Abstract
The role of circulating cytokines and chemokines (C&Ckine) in activating signal transduction in leukemic cells is incompletely defined. We hypothesized that comprehensive profiling of C&Ckine expression in leukemia would provide greater insight compared with individual analyses. We used multiplex array technology to simultaneously measure the level of 27 C&Ckines in serum from 176 acute myelogenous leukemia (AML) and 114 myelodysplastic syndrome (MDS) patients and 19 normal controls. C&Ckine levels in AML and MDS differed significantly from normal controls (5 higher, 13 lower) but were similar to each other for 24 of 27 analytes, with interleukin-8 and interleukin-13 higher in AML and vascular endothelial growth factor A higher in MDS. Levels did not correlate with age, gender, infection, or blood counts; however, 3 correlated with specific cytognetic abnormalities in AML. Individually, few cytokines had any correlation with response or survival. In newly diagnosed AML, 8 C&Ckine signatures, distinct from the normal control signature, were observed. These signatures had prognostic impact, affecting remission, primary resistance, relapse rates, and overall survival, individually (P = .003) and in multivariable analysis (P = .004). These patterns suggest specific therapeutic interventions to investigate in subsets of AML patients. In conclusion, C&Ckine expression in AML and MDS differs from normal, is similar with one another, and forms recurrent patterns of expression with prognostic relevance.
Introduction
Inflammation is thought to promote tumor development and progression via direct or indirect effects of cytokines and chemokines (C&Ckines), and growth factors on tumor cells or their environment.1-3 In normal hematopoietic cells, activation of cell surface receptors by C&Ckines and growth factors regulates signal transduction pathway (STP) activity.4 Abnormalities in signaling through STP are common events in leukemia and are thought to contribute to leukemogenesis.5,6 Mutations in the receptor or STP components were shown to induce oncogenesis7 as the insertion of the mutated gene often recapitulates leukemia development. The frequency of activation of STP in acute myelogenous leukemia (AML)8 vastly exceeds the frequency of mutations or genetic alterations found in the receptors and STP components,9,10 suggesting that abnormal stimulation by extracellular signals, such as C&Ckines and growth factors, occurring through genetically intact receptors and STP, may be a frequent event in leukemia that provides leukemic cells with proliferative and survival advantages by inhibiting apoptosis and blocking differentiation.11 C&Ckine signaling could thus contribute to leukemogenesis through diverse mechanisms independent of mutational aberrancy within the pathways they trigger.
Evidence for deregulation of C&Ckine and growth factor expression in AML and myelodysplastic syndrome (MDS), and for abnormal responsiveness to them, is well documented.5 Levels of interleukin-3 (IL-3), IL-6, IL-8, thrombopoietin, tumor necrosis factor-α (TNF-α), CSF2 (granulocyte-macrophage colony-stimulating factor), interferon-γ (IFN-γ), and stem cell factor have been shown to be elevated in leukemia patients compared with healthy controls.12-16 We have shown that IL-1, IL-2, or CCL3 (MIP1-α) stimulate leukemia cell proliferation.17-20 Overexpression of cytokines in leukemia patients often declines after chemotherapy administration or remission attainment.13 The response of leukemic cells to perturbations by C&Ckines provides useful classification schemes that are of prognostic significance.21,22 In these studies, IL-2, sIL-2RA, and IL-6 levels were higher in patients progressing from myelofibrosis to AML, whereas high IL-10 serum levels were associated with a fatal outcome after bone marrow transplantation.23 AML patients with lower serum concentration of hepatocyte growth factor had better leukemia-free survival rates compared with patients with higher levels.24 In AML and myelodysplasia (MDS) patients, higher levels of TNF-α negatively affected overall survival,16 whereas increased vascular endothelial growth factor A (VEGFA) levels correlated with reduced survival in AML but not MDS patients.25
Most studies of C&Ckines have measured the level of an individual analyte or a few cytokines. However, because there is tremendous redundancy in C&Ckine actions, with many capable of activating a single STP, and because abnormal serum levels of multiple C&Ckines could be present simultaneously, a complex interaction of numerous C&Ckines probably exists. This makes it difficult to attribute the activation of a given STP to a single cytokine. We hypothesized that comprehensive profiling of the serum levels of multiple C&Ckine expression in leukemia would provide greater insight to their role in signaling compared with individual analyses. Newer multiplex technology makes it possible to simultaneously measure broad panels of cytokines using small volumes of material.26 The utility of multiplex analysis of C&Ckines for cancer prediction and screening has been shown in several solid tumors.27-29 In this study, we present data on C&Ckine profiling using a 27-component panel in serum samples from 176 AML and 114 MDS patients.
Methods
Patient population
Peripheral blood specimens were collected from 176 patients with AML, at initial diagnosis (n = 138), or during relapse (n = 38) and 114 MDS patients including 97 collected at diagnosis. This represented all available serum samples in the M. D. Anderson Cancer Center (MDACC) Leukemia Sample Bank. As discussed below for quality control reasons, 28 (14 AML, 14 MDS) were excluded. The demographics of the included patients are listed in Table 1. The ethnic diversity of patients referred to MDACC mirrors the general population, and all patients were approached for consent; unfortunately, Hispanic and black patients declined to participate at greater frequency, resulting in their underrepresentation in this cohort. Patients with acute promyelocytic leukemia were not included. All patients were used for range determinations but only newly diagnosed patients are used for outcomes analyses. Samples were collected for the Leukemia Sample Bank at the University of Texas M. D. Anderson Cancer Center between 1997 and 2006, on Institutional Review Board-approved protocol (Lab01-473), and consent was obtained according to the Declaration of Helsinki. Samples were analyzed under a separate Institutional Review Board-approved laboratory protocol (Lab05-0654).
Patient demographics
| Category . | AML . | MDS . | |
|---|---|---|---|
| New . | Relapsed . | ||
| No. of cases | 125 | 37 | 100 |
| Sex, female/male | 65/60 | 14/23 | 26/74 |
| Median age (range), y | 62.18 (21-86) | 55.48 (19-73) | 64.45 (21-83) |
| Race | |||
| American Indian | 0 | 1 | 0 |
| Asian | 1 | 1 | 2 |
| Black | 5 | 0 | 2 |
| Hispanic | 9 | 5 | 6 |
| White | 110 | 30 | 90 |
| FAB | |||
| Unknown | 2 | 8 | 2 (unknown) |
| M0 | 7 | 1 | 11 (CMML) |
| M1 | 12 | 6 | 17 (RA) |
| M2 | 50 | 10 | 10 (RARS) |
| M4 | 25 | 9 | 44 (RAEB) |
| M4Eos | 6 | 0 | 16 (RAEBT) |
| M5 | 12 | 2 | |
| M6 | 5 | 1 | |
| M7 | 4 | 0 | |
| Status | |||
| New | 125 | 0 | 83 |
| Post biologic | 0 | 0 | 12 |
| Relapsed | 0 | 36 | 2 |
| Relapsed resistant | 0 | 1 | 3 |
| Cytogenetics | |||
| t(8:21) | 1 | 1 | 0 |
| Inversion16 | 6 | 0 | 1 |
| Diploid | 55 | 7 | 39 |
| Insufficient metaphases | 0 | 1 | 3 |
| 5q− | 5 | 1 | 11 |
| −5 | 2 | 0 | 4 |
| −5, −7 | 4 | 0 | 9 |
| −7, 7q− | 7 | 6 | 6 |
| −5, −7, +8 | 3 | 0 | 1 |
| +8 | 7 | 5 | 10 |
| 11q23 | 3 | 1 | 0 |
| Miscellaneous | 28 | 8 | 13 |
| t(6:9) | 1 | 0 | 0 |
| +21 | 0 | 4 | 3 |
| Not done | 3 | 3 | 0 |
| AHD > 2 months, % | 38 | 5 | NA |
| Treatment outcomes | |||
| Not treated | 118 | 27 | 43 |
| Remission, % | 58 | 11 | 47 |
| Relapse, % | 50 | 66 | 70 |
| Alive, % | 35 | 7 | 17 |
| Category . | AML . | MDS . | |
|---|---|---|---|
| New . | Relapsed . | ||
| No. of cases | 125 | 37 | 100 |
| Sex, female/male | 65/60 | 14/23 | 26/74 |
| Median age (range), y | 62.18 (21-86) | 55.48 (19-73) | 64.45 (21-83) |
| Race | |||
| American Indian | 0 | 1 | 0 |
| Asian | 1 | 1 | 2 |
| Black | 5 | 0 | 2 |
| Hispanic | 9 | 5 | 6 |
| White | 110 | 30 | 90 |
| FAB | |||
| Unknown | 2 | 8 | 2 (unknown) |
| M0 | 7 | 1 | 11 (CMML) |
| M1 | 12 | 6 | 17 (RA) |
| M2 | 50 | 10 | 10 (RARS) |
| M4 | 25 | 9 | 44 (RAEB) |
| M4Eos | 6 | 0 | 16 (RAEBT) |
| M5 | 12 | 2 | |
| M6 | 5 | 1 | |
| M7 | 4 | 0 | |
| Status | |||
| New | 125 | 0 | 83 |
| Post biologic | 0 | 0 | 12 |
| Relapsed | 0 | 36 | 2 |
| Relapsed resistant | 0 | 1 | 3 |
| Cytogenetics | |||
| t(8:21) | 1 | 1 | 0 |
| Inversion16 | 6 | 0 | 1 |
| Diploid | 55 | 7 | 39 |
| Insufficient metaphases | 0 | 1 | 3 |
| 5q− | 5 | 1 | 11 |
| −5 | 2 | 0 | 4 |
| −5, −7 | 4 | 0 | 9 |
| −7, 7q− | 7 | 6 | 6 |
| −5, −7, +8 | 3 | 0 | 1 |
| +8 | 7 | 5 | 10 |
| 11q23 | 3 | 1 | 0 |
| Miscellaneous | 28 | 8 | 13 |
| t(6:9) | 1 | 0 | 0 |
| +21 | 0 | 4 | 3 |
| Not done | 3 | 3 | 0 |
| AHD > 2 months, % | 38 | 5 | NA |
| Treatment outcomes | |||
| Not treated | 118 | 27 | 43 |
| Remission, % | 58 | 11 | 47 |
| Relapse, % | 50 | 66 | 70 |
| Alive, % | 35 | 7 | 17 |
Outcomes data are only for patients treated at MDACC.
FAB indicates French-American-British classification system; CMML, chronic myelomonocytic leukemia; RA, refractory anemia; RARS, refractory anemia with ringed sideroblasts; RAEB, refractory anemia with excess blasts; RAEBT, refractory anemia with excess blasts in transformation; AHD, antecedent hematologic disorder ≥ 2 months; and NA, not applicable.
Various treatment regimens were used for the 138 newly diagnosed, previously untreated AML patients and 97 previously untreated MDS cases, including: anthracycline and high-dose cytosine arabinoside (Ara-C) (HDAC)-based regimens (AML = 71, MDS = 5), HDAC plus nonanthracycline chemotherapy regimens (eg, Fludarabine, Ara-C, G-CSF [FLAG], FA, clofarbine, AML = 34, MDS = 28), other non–HDAC-based chemotherapy (AML = 7), targeted agents (AML = 9, MDS = 4), and demethylating or histone deacetylating agents (AML = 10, MDS = 5). One AML patient received low-dose Ara-C, and 7 AML and 52 MDS patients were not treated at MDACC. Excluding 2 AML and 1 MDS patients lost to follow-up, the minimum follow-up is 109 weeks and the median follow up is more than 5 years.
Serum collection
Peripheral blood was collected in serum separating (red top) tubes and refrigerated until transport to the laboratory (5 times per day) and processed within 2 hours of collection by centrifugation at 314.4g for 5 minutes. Serum was aliquoted and stored at −80°C. Serum controls were also collected from 19 normal, healthy persons, 11 males, 8 females, of diverse ethnicity (11 white, 3 Chinese, 2 black, 2 South American, 1 from India) and ages (range, 24-57 years).
Multiplex cytokine analysis
Multiplex analysis was performed using the Luminex system on a Bioplex (Bio-Rad) cytometer. Procedural and full technical details, including sensitivity, dynamic range, and intra-assay and inter-assay reproducibility, are given at www.bio-rad.com (bulletins 3157 and 5560). Briefly 5.6-micron polystyrene spheres are internally dyed with red and infrared fluorophores in different ratios. A capture antibody specific for one cytokine is bound to beads with a specific dye mixture. Panels can be formed by mixing beads with different dye mixtures and antibodies together. In this study, we used the Bio-Plex Human Cytokine 27-Plex Panel, which contains beads for the following C&Ckines: IL-1RA, IL-1B, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12, IL-13, IL-15, IL-17, CCL11 (Eotaxin), FGF2 (FGF-basic), CSF3 (G-CSF), CSF2 (granulocyte-macrophage colony-stimulating factor), IFN-γ, CXCL10 (IP-10), CCL2 (MCP-1), CCL3 (MIP-1α), CCL4 (MIP-1β), PDGF-BB, CCL5 (Rantes), TNF-α, and VEGFA. Patient serum was diluted 1:4 with serum diluent, mixed with beads for 30 minutes, washed 3 times in wash buffer, exposed to biotinylated detection antibodies for 30 minutes, washed 3 times in wash buffer then incubated with streptavidin-phycoerythrin for 10 minutes, and washed 3 times in assay buffer and assayed according to manufacturer's instructions at room temperature. Samples were run in duplicate. Eight trays were required to run this sample set. The samples were allocated to trays to balance each tray for diagnosis (AML or MDS) and the year of sample acquisition (1998-2006).
Statistical analysis
Data were analyzed using Bioplex Manager (Version 4.0) software. Duplicate wells containing negative controls were used to estimate background intensity in each tray. Duplicate measurements of an 8-step, 4-fold dilution series of known standards were used to fit a 5-parameter logistic curve.30 Average observed intensities of the duplicates of experimental samples were mapped to the fitted curve to interpolate concentration levels; observed intensities outside the range of the standards were flagged as out-of-range (OOR) high or low. Values were expressed as picograms per milliliter. One case had the maximal value for 19 of the 27 cytokines raising concern about loading, and 27 cases had OOR low values for at least 33% of cytokines, raising questions about sample quality, so these cases were excluded from analysis. These 28 cases (15 AML, 13 MDS) had no significant associations with clinical or laboratory characteristics. Cytokine expression values were normalized by dividing by the one-third trimmed mean (to adjust for variation in loading) and multiplying by the overall mean (to maintain the natural scale of cytokine expression). Values flagged as OOR low were replaced with zero; values flagged as OOR high were replaced by 110% of the maximum observed value. Results were transformed by adding 0.25 and taking the base-2 logarithm. The constant 0.25 was chosen to ensure that a real 2-fold change is still estimated at 90% of that level at a normalized intensity of 1.
Additional quality control was applied to data from the normal controls by performing principal component (PC) analysis. The first 4 PCs explained more than 80% of the variation in the cytokine expression of normal controls. Because the Mahalanobis distance of samples from the center of d-dimensional PC space follows a χ2 distribution with d degrees of freedom,31 we used this measure to identify outliers. One sample had χ2 = 114.5 (P < .001) and was removed from the dataset. We then defined the “normal range” of expression, based on the normal samples that passed quality control, to be the interval centered at the mean and extending 1.96 SD in both directions, which would be expected to contain 95% of normal values.
Statistical analysis was performed using the R statistical programming environment, Version 2.9.1 (http://www.r-project.org). Summary statistics were computed to describe the expression of cytokines in normal controls. Two-sample t tests were used to estimate differences between disease and normal or between AML and MDS; although we report the nominal P values, we also report the changes that remain significant after Bonferroni correction to account for multiple testing. Pearson correlation for continuous variables, t tests for dichotomous variables, F tests from one-way analysis of variance for categorical variables, and Cox proportional hazards models with log-rank tests for time-to-event variables were used to test the association of C&Ckine expression with clinical covariates or outcomes.
We used the bimodality index to identify C&Ckines with bimodal expression patterns.32 We performed hierarchical clustering with distances defined by Pearson distance and Ward linkage. To test the consistency of clusters, we repeatedly clustered random subsets of the patient samples by adding Gaussian noise and using a variety of different algorithms (k-means, model-based clustering, partitioning around medoids, and hierarchical clustering with either Pearson or Euclidean distance and either Ward or average linkage), recording for each pair of samples the probability that they clustered together. We used the silhouette width to estimate the number of clusters and to determine whether individual samples were correctly clustered.
Univariable and multivariable (Cox proportional hazards models) were also performed using R statistical programming environment, Version 2.9.1. For the univariable analysis, 27 variables, including demographics (age, gender, and race) clinical features (French-American-British, antecedent hematologic disorders, etiology, performance status, and infection status), laboratory findings (cytogenetics, Fms-like tyrosine kinase 3-internal tandem duplication status, surface marker expression, hematology, and serum chemistries), and cytokine signature group, were analyzed. For the multivariable analysis, all variables associated with the endpoint (P < .10 for overall survival or remission duration) were included in the initial model. In an iterative process, variables were removed or added using the Akaike Information Criterion.
Results
Sample stability
To ensure that expression was consistent across the 8 trays, we compared the range of expression of each analyte across the 8 trays. This revealed no statistically significant variance between trays (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). We were also concerned that sample storage at −80°C for up to 9 years could affect the level of expression of some analytes. We looked at the range of expression of each C&Ckine by year of acquisition; there was no evidence of diminution or variance of signal over time for any of the 27 panel components (supplemental Figure 1B).
Comparison of expression in normal samples with multiplex analysis versus the literature
Most prior studies of C&Ckine expression in normal and leukemic samples have used enzyme-linked immunosorbent assay methodology. Head-to-head comparisons between these methods using nonleukemia samples have shown excellent correlation.26 To determine whether the multiplex-based assay was providing results comparable with those of enzyme-linked immunosorbent assay or other platforms, we compared the results obtained with the Bioplex platform with those from a search of the literature for normal and leukemic samples (supplemental Tables 1, 2).33,34 The data for normal samples are presented in supplemental Table 1. There was heterogeneity of results within the reviewed literature on what the “normal” population value is, perhaps reflecting different methodologies or their sensitivity. A full statistical comparison between expression values in the literature versus those obtained with the Bioplex kit cannot be performed based on the limited data available in the literature. For most analytes, results obtained with the Bioplex system in this report were in general agreement with the prior studies, although levels of IL-5, IL-8, IL-10, IL-13, CSF2, CCL2, and CCL3 were lower and levels of IL-6, IL-9, IFN-γ, and CXCL10, were higher in normal compared with the existing literature.
Comparison of expression in AML and MDS samples with multiplex analysis versus the literature
We also searched the literature for prior studies that measured the level of expression of these C&Ckines in AML or MDS. For many of these C&Ckines, there were no publications with data on expression in AML or MDS. A comparison of the results obtained from the multiplex system versus those found in the literature for AML and MDS is shown in supplemental Table 2. Results obtained with the multiplex assay in AML and MDS were generally similar to, or higher than, the published literature.
Comparison of expression in AML, MDS, and normal samples using multiplex analysis
The median level and range of expression, for newly diagnosed AML, relapsed AML, and MDS are shown in Table 2. The distribution of expression of the normalized values for each C&Ckine in AML and MDS relative to the normal samples is shown in Figure 1 and supplemental Figure 2. With the exception of CCL2 and IL-2, C&Ckine expression was generally shifted in only one direction (ie, values were normal and higher, or normal and lower). The mean levels of expression of IL-4, IL-6, IL-7, IL-10, IFN-γ, CCL3, and PDGF-BB were all significantly lower in AML and MDS compared with expression in normal samples, with t test P values below the Bonferroni-corrected P (ie, nominal P < .0018). Levels of FGF2, IL-13, and CCL5 were also lower, with nominal P values less than .004, which corresponds to a Bonferroni-corrected P < .108. Likewise, the mean expression of CSF3, IL-1RA, IL-8, IL-12, IL-15, and CXCL10 was significantly higher than normal in AML and MDS at the Bonferroni cutoff (nominal P< .0018), and TNF-α and IL-17 were also higher with nominal P values less than .006 (Bonferroni-corrected P < .162). The expression patterns of IL-1B, IL-2, IL-5, IL-9 CCL11, CSF3, CCL4, CCL2, and VEGFA were not statistically different in AML and MDS compared with the normal controls. There were no significant differences between levels in samples obtained at diagnosis compared with those collected at relapse.
C&Ckines with differential expression by cytogenetic risk category in AML patients
| C&Ckines . | F statistic . | P . | Relative fold change in expression . | ||
|---|---|---|---|---|---|
| Favorable . | Intermediate . | Unfavorable . | |||
| CCL2 | 4.67 | .011 | 1 | 5 | 6.67 |
| IL-7 | 4.3 | .015 | 3.37 | 1 | 1.39 |
| PDGF-BB | 3.37 | .037 | 1 | 2.7 | 2.03 |
| C&Ckines . | F statistic . | P . | Relative fold change in expression . | ||
|---|---|---|---|---|---|
| Favorable . | Intermediate . | Unfavorable . | |||
| CCL2 | 4.67 | .011 | 1 | 5 | 6.67 |
| IL-7 | 4.3 | .015 | 3.37 | 1 | 1.39 |
| PDGF-BB | 3.37 | .037 | 1 | 2.7 | 2.03 |
Percentages of AML (left) and MDS (right) samples that are below, within, or above the range of cytokine expression observed in the 19 samples from normal persons.
Percentages of AML (left) and MDS (right) samples that are below, within, or above the range of cytokine expression observed in the 19 samples from normal persons.
Because MDS is increasingly recognized to have a different pathophysiology from AML, we next asked whether C&Ckine expression varied between these 2 diseases. After applying the Bonferroni correction, the expression profiles of AML and MDS were statistically indistinguishable for 24 of the 27 C&Ckines with only IL-8 (nominal P = .0013; Bonferroni P = .035) and IL-13 (nominal P = .0008; Bonferroni P = .022), which were higher in AML, and VEGFA (nominal P < .00001; Bonferroni P < .0003), which was higher in MDS being significantly different.
Cytokine levels correlate with cytogenetics but not clinical features
The level of expression of the individual C&Ckines showed no correlation with age, gender, or hemoglobin at the time of diagnosis. The presence of infection was correlated with IL-8 levels (nominal P = .01), but this was not significant after Bonferroni correction. The white blood cell count correlated with IL-1RA and IL-1B (R = 0.2, for each and nominal P = .001 and .0009, respectively) and the platelet count with PDGF-BB (R = .55, nominal P = .0001). Among newly diagnosed AML patients, expression of CCL2, PDGF-BB, and IL-7 (nominal P < .03) was significantly associated with cytogenetics (Table 2), although none was significant after Bonferroni correction. However, having 3 with significant differences (P < .05) exceeded expectations (expectation = .05 × 27 = 1.35 events) for this cutoff. Platelet-derived cytokine release could have contributed to PDGF levels, but we observed no evidence of correlation with platelet count.
Association between individual cytokine expression and outcome
Among all the AML cases, patients who attained remission were more likely to have higher levels of CCL5, IL-4, IL-10, IL-2, and IL-5 (nominal P = .0015, .0019, .007, .02, and .03, respectively) and lower levels of CCL2 and TNF-α (nominal P = .014 and .05, respectively), but only CCL5 was significant after Bonferroni correction. No individual cytokine was predictive of remission attainment in MDS. Higher levels of CCL5, IL-2, and IL-5, and lower levels of IL-8 and CCL4 were associated with longer survival in AML, and higher levels of IL-4 and CCL3 were associated with longer survival in MDS (Table 3). CCL5, IL-8, and IL-4 retained significance after Bonferroni correction. Notably, TNF-α, which was prognostic for outcome in AML in another study,16 did not achieve statistical significance in this dataset (nominal P = .12)
Cytokines predictive of survival individually
| Disease/cytokine . | Log-rank . | P . | Hazard ratio . |
|---|---|---|---|
| AML | |||
| CCL5 | 9.52 | .002 | 0.88 |
| IL-8 | 7.11 | .007 | 1.14 |
| IL-2 | 5.39 | .02 | 0.89 |
| CCL4 | 4.07 | .04 | 1.21 |
| IL-5 | 3.67 | .05 | 0.88 |
| MDS | |||
| IL-4 | 6.99 | .008 | 0.69 |
| CCL3 | 4.65 | .03 | 0.84 |
| Disease/cytokine . | Log-rank . | P . | Hazard ratio . |
|---|---|---|---|
| AML | |||
| CCL5 | 9.52 | .002 | 0.88 |
| IL-8 | 7.11 | .007 | 1.14 |
| IL-2 | 5.39 | .02 | 0.89 |
| CCL4 | 4.07 | .04 | 1.21 |
| IL-5 | 3.67 | .05 | 0.88 |
| MDS | |||
| IL-4 | 6.99 | .008 | 0.69 |
| CCL3 | 4.65 | .03 | 0.84 |
C&Ckines form recurrent patterns of expression that correlate with outcome.
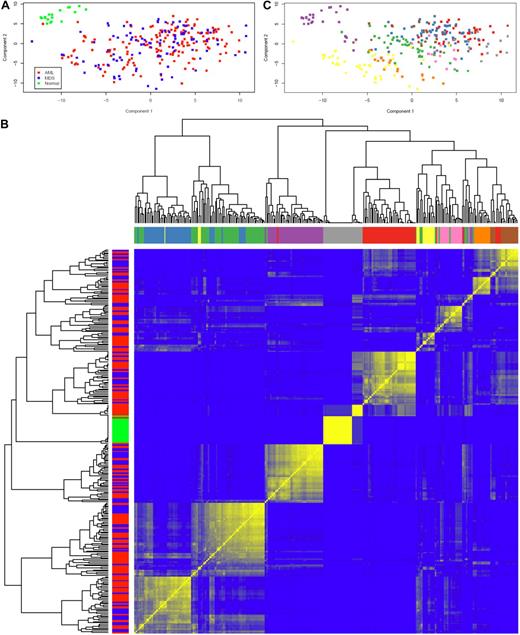
Having evaluated the cytokines individually, we next asked whether there were patterns of cytokine expression in AML and MDS. In the PC plot in Figure 2A, normal samples form a distinct cluster separate from AML and MDS, which were indistinguishable. This finding was supported by repeated hierarchical clustering (with distances defined by Pearson correlation and Ward linkage) after perturbing the data by adding normal noise (Figure 2B). Based on silhouette width on the original clustering and the perturbation reclustering, there appear to be 9 C&C “signatures” as shown in Figure 2C. To determine which C&Ckines were more informative for clustering, we performed one-way analysis of variance for each cytokine using a β-uniform model with a false discovery rate of 1%. This identified 13 C&Ckines as informative: IL-1RA, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12, IL-13, IL-15, IL-17, FGF2, CXCL10, and CCL4. We also used the “bimodality index”32 to identify C&Ckines that separate samples into at least 2 different normally distributed classes. This identified 11 bimodal C&Ckines, 6 of which were also in the informative group (IL-2, IL-4, IL-12, IL-15, IL-17, and FGF2) and 5 others that were not (CSF2, TNF-α, CCL5, CCL3, and PDGF-BB). We used 4 sets of cytokines “all,” “interesting,” “bimodal,” and the union of these 2 sets (n = 27, 13, 11, and 18 C&Ckines) and tested the stability of clustering using different clustering algorithms and subsets of patients. Based on silhouette widths and a computed average confidence as a measure of how often the same samples clustered together, we concluded that the most robust clustering of the samples was the one defined by the bimodal cytokines. Very few patients move from one signature group to another between “bimodal,” “interesting,” and their union. The expression of each C&Ckine for each patient is shown in supplemental Figure 3, and the expression for each patient based on the 11 bimodal C&Ckines is shown in Figure 3. The median expression of the 11 informative C&Ckines in the 9 signatures is shown in Figure 4. Cases of AML and MDS were equally distributed in the 9 signatures with the exception of group 7 (MDS rich) and group 8 (AML only, no MDS cases) (Table 4). Signatures did not differ with respect to age or performance status. Cytogenetics were generally evenly distributed across the C&C signatures (Table 4), with the exceptions that the favorable patients were all confined within bimodal groups 1, 3, and 4 and bimodal group 8 had a very high percentage (11 of 14) with unfavorable cytogenetics.
Principal component analysis and unsupervised hierarchical clustering of C&Ckines. (A) Plot of the first 2 PCs of the C&Ckine data. Normal samples (green) form a separate cluster, but MDS (blue) and AML (red) are indistinguishable. (B) Heatmap showing robustness of clustering in the presence of noise with SD = 1. For each of the 9 sample clusters, each sample clustered with almost all of its neighbors in the assigned cluster more than 50% of the time (and many samples did so > 80% of the time). Based on the silhouette width, more than 90% of samples were correctly clustered. Color bars along the y-axis show diagnosis (green represents normal; red, AML; and blue, MDS) and along the x-axis indicate the 9 highest branches in the dendogram. (C) Clustering of samples based on the dendogram in panel C. Unsupervised hierarchical clustering of C&Ckines based on the Pearson correlation of log intensity across samples using average linkage is shown by the dendogram across the top and left side. Results of a bootstrap cluster test of the reproducibility of hierarchical clustering of cytokines is shown with the color indicating the percentage of times that each pair of samples clustered together, with pure blue = 0% and pure yellow = 100%.
Principal component analysis and unsupervised hierarchical clustering of C&Ckines. (A) Plot of the first 2 PCs of the C&Ckine data. Normal samples (green) form a separate cluster, but MDS (blue) and AML (red) are indistinguishable. (B) Heatmap showing robustness of clustering in the presence of noise with SD = 1. For each of the 9 sample clusters, each sample clustered with almost all of its neighbors in the assigned cluster more than 50% of the time (and many samples did so > 80% of the time). Based on the silhouette width, more than 90% of samples were correctly clustered. Color bars along the y-axis show diagnosis (green represents normal; red, AML; and blue, MDS) and along the x-axis indicate the 9 highest branches in the dendogram. (C) Clustering of samples based on the dendogram in panel C. Unsupervised hierarchical clustering of C&Ckines based on the Pearson correlation of log intensity across samples using average linkage is shown by the dendogram across the top and left side. Results of a bootstrap cluster test of the reproducibility of hierarchical clustering of cytokines is shown with the color indicating the percentage of times that each pair of samples clustered together, with pure blue = 0% and pure yellow = 100%.
Heatmap of the cytokine data for individual cases. C&Ckine expression for each of the 11 “bimodal” C&Ckines (y-axis) is shown for all cases (x-axis) with the cases sorted into 8 signatures (color boxes along left side x-axis). For display purposes, columns have been standardized to mean of zero and SD = 1. Bimodal cytokines were clustered using Ward linkage and Spearman rank correlation to define distance.
Heatmap of the cytokine data for individual cases. C&Ckine expression for each of the 11 “bimodal” C&Ckines (y-axis) is shown for all cases (x-axis) with the cases sorted into 8 signatures (color boxes along left side x-axis). For display purposes, columns have been standardized to mean of zero and SD = 1. Bimodal cytokines were clustered using Ward linkage and Spearman rank correlation to define distance.
Median expression heatmap for 8 C&Ckine expression signatures. The median expression of each analyte in each signature is shown.
Median expression heatmap for 8 C&Ckine expression signatures. The median expression of each analyte in each signature is shown.
Clinical features and response data for C&Ckine signatures based on bimodal C&Ckines
| Category . | B1 . | B2 . | B3 . | B4 . | B5 . | B6 . | B7 . | B8 . | B9 . |
|---|---|---|---|---|---|---|---|---|---|
| All cases | |||||||||
| AML | 21 | 38 | 20 | 38 | 11 | 8 | 9 | 15 | 2 |
| MDS | 23 | 18 | 14 | 20 | 4 | 8 | 13 | 0 | 1 |
| Normal | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 19 |
| B1 | B2 | B3 | B4 | B5 | B6 | B7 | B8 | Total | |
| New AML | |||||||||
| No. | 17 | 19 | 16 | 31 | 10 | 7 | 8 | 14 | 122 |
| AHD/secondary AML | 23.5% | 47.4% | 43.8% | 35.5% | 50.0% | 42.9% | 37.5% | 50.0% | 40.2% |
| Favorable cytogenetics | 5.9% | 0.0% | 18.8% | 9.7% | 0.0% | 0.0% | 0.0% | 0.0% | 5.7% |
| Intermediate cytogenetics | 41.2% | 52.6% | 25.0% | 51.6% | 50.0% | 71.4% | 62.5% | 21.4% | 45.1% |
| Unfavorable cytogenetics | 52.9% | 47.4% | 56.3% | 38.7% | 50.0% | 28.6% | 37.5% | 78.6% | 49.2% |
| FLT3 mutation | 25.0% | 15.4% | 0.0% | 12.5% | 28.3% | 50.0% | 33.0% | 22.0% | 18.8% |
| Treated new AML | |||||||||
| Treated | 17 | 18 | 16 | 28 | 10 | 6 | 7 | 12 | 114 |
| CR | 76.5% | 33.3% | 68.8% | 71.4% | 50.0% | 50.0% | 28.6% | 58.3% | 58.8% |
| Resistant | 23.5% | 38.9% | 25.0% | 25.0% | 20.0% | 50.0% | 71.4% | 25.0% | 30.7% |
| Fail | 0.0% | 27.8% | 6.3% | 3.6% | 30.0% | 0.0% | 0.0% | 16.7% | 10.5% |
| Relapse | 76.9% | 66.7% | 36.4% | 55.0% | 80.0% | 66.7% | 50.0% | 57.1% | 59.7% |
| Alive | 23.5% | 5.6% | 18.8% | 25.0% | 10.0% | 16.7% | 14.3% | 25.0% | 18.4% |
| Category . | B1 . | B2 . | B3 . | B4 . | B5 . | B6 . | B7 . | B8 . | B9 . |
|---|---|---|---|---|---|---|---|---|---|
| All cases | |||||||||
| AML | 21 | 38 | 20 | 38 | 11 | 8 | 9 | 15 | 2 |
| MDS | 23 | 18 | 14 | 20 | 4 | 8 | 13 | 0 | 1 |
| Normal | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 19 |
| B1 | B2 | B3 | B4 | B5 | B6 | B7 | B8 | Total | |
| New AML | |||||||||
| No. | 17 | 19 | 16 | 31 | 10 | 7 | 8 | 14 | 122 |
| AHD/secondary AML | 23.5% | 47.4% | 43.8% | 35.5% | 50.0% | 42.9% | 37.5% | 50.0% | 40.2% |
| Favorable cytogenetics | 5.9% | 0.0% | 18.8% | 9.7% | 0.0% | 0.0% | 0.0% | 0.0% | 5.7% |
| Intermediate cytogenetics | 41.2% | 52.6% | 25.0% | 51.6% | 50.0% | 71.4% | 62.5% | 21.4% | 45.1% |
| Unfavorable cytogenetics | 52.9% | 47.4% | 56.3% | 38.7% | 50.0% | 28.6% | 37.5% | 78.6% | 49.2% |
| FLT3 mutation | 25.0% | 15.4% | 0.0% | 12.5% | 28.3% | 50.0% | 33.0% | 22.0% | 18.8% |
| Treated new AML | |||||||||
| Treated | 17 | 18 | 16 | 28 | 10 | 6 | 7 | 12 | 114 |
| CR | 76.5% | 33.3% | 68.8% | 71.4% | 50.0% | 50.0% | 28.6% | 58.3% | 58.8% |
| Resistant | 23.5% | 38.9% | 25.0% | 25.0% | 20.0% | 50.0% | 71.4% | 25.0% | 30.7% |
| Fail | 0.0% | 27.8% | 6.3% | 3.6% | 30.0% | 0.0% | 0.0% | 16.7% | 10.5% |
| Relapse | 76.9% | 66.7% | 36.4% | 55.0% | 80.0% | 66.7% | 50.0% | 57.1% | 59.7% |
| Alive | 23.5% | 5.6% | 18.8% | 25.0% | 10.0% | 16.7% | 14.3% | 25.0% | 18.4% |
B1 through B8 indicates C&Ckine signature number based on bimodal analytes.
AHD/second indicates antecedent hematologic disorder ≥ 2 months or secondary AML; FLT3 mutation, positive test for an FLT-internal tandem duplication or D835 mutation; and CR, complete remission.
Cytokine signatures form 3 groups prognostic of response and survival
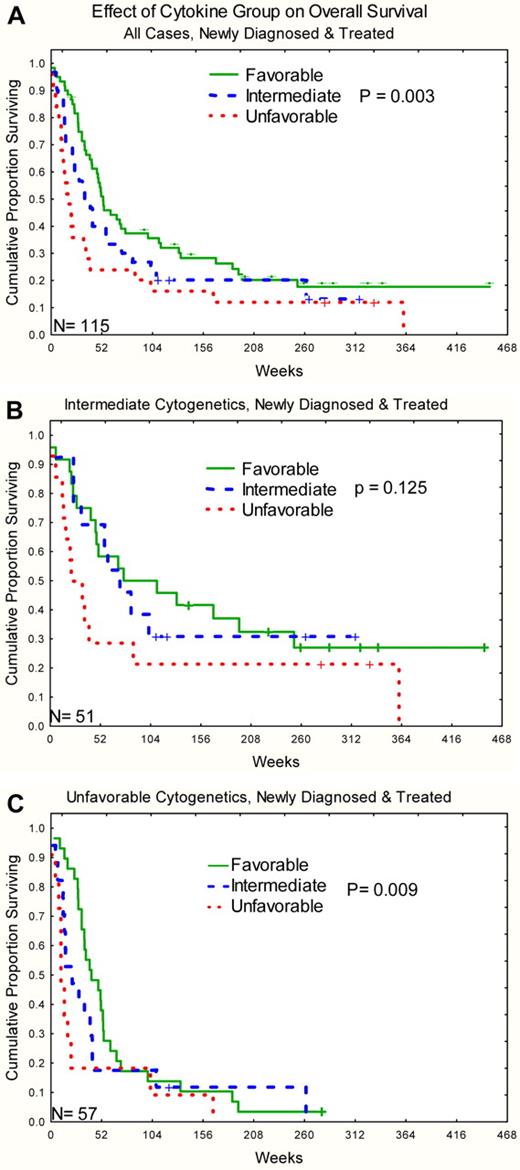
For outcomes analysis, the 8 signatures from the bimodal clustering were used with the omission of the 3 cases that clustered with the normal cluster 9. Response rates by disease and signature for bimodal C&C are shown in Table 4. Bayesian analysis was performed and supported the idea that there were 3 different groupings of signatures based on “CR” outcomes based on posterior probabilities of a 10% difference in CR rates. The favorable group consisted of signatures B1, B3, and B4, an intermediate group composed of signatures B5, B6, and B8, and an unfavorable group composed of signatures B2 and B7. Distribution of selected clinical features and outcomes for these 3 groups is shown in Table 5. Patients in the favorable group had higher complete remission rates and lower primary resistance rates than those in the unfavorable group. Relapse rates and remission durations did not differ between the 3 groups. Overall survival was significantly different with median survival of 52, 32, and 17 weeks, respectively, for the favorable, intermediate, and unfavorable groups (P = .003; Figure 5A). Similar patterns were observed for both intermediate and unfavorable cytogenetic subsets (P = .12 and .009), with there being too few favorable cytogenetic cases to test this effect (Figure 5B-C). Likewise, similar patterns were observed in HDAC and non–HDAC-treated cases (P = .09 and .03, respectively) Multivariable analysis was performed, including traditional AML prognostic features (as listed in “Methods”) and C&Cgroup with overall survival as the outcome. The final model included age, cytogenetics, lactate dehydrogenase, creatinine, hemoglobin, and C&C group (Table 6).
Clinical features and response data for the 3 cytokine signatures based cohorts based on bimodal cytokines
| Category . | Good . | Intermediate . | Bad . | P . |
|---|---|---|---|---|
| All cases | ||||
| AML | 79 | 34 | 47 | — |
| MDS | 57 | 12 | 31 | — |
| New AML | ||||
| No. | 64 | 31 | 27 | — |
| AHD/second | 34.4% | 48.4% | 44.4% | .37 |
| Favorable cytogenetics | 10.9% | 0.0% | 0.0% | .09 |
| Intermediate cytogenetics | 42.2% | 41.9% | 55.6% | |
| Unfavorable cytogenetics | 46.9% | 58.1% | 44.4% | |
| FLT3 mutation | 11.9% | 29.2% | 21.1% | .22 |
| Treated new AML | ||||
| Treated | 61 | 28 | 25 | — |
| Complete remission | 72.1% | 53.6% | 32.0% | .005 |
| Resistant | 24.6% | 28.6% | 48.0% | |
| Fail | 3.3% | 17.9% | 20.0% | |
| Relapse | 56.8% | 66.7% | 62.5% | .78 |
| Alive | 23.0% | 17.9% | 8.0% | — |
| Median survival, wk | 52.3 | 37.4 | 16.8 | .003 |
| Category . | Good . | Intermediate . | Bad . | P . |
|---|---|---|---|---|
| All cases | ||||
| AML | 79 | 34 | 47 | — |
| MDS | 57 | 12 | 31 | — |
| New AML | ||||
| No. | 64 | 31 | 27 | — |
| AHD/second | 34.4% | 48.4% | 44.4% | .37 |
| Favorable cytogenetics | 10.9% | 0.0% | 0.0% | .09 |
| Intermediate cytogenetics | 42.2% | 41.9% | 55.6% | |
| Unfavorable cytogenetics | 46.9% | 58.1% | 44.4% | |
| FLT3 mutation | 11.9% | 29.2% | 21.1% | .22 |
| Treated new AML | ||||
| Treated | 61 | 28 | 25 | — |
| Complete remission | 72.1% | 53.6% | 32.0% | .005 |
| Resistant | 24.6% | 28.6% | 48.0% | |
| Fail | 3.3% | 17.9% | 20.0% | |
| Relapse | 56.8% | 66.7% | 62.5% | .78 |
| Alive | 23.0% | 17.9% | 8.0% | — |
| Median survival, wk | 52.3 | 37.4 | 16.8 | .003 |
AHD/second indicates antecedent hematologic disorder ≥ 2 months or secondary AML; FLT3 mutation, positive test for an FLT-internal tandem duplication or D835 mutation; and —, not applicable.
Kaplan-Meier survival curve for overall survival stratified by C&Ckine signature group. (A) All cases. (B) Intermediate cytogenetics. (C) Unfavorable cytogenetics. The number of cases is shown in the lower left corner of each graph.
Kaplan-Meier survival curve for overall survival stratified by C&Ckine signature group. (A) All cases. (B) Intermediate cytogenetics. (C) Unfavorable cytogenetics. The number of cases is shown in the lower left corner of each graph.
Multivariable analysis
| Variable . | β . | SE . | z-value . | HR . | LRT . | P* . |
|---|---|---|---|---|---|---|
| Age | 0.0545 | 0.0111 | 4.923 | 1.056 | 25.389 | < .00001 |
| Intermediate cytogenetics | −1.0895 | 0.778 | −1.400 | 0.336 | 7.88 | .01942 |
| Unfavorable cytogenetics | −0.4097 | 0.764 | −0.536 | 0.664 | — | — |
| LDH | 0.0003 | 0.000066 | 4.754 | 1.0003 | 12.37 | .00044 |
| Hemoglobin | −0.1420 | 0.0072 | −1.972 | 0.868 | 4.066 | .04376 |
| Creatinine | 0.3753 | 0.172 | 2.178 | 1.455 | 3.335 | .06781 |
| C&C group intermediate | −0.4509 | 0.333 | −1.354 | 0.637 | 12.495 | .00193 |
| C&C group good | −1.0360 | 0.297 | −3.491 | 0.355 | — | — |
| Variable . | β . | SE . | z-value . | HR . | LRT . | P* . |
|---|---|---|---|---|---|---|
| Age | 0.0545 | 0.0111 | 4.923 | 1.056 | 25.389 | < .00001 |
| Intermediate cytogenetics | −1.0895 | 0.778 | −1.400 | 0.336 | 7.88 | .01942 |
| Unfavorable cytogenetics | −0.4097 | 0.764 | −0.536 | 0.664 | — | — |
| LDH | 0.0003 | 0.000066 | 4.754 | 1.0003 | 12.37 | .00044 |
| Hemoglobin | −0.1420 | 0.0072 | −1.972 | 0.868 | 4.066 | .04376 |
| Creatinine | 0.3753 | 0.172 | 2.178 | 1.455 | 3.335 | .06781 |
| C&C group intermediate | −0.4509 | 0.333 | −1.354 | 0.637 | 12.495 | .00193 |
| C&C group good | −1.0360 | 0.297 | −3.491 | 0.355 | — | — |
HR indicates hazard ratio (the exponential of the β coefficient); LRT, likelihood ratio test statistic; and —, not applicable.
P value for significance of LRT.
Discussion
C&Ckines have both pathogenic and therapeutic implication in leukemias.3 Numerous studies have documented that leukemic blasts show abnormal degrees of responsiveness to cytokine stimulation and that leukemic blasts are often the source of cytokine production, potentially initiating or sustaining paracrine or autocrine loops. Despite this evidence of relevance to leukemia biology, the expression levels of most C&Ckines has been incompletely documented in AML and MDS, and no study has looked for patterns of expression. This analysis addressed this deficiency by simultaneously studying the expression of 27 different C&Ckines in the serum in a large series of AML and MDS patient samples using the Bioplex multiplex technology. This is the first published report of expression levels for half of these analytes in AML and for nearly all in MDS. There are numerous findings from this analysis.
First, the Bioplex technology gave results comparable with those obtained with other methodologies (typically enzyme-linked immunosorbent assay). In many cases, the average level was lower with the Bioplex, perhaps reflecting better sensitivity for lower amounts of those analytes compared with other technologies. The methodology was robust and reliable. There was no evidence of sample degradation, despite up to 9 years of storage for some samples. Likewise, there was insignificant tray-to-tray variance, ascertained both by the reproducibility of the standards and the equality of the median and range of expression for each cytokine across each tray. Thus, we observed no evidence of a batch effect. Consequently, our confidence in the reliability for the methodology is very high.
The second finding is that C&Ckine expression was largely independent of most clinical features, such as age, gender, or hemoglobin. The white blood cell count was correlated with IL-1RA and IL-1B and the platelet count with PDGF-BB. The implication is that inclusion of information from cytokine profiling will be simplified by not needing to take these variables into account.
The normal controls had highly similar expression patterns that clustered together tightly, distinct from nearly all cases of AML and MDS. The levels of most C&Ckines in AML and MDS differed significantly from normal with 6 being lower and 7 higher than normal. Although they are clinically distinct diseases, C&Ckine expression in AML and MDS is extremely similar. Differential expression between them was limited to higher level of IL-8 and IL-13 in AML and higher levels of VEGFA in MDS. Exposure of AML blasts to IL-13 was reported to result in increased proliferation and decreased levels of IL-1RA, IL-1B, IL-6, TNF-α, CSF2, and CSF3,35,36 and we saw a similar pattern in signature B8. The VEGFA data support findings by others that microvascular density, promoted by the angiogenesis inducing effects of VEGFA, is higher in MDS and AML than in normal marrow and confers an adverse prognosis.25,37 C&Ckine levels were relatively similar in samples from newly diagnosed and relapsed patients.
To our surprise, 5 cytokines, more than would be predicted by chance, appeared to have prognostic relevance for remission attainment or survival in AML when considered individually. In our data, we observed that expression of some C&Ckines was strongly correlated with certain cytogenetic abnormalities. Thus, these C&Ckines could be produced by the leukemic cells in an autocrine stimulation loop, or the leukemic cells could be inducing production by other cells to establish paracrine loops. Notably, IL-7, CCL2, and PDGF-BB were much more highly expressed in cases with intermediate and unfavorable prognosis cytogenetics. TNF-α expression was highest in unfavorable signatures B2 and B7 and lowest in favorable signatures B1 and B3. A similar association between adverse cytogenetics and higher TNF-α mRNA expression was observed in MDS with unfavorable cytogenetic abnormalities (11 of 14 cases) compared with AML with unspecified cytogenetics (0 of 6) or controls (0 of 8).38 In a study by Vinante et al,39 levels of soluble TNF-α receptor were higher than normal in the serum of 62 AML patients and seemed to derive from the AML blasts, and, similar to this study, was associated with intermediate and unfavorable cytogenetics and an adverse outcome. Tsimberidou et al also observed an adverse prognostic impact of high TNF-α expression in AML.16 In patients with myelodysplasia, several studies have demonstrated high levels of TNF-α in the bone marrow, either in the neoplastic cells or the monocytes and macrophages.38,40,41
The final observation was that expression of these C&Ckines formed 8 recurrent patterns of expression or “cytokine signatures” with prognostic implications independent of age, cytogenetics, performance status, and antecedent hematologic disorders. Furthermore, these 8 signatures combine to form 3 groups with favorable, intermediate, and unfavorable prognostic implications for remission attainment and overall survival. The observation that recurrent signatures are formed suggests that there are interactions between leukemic cells in AML and MDS and their environments that lead to specific patterns of expression. This is reinforced by the association between specific C&C groups and particular protein expression signatures, which suggests that cytokine expression patterns have a recurrent effect on protein expression signatures.
These patterns suggest a means to rationally use agents that interact (blocking or augmenting) with a particular C&Ckine only when it is relevant to the biology of the leukemic cell. Signature B2, B4, and B5 with high TNF-α and CSF2 provides such an opportunity. TNF-α has different functions, either predisposing cells to apoptosis or promoting growth and survival,42 possibly in a NF-κB dependent manner,43 but it is not clear which predominates in AML. TNF-α has also been shown to inhibit CSF3 receptor expression in AML blasts, thereby inhibiting proliferation,44 and this could decrease sensitivity to cycle specific chemotherapy, such as Ara-C. There are 2 clinically approved agents that target TNF-α: Etanercept (Enbrel), a recombinant dimeric molecule composed of the soluble extracellular TNF-α binding portion of the TNF receptor linked to the Fc portion of IgG1, and infliximab (Remicade), a chimeric monoclonal anti-TNF-α antibody that binds both soluble and membrane-bound TNF-α. Both agents have been tested in small clinical trials in MDS, and some hematologic responses were observed. Anti-TNF-α therapy was not tried in combination with chemotherapy, in MDS or AML.45 There are also reports of trials with anti-CSF2 antibodies in autoimmune diseases, and there are ample data that CSF2 and TNF-α act synergistically to drive inflammation.46 We hypothesize that this combination of C&Ckines present in signatures B2, B4, and B5 results in resistance of AML cells to chemotherapy. If verified, therapeutic strategies that interfere with this by blocking TNF-α alone, or in combination with CSF2 blockade, might restore sensitivity to chemotherapy. This signature was observed in approximately 50% of cases of AML, so clinical application of this therapy could be used in a significant portion of AML patients.
This study is limited by the composition of the 27 C&Ckines on the available panel, which omits some C&Ckines that are probably relevant in AML. Notably missing are IL-3 and stem cell factor. Another kit, with 23 additional analytes, is now available, and analysis using both kits is underway in a separate cohort of patients. We used serum for this study, but it has been observed that levels of 44 of 80 cytokines, chemokines, and growth factors were significantly different between serum and plasma.33 Levels might also be different if “marrow juice,” which may be more representative of the intra-marrow milieu, was used instead of serum. Conceivably, functionally important levels within the marrow might be insufficient to impact systemic levels. We initially attempted to collect marrow juice but abandoned this over concerns about standardization. Specifically, we collect our marrow in 3 mL of anticoagulant, but the amount of marrow that we receive in the MDACC Leukemia Sample Bank varies from 1 to 7 mL, so variance in the degree of dilution creates one problem, although careful monitoring of volume received could account for this. More vexing, because it cannot be accounted for, is the fact that research samples are drawn on the fourth pull after the samples for clinical tests are collected (morphology, cytogenetics, and flow cytometry, and other molecular studies are drawn first), and there is inevitably some unaccountable degree of dilution of marrow with peripheral blood.
Fully understanding the relationship of C&Ckines to their effect on leukemia cell biology requires additional information. The level of expression of the cell surface receptors, and/or the presence of receptor mutations, would modulate the effect of serum or marrow cytokine levels. Likewise, abnormalities in STP activation, with cross-talk leading to activation independent of cytokine level or ligand-receptor engagement, or mutations leading to constitutive activation (eg, Fms-like tyrosine kinase 3-internal tandem duplication) independent of cytokine level, could generate a similar physiologic effect within a cell regardless of the level of cytokine expression outside of the cell. In addition, if the leukemic cell is producing the cytokine, or inducing local production by nearby stroma, then autocrine or paracrine loops could be active without affecting levels in the periphery.47 We did not examine whether the cytokines come from the leukemia cells, the stroma, or elsewhere. There is evidence that spontaneously cytokine production is higher in lymphocytes from AML patients compared with lymphocytes from normal persons.48 In future studies, we plan to use these same assays to measure the level of intracellular cytokines to address the autocrine loop question. In separate research, we have used reverse-phase arrays to measure protein and phosphoprotein levels from STP, apoptosis, and cell cycle regulating pathways and their downstream targets8 and are expanding our validated antibody repertoire to include cytokine receptors. Integration of these datasets will enable us to look at the level of cytokines, their receptors, and the activation status of the STP they activate. We think that most of these pathways will behave according to the canonical pathways but that alternate means of activation will be observed and that these will result in common physiologies despite diverse etiologies. Understanding the role of C&Ckines in STP activation on an individualized basis will determine whether strategies designed to interfere with cytokine-based signaling probably work, or be frustrated by, the biology of the leukemic blast. Thus the rational use of C&Ckine effect modulating agents based on this strategy would result in improved response and outcome and could minimize the potential toxicity associated with C&Ckine inhibitors by their selective application to circumstances where that particular cytokine is relevant to the biology of the leukemia cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.M.K. designed the research, performed the research, performed data analysis, and wrote the manuscript; D.M. and N.S. performed the analysis and assisted with data analysis and the manuscript; W.C. generated the database for the analysis, participated in the data analysis, and assisted with the manuscript; Z.E. assisted with research design, data interpretation, and the manuscript; and K.R.C. participated in research design, performed the statistical analysis, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Steven M. Kornblau, Section of Molecular Hematology and Therapy, Unit 448, University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030-4095; e-mail: skornblau@mdanderson.org.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal