell-cycle regulators expression in SrfΔ/Δ;Mx1-Cre LSKs. Expression levels were normalized to Gapdh, and results were expressed relative to the level of expression measured in Srf+/+;Mx1-Cre LSKs (set to 1). Values shown are mean ± SEM from 3 independent experiments.
Loss of Srf does not induce persistent changes in stem and progenitor cell cycle. (A) BrdU incorporation following a 24 hours in vivo pulse in Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre mice. Cell cycle indices (G0-G1 and S) of BM LSKs, LT-HSCs, ST-HSCs, MPPs, and MPs are shown for each genotype at 5 (top) and 35 (bottom) days after pIpC injection (n = 4 mice per genotype *P < .05 by Mann-Whitney test). Box-and-whisker plots show medians, quartiles, highest and lowest values. (B) In vitro growth of LSKs isolated from Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre mice 35 days after pIpC treatment. Values shown are mean ± SEM (n = 3 per genotype). (C) qRT-PCR analysis of cell-cycle regulators expression in SrfΔ/Δ;Mx1-Cre LSKs. Expression levels were normalized to Gapdh, and results were expressed relative to the level of expression measured in Srf+/+;Mx1-Cre LSKs (set to 1). Values shown are mean ± SEM from 3 independent experiments.
Srf loss affects retention but not homing of BM cells
We then examined the impact of Srf deletion on the ability of HSCs to provide long-term hematopoietic reconstitution using competitive transplantations of unfractionated Srf+/+;Mx1-Cre or SrfΔ/Δ;Mx1-Cre CD45.2 BM cells into lethally irradiated CD45.1 congenic recipients. Several ratios of CD45.2 SrfΔ/Δ;Mx1-Cre to Srf+/+;Mx1-Cre donor cells (4 × 106 total cells) were transplanted and analyzed for their hematopoietic contribution up to 24 weeks after transplantation. Examination of donor chimerism in the PB indicated that Srf-null BM cells were rapidly out-competed by wild-type cells, even when injected in 3-fold excess (Figure 4A left and middle panels). However, in noncompetitive conditions, SrfΔ/Δ;Mx1-Cre BM cells (4 × 106) conserved the capacities of a sustained long-term multilineage reconstitution, although they were severely compromised compared with Srf+/+;Mx1-Cre BM cells (Figures 4A right panel and B). BM analyses performed at 24 weeks posttransplantation indicated the presence of CD45.2 SrfΔ/Δ;Mx1-Cre cells in all the investigated compartments including LSKs, MPs, megakaryocytes, granulocytes, and T- and B-lymphocytes, and PCR genotyping confirmed the absence of wild-type Srf in all these isolated populations (not shown). These results indicate that Srf-null stem and progenitor cells, while still being functional, are severely impaired in their engraftment efficiency. As decreased engraftment could be the direct consequence of improper homing to the BM, we performed short-term in vivo homing assays (Figure 4C). Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre LSKs (5 × 104) were labeled with CFSE and transplanted into lethally irradiated hosts, and 24 hours after injection, the numbers of CFSE+ cells present in the BM were quantified by flow cytometry. Similar numbers of control and Srf-null LSK were found in the BM indicating that Srf-null LSK do not have significantly compromised homing capacity after transplantation. To address the possibility of impaired retention, we analyzed the blood of Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre mice to quantify the numbers of circulating stem and progenitor cells 5 weeks after pIpC induction (Figure 4D). Strikingly, we observed a considerable increase in the numbers of LSKs (∼ 14-fold) and MPs (∼ 9-fold) circulating in the bloodstream of Srf-null mice compared with control mice. In addition, Srf-null blood cells generated 6-fold more colonies in methylcellulose than control blood cells (Figure 1D), hence confirming at the functional level the increase in circulating stem and progenitor cells. Finally, SrfΔ/Δ;Mx1-Cre mice displayed a considerable increase in the numbers of LSKs (∼ 20- to 4-fold, from day 10 to day 35 postinduction, respectively) and MPs (∼ 5- to 1.5-fold) present in the spleen, which are likely to reflect their egress from the BM and to mediate the extramedullary hematopoiesis observed in these animals (Figure 4E). Finally we tested the ability of the mobilizing agent G-CSF to induce Srf-null HSC/progenitor migration from the BM into the bloodstream. In vitro CFCs were monitored in PB (Figure 4F). G-CSF injection during 5 days induced a significant increase in the numbers of circulating HSCs/progenitors from both treated groups (control and Srf-null animals), and we confirmed the colonies to be all SrfΔ/Δ from the Srf-null animals (not shown). This indicates that Srf loss only impairs HSC/progenitor BM retention, but it does not inhibit it. Taken together, these findings suggest that Srf loss affects HSC function by decreasing HSC retention in the BM, leading to their sustained spontaneous mobilization.
Loss of Srf profoundly affects HSC engraftment ability. (A) Competitive and noncompetitive BMT assays. CD45.2 donor BM cells (4 × 106) isolated from Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre mice 5 weeks after pIpC treatment were transplanted into lethally irradiated CD45.1 congenic recipients. Donor cells were either equivalent in number to competitor CD45.1 BM cells (1:1) (left panel) or outnumbered competitor cells by 3-fold (3:1) (middle panel). Noncompetitive BMT were performed without competitor (1:0) (right panel). The percentage of donor chimerism (CD45.2) in the PB is given at the indicated time points posttransplantation (n = 4 mice per cohort). (B) Percentage of CD45.2 chimerism in BM subpopulations from the noncompetitive BMT 24 weeks posttransplantation. LSKs, MPs, megakaryocytes (Mks, CD41highCD42bhigh), myelo-monocytic cells (CD11b+Gr-1+). Values shown are mean ± SEM (n = 4 mice per cohort). (C) Short-term in vivo homing assay. LSKs at 5 × 104 and isolated from Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre mice (5 weeks after pIpC treatment) were stained with CFSE prior transplantation into lethally irradiated recipients. The percentage of CFSE+ LSKs present in the PB, BM, and spleen was scored by flow cytometry 24 hours after injection (n = 4 mice per cohort). The percentage of homing was determined as reported.28 (D) Absolute number of LSKs and MPs in total PB of Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre mice 35 days after pIpC treatment. Values shown are mean ± SEM (n = 5 mice per group, *P < .05). (E) Percentage of LSKs and MPs in spleen of Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre mice 35 days after pIpC treatment. Values shown are mean ± SEM (n = 5 mice per group, *P < .05 by Mann-Whitney test). (F) G-CSF mobilization assay on Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre mice 35 days after pIpC treatment. Mice were daily treated by G-CSF (200 μg/kg/d†) or NaCl during 5 days. At day 6, 100 μL PB were collected by retro-orbital plexus and cultured in methylcellulose medium. Scatter plots show the numbers of colonies scored after 7 days of culture (n = 4 mice per group, *P < .05 by Mann-Whitney test).
Loss of Srf profoundly affects HSC engraftment ability. (A) Competitive and noncompetitive BMT assays. CD45.2 donor BM cells (4 × 106) isolated from Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre mice 5 weeks after pIpC treatment were transplanted into lethally irradiated CD45.1 congenic recipients. Donor cells were either equivalent in number to competitor CD45.1 BM cells (1:1) (left panel) or outnumbered competitor cells by 3-fold (3:1) (middle panel). Noncompetitive BMT were performed without competitor (1:0) (right panel). The percentage of donor chimerism (CD45.2) in the PB is given at the indicated time points posttransplantation (n = 4 mice per cohort). (B) Percentage of CD45.2 chimerism in BM subpopulations from the noncompetitive BMT 24 weeks posttransplantation. LSKs, MPs, megakaryocytes (Mks, CD41highCD42bhigh), myelo-monocytic cells (CD11b+Gr-1+). Values shown are mean ± SEM (n = 4 mice per cohort). (C) Short-term in vivo homing assay. LSKs at 5 × 104 and isolated from Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre mice (5 weeks after pIpC treatment) were stained with CFSE prior transplantation into lethally irradiated recipients. The percentage of CFSE+ LSKs present in the PB, BM, and spleen was scored by flow cytometry 24 hours after injection (n = 4 mice per cohort). The percentage of homing was determined as reported.28 (D) Absolute number of LSKs and MPs in total PB of Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre mice 35 days after pIpC treatment. Values shown are mean ± SEM (n = 5 mice per group, *P < .05). (E) Percentage of LSKs and MPs in spleen of Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre mice 35 days after pIpC treatment. Values shown are mean ± SEM (n = 5 mice per group, *P < .05 by Mann-Whitney test). (F) G-CSF mobilization assay on Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre mice 35 days after pIpC treatment. Mice were daily treated by G-CSF (200 μg/kg/d†) or NaCl during 5 days. At day 6, 100 μL PB were collected by retro-orbital plexus and cultured in methylcellulose medium. Scatter plots show the numbers of colonies scored after 7 days of culture (n = 4 mice per group, *P < .05 by Mann-Whitney test).
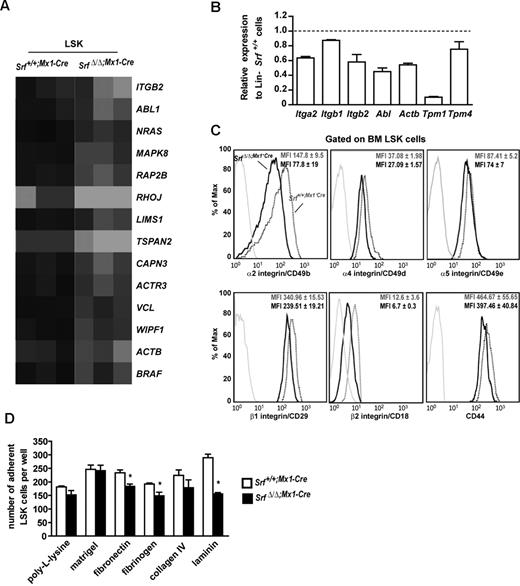
Srf loss affects the integrin network and other molecules associated with HSC–cell and –matrix interactions
To gain insights on the implication of Srf in HSC biology, we investigated the expression of proteins known to be involved in HSC mobilization and retention in the BM. We did not observe any differences between control and Srf-null LSK for the expression level and membrane staining of CD44, N-cadherin, and chemokine CXC motif receptor 4 (CXCR4; Figure 5C and not shown). To gain broader insights, we compared the gene expression profiles of age-matched SrfΔ/Δ;Mx1-Cre and Srf+/+;Mx1-Cre LSK by micro arrays and identified a set of 790 differentially expressed genes (378 down-regulated and 412 unregulated) in Srf-null cells. First, we evaluated whether our results were consistent with the available knowledge on Srf target genes and confirmed down-regulation of actin-related protein 3 (Arp3/Actr3), coronin actin-binding1A (CoroA1), beta actin (Actb), and members of the Tropomyosin family2,31 in Srf-null LSKs (Figure 5B and not shown). We also identified the GTPase RhoJ/TC10-like protein (Tcl) and the effector of adherent junction formation, N-Myc down-regulated gene-1 (Ndrg1), which possesses Srf-binding motifs within their regulatory regions as down-regulated in Srf-null LSKs. In line, defective expression of adherent junction molecules has been reported in Srf-deficient backgrounds.26,32,33 Using Gene Set Enrichment Analysis, we found that the SrfΔ/Δ;Mx1-Cre LSK transcriptional profile displayed enrichment for markers associated with LT-HSC identity34,35 and for markers of human mobilized HSCs36 (not shown). We also searched for functional gene categories that were the most differentially represented and found a large number of differentially expressed transcripts that encode cytoplasmic proteins associated with actin filament growth and cytoskeleton dynamics and organization. Ingenuity Pathway analysis indicated top-scoring networks centered on ephrin (P = .00355) and integrin (P = .01561) pathways (Figure 5A; supplemental Table 2), both including a high proportion of bona fide Srf target genes in nonhematopoietic cells.2,31,32,37 Genes containing putative Srf binding sites were found at a higher rate (> 50%) in the first 5 modified pathways (data not shown). Several of the genes identified by our microarray experiments also fitted into pathways and protein complexes involved in LT-HSC–cell and –matrix interactions and include up-regulation of the junctional adhesion molecule 3 (Jam3), the tight junction protein 1 (Tjp1/ZO-1) and the transglutaminase 2 (Tgm2) in Srf-null LSKs.34
Loss of Srf deregulates expression of the integrin network and impairs stem cell and MPP adhesion to synthetic substrates. (A) Heat map representation of the transcripts abundance of genes associated to the integrin signaling pathway identified in SrfΔ/Δ;Mx1-Cre LSKs. Black indicates high abundance and gray/white low abundance of each transcript. Gene names are indicated. Map was obtained using the Ingenuity Pathway Analysis. (B) qRT-PCR analysis of the expression levels of genes associated to the integrin signaling pathway or encoding for actomyosin cytoskeleton components, in Lineage (Lin)-negative cell populations from SrfΔ/Δ;Mx1-Cre mice. Expression levels were normalized to Gapdh, and results were expressed relative to gene expression levels in Srf+/+;Mx1-Cre mice (set to 1). Values shown are the mean ± SEM from 2 independent experiments. (C) Representative FACS profile of α2, α4, α5, β1, β2 integrins and CD44 expression on the surface of Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre LSK (n = 4 per genotype; 5 weeks after pIpC treatment). The mean fluorescence intensity (MFI) ± standard deviation is indicated. (D) Ex vivo adhesion properties of Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre LSKs. Cells were counted 2 hours after plating onto synthetic matrix-coated wells. Background adhesion of LSKs on uncoated slides was subtracted from each value. Two independent experiments were performed, and each condition was analyzed in triplicate or more; data represent means ± SEM (n = 4 for each genotype, *P < .05 by Mann- Whitney test).
Loss of Srf deregulates expression of the integrin network and impairs stem cell and MPP adhesion to synthetic substrates. (A) Heat map representation of the transcripts abundance of genes associated to the integrin signaling pathway identified in SrfΔ/Δ;Mx1-Cre LSKs. Black indicates high abundance and gray/white low abundance of each transcript. Gene names are indicated. Map was obtained using the Ingenuity Pathway Analysis. (B) qRT-PCR analysis of the expression levels of genes associated to the integrin signaling pathway or encoding for actomyosin cytoskeleton components, in Lineage (Lin)-negative cell populations from SrfΔ/Δ;Mx1-Cre mice. Expression levels were normalized to Gapdh, and results were expressed relative to gene expression levels in Srf+/+;Mx1-Cre mice (set to 1). Values shown are the mean ± SEM from 2 independent experiments. (C) Representative FACS profile of α2, α4, α5, β1, β2 integrins and CD44 expression on the surface of Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre LSK (n = 4 per genotype; 5 weeks after pIpC treatment). The mean fluorescence intensity (MFI) ± standard deviation is indicated. (D) Ex vivo adhesion properties of Srf+/+;Mx1-Cre and SrfΔ/Δ;Mx1-Cre LSKs. Cells were counted 2 hours after plating onto synthetic matrix-coated wells. Background adhesion of LSKs on uncoated slides was subtracted from each value. Two independent experiments were performed, and each condition was analyzed in triplicate or more; data represent means ± SEM (n = 4 for each genotype, *P < .05 by Mann- Whitney test).
Srf loss impairs HSC and MPP adhesion properties
Based on the Ingenuity pathway analysis, we next compared by flow cytometry the cell surface expression of α4 (CD49d/Itga4), α5 (CD49e/Itga5), α2 (GpIIa/CD49b/Itga2), β1 (CD29/Itgb1) and β2 (CD18/Itgb2) integrin subunits on SrfΔ/Δ;Mx1-Cre and Srf+/+;Mx1-Cre LSK (Figure 5C). Expression of α2 and β2 subunits was markedly decreased on Srf-null cells (∼50% decrease), while expression of α4 and β1 were reduced to a lesser extent (∼30% decrease) and α5 was almost not affected (∼15% decrease). Altogether, these results support an important role for Srf in controlling LSK adhesion through the regulation of the integrin receptor expression.
To address whether reduced integrin expression affects LSK adhesion, we performed ex-vivo adhesion experiments onto synthetic matrix-coated slides using SrfΔ/Δ;Mx1-Cre and Srf+/+;Mx1-Cre LSK (Figure 5D). Plating of Srf-null cells onto laminin-coated slides, which engage multiple β1 integrins including the VLA2/α2β1, resulted in approximately 50% decreased adhesion after 2 hours of incubation. Srf-null LSK also showed significant decreased adhesive functions when plated onto fibronectin- and fibrinogen-coated slides, which mainly act through VLA4/α4β1 and VLA5/α5β1 for the former and Mac/αMβ2 and αXβ2 for the latter. In contrast, no significant difference between control and Srf-null cells were found on collagen IV that engage several β1 receptors, or on laminin/collagen IV/heparan sulfate proteoglycan (Matrigel) and poly-L-lysine adhesive surfaces. Together, these results indicate that a subset of adhesion receptors is functionally impaired by Srf deficiency. They also imply that disruption of this integrin regulatory network and defective adhesion are likely to be responsible for the expansion/BM egress of this compartment in Srf-null mice and the defective engraftment of Srf-null BM cells.
Discussion
Here, we establish that Srf has multiple functions in the adult hematopoietic system. The loss of Srf has broad consequences for the development of all the hematopoietic lineages, leading to severe thrombocytopenia, lymphocytopenia and increased production of myelo-monocytic cells, and negatively impacts HSC function in competitive transplantation assays. Srf deletion leads to the accumulation of HSCs that have impaired adhesion to the extracellular matrix due, in part, to defects in integrin expression.
Srf ablation induced a 3-fold increase in the stem cell and MPP compartment (LT-HSCs, ST-HSCs, and MPPs), and also leads to an increase in the numbers of stem and progenitor cells in the bloodstream and the spleen. However, Srf hematopoietic deficiency did not permanently affect cycling or apoptosis parameters, as the numbers and frequencies of stem and progenitor cells transiting through the cell cycle or undergoing apoptosis were similar in both control and Srf-null mice 5 weeks after Cre-mediated Srf inactivation. This phenotype sharply contrasts with the quick exhaustion of the HSC compartment often observed for important regulators of HSC homeostasis, as β-catenin,38,39 phosphate and tensin homolog,40,41 and Cdc4242 in similar models. Genes like c-Myc, p53, Gfi-1 and Mef/Elf4 have been shown to regulate quiescence and proliferation concomitantly (reviewed in van Os et al43 ). Srf-null stem cells and va Os et al progenitors undergo amplification early after Srf inactivation. One obvious explanation to this phenomenon is that the partial defect in adhesion to BM niches releases a control in cell-cycle progression and allows proliferation. We did not evidence a microenvironment-dependent effect of Srf loss on LSK expansion (supplemental Figure 6). Srf-null HSCs do not show marked proliferation abnormalities while their adhesion properties and expression of integrin receptors are affected. Srf loss differs from c-Myc deficiency, as it leads to a broad down-regulation of adhesive molecules and to HSC expansion/migration, whereas c-Myc ablation induces over expression of N-cadherin and integrins and loss of HSC self-renewal activity at the expense of differentiation.44 We observed a down-regulation of Egr-1, a known Srf target, that might contribute to the amplification of LSK.45 Of note, loss of Srf did not impact the HSC and MPP developmental potential as SrfΔ/Δ;Mx1-Cre mice displayed normal numbers of lineage-committed CMPs and CLPs.
It is now well established that regulation of HSC numbers and function (ie, self-renewal vs differentiation and proliferation vs quiescence) is controlled in part by adhesion to their local BM microenvironment. Our microarray analyses as well as our functional data support a contribution of Srf to HSC adhesion properties and retention/egress from the BM. In keeping, increased numbers of stem and progenitor cells were observed in the bloodstream and spleen of invalidated mice, suggesting that an adhesion decrease of LSKs allows for their sustained spontaneous mobilization. However, as underscored by our G-CSF–induced mobilization experiments, Srf loss only impairs HSC/progenitor BM retention but does not inhibit it. Stromal cell–derived (SDF-1)/CXCR4 interactions are involved in the retention of HSCs/progenitors within the BM microenvironment; however, the level of CXCR4 expression was unchanged in LSKs after Srf loss (not shown). Gene expression profiling identified the deregulated expression of known components of integrin networks and matrix-cell adhesion complexes, and in vitro adhesion experiments showed altered capacities of Srf-null HSCs and MPPs to firmly adhere to various extracellular matrix molecules, a defect that appears as an unifying theme for Srf-deficient cells.30,32,33,37 In line, our FACS analysis shows a marked decreased staining for the α2 and β2 integrins and to a lesser extent for β1 and α4, on the cell surface of Srf-null LSKs. Both β1 and α4 contribute to HSC homing, and β1−/− mice showed HSC abnormalities that result from the dual role of this subunit in HSC and stromal cells.46 The striking inability of Srf-null LSKs to efficiently compete in BM transplantation (BMT) assays is likely due to the simultaneous loss of several integrin receptors, as individual integrin deficiency showed limited effect on HSC homing and/or retention in mouse genetic studies, presumably due to alternative or compensatory mechanisms. Although α2 and β2 transcript accumulation was rapidly achieved upon serum stimulation in murine embryonic fibroblasts (not shown), the putative CArG elements that we identified in the 2 loci did not coprecipitate with Srf in chromatin immunoprecipitation experiments (not shown). These 2 subunits participate to cellular receptors that are broadly expressed by hematopoietic cells, such as very late activation antigen 2 (VLA-2)/α2β1, lymphocyte function-associated antigen (LFA-1)/αLβ2, and Mac-1/αMβ2. The BM of Srf-null mice showed excess granulocytes and monocytes likely linked to defective LFA-1 functions as loss of β2 expression in neutrophils also leads to granulocytosis and splenomegaly.47 As VLA-2 interacts with molecules (laminin, collagen, and fibronectin) present on endothelial and stromal cells, and LFA-1 participates in adhesive interactions with the intercellular adhesion molecule 1 (ICAM1), both α2 and β2 might support HSC migration and homing. In addition, the presence of a large population of circulating stem and progenitor cells in Srf-null mice argues for a role of Srf-controlled signaling in mediating mobilization. Srf and its coactivator Mal have been recently linked to the regulation of Itgb1 expression in nonhematopoietic cells.4 Moreover, the Srf/Mal signaling network regulates the expression of proteins essential to the connection of integrin receptors to the actin network and/or the network dynamics. Srf/Mal has been involved in the control of cell adhesion, polarization, spreading, and migration in multiple cellular systems. It participates in the murine neurite outgrowth and axon guidance37 and contributes to the expression of the myosin heavy chain 9 (Myh9) and myosin light chain 9 (Myl9)/Mlc2 genes, which encode components of the actomyosin contractile apparatus in human and murine cells.5,48 In agreement, decreased expression of genes encoding actomyosin cytoskeleton components and/or proteic partners was observed in Srf-deficient LSKs and in Lin- progenitors (supplemental Table 2; Figure 5B) as in SrfΔ/Δ megakaryocytes16 ; such an impaired expression is likely to alter the contractile apparatus integrity. Evidence therefore points to Srf as a general contributor to cell architecture and integrin functions in hematopoietic cells.
In summary, our results demonstrate that Srf-controlled signals participate in HSC adhesion and function through cell-matrix interactions and integrin signaling. A contribution of Srf/Mrtf signaling to the in vivo invasive behavior of tumor cells and metastasis has been shown,4,5 and deregulated Mrtf-A/Mal activity is associated with a leukemic process.7,6 Whether the modification of this Srf function participates in tumor initiation and/or progression should be investigated.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank J. Megret and C. Cordier-Garcia (IFR94-IRNEM, Paris, France) for cell sorting and members of the LEAT (Faculté de Médecine Necker-Enfants Malades, Paris, France) for mouse husbandry. Mx1-Cre transgenic mice were kindly provided by J. Di Santo (Institut Pasteur, Paris, France).
This work was supported by funding from Inserm, ARC (Association pour la Recherche sur le Cancer), LNCC (Ligue Nationale Contre le Cancer), and INCa (Institut National du Cancer). C.R. was funded by a fellowship from the FRM (Fondation pour la Recherche Medicale) and the SFH (Société française d'hématologie).
Authorship
Contribution: C.R. and G.E. performed experiments and analyzed data; E.M. performed G-CSF mobilization assays; N.C. and C.O. analyzed micro arrays data; E.P., D.D., and W.V. provided valuable insights into experimental approaches and data interpretation; O.A.B. and V.P-L. conceived the project and designed, supervised, and interpreted the experiments; and O.A.B., V.P-L., and C.R. wrote the manuscript, with additional input from C.O., E.P., D.D., and W.V.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Virginie Penard-Lacronique, Inserm U985, IGR, 39 rue Camille Desmoulins, Villejuif, F-94805, France; e-mail: virginie.penard-lacronique@inserm.fr.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal