Abstract
We have studied the effect of a 13-bp deletion in the promoter of the von Willebrand factor (VWF) gene in a patient with type 1 von Willebrand disease. The index case has a VWF:Ag of 0.49 IU/mL and is heterozygous for the deletion. The deletion is located 48 bp 5′ of the transcription start site, and in silico analysis, electrophoretic mobility shift assays, and chromatin immunoprecipitation studies all predict aberrant binding of Ets transcription factors to the site of the deletion. Transduction of reporter gene constructs into blood outgrowth endothelial cells showed a 50.5% reduction in expression with the mutant promoter (n = 16, P < .001). A similar 40% loss of transactivation was documented in transduced HepG2 cells. A similar marked reduction of transgene expression was shown in the livers of mice injected with the mutant promoter construct (n = 8, P = .003). Finally, in studies of BOEC mRNA, the index case showed a 4.6-fold reduction of expression of the VWF transcript associated with the deletion mutation. These studies show that the 13-bp deletion mutation alters the binding of Ets (and possibly GATA) proteins to the VWF promoter and significantly reduces VWF expression, thus playing a central pathogenic role in the type 1 von Willebrand disease phenotype in the index case.
Introduction
von Willebrand factor (VWF) is a multimeric adhesive glycoprotein that plays several key roles in hemostasis. It mediates platelet adhesion and aggregation at the sites of vascular injury, and acts as a carrier for coagulation factor VIII to protect it from proteolytic degradation.1,2 VWF plasma levels below 50% are found in the most common type of von Willebrand disease (VWD), type 1 VWD, which is characterized by a mild/moderate deficiency of functionally normal VWF. The understanding of the genetic basis and molecular pathophysiology of type 1 VWD has been a challenge, and a heterogeneous array of VWF gene mutations, located throughout the gene, have been documented in these patients. The details of these mutations are listed and updated on the ISTH VWF SSC Database (http://www.sheffield.ac.uk/VWF/index.html).
Three large population studies of type 1 VWD patients have detailed the spectrum of VWF gene mutations in several European Countries and in Canada. However, candidate pathogenic VWF mutations were not found in 27%,3 in 36%,4 and in 28%5 of index cases diagnosed with type 1 VWD. In the Canadian study, 8 sequence variations within the 5′ regulatory region of the VWF gene were found after sequencing DNA from 123 index cases with type 1 VWD. These 5′ regulatory sequence changes represented 16% of the total candidate mutations found in this study. However, to date, there has been no functional evidence to support the pathogenic nature of any of these sequence variations.
Earlier VWF promoter studies have indicated that, in vitro, the basal transcription of the human VWF gene is mediated through a region in the promoter located between base pairs −89 and +19 (relative to transcription start site, TSS +1). This region was functional in both endothelial and nonendothelial cell types.6 In vivo studies demonstrated that VWF sequences between −487 to +247 are responsible for promoter activation in brain vascular endothelial cells.7
The −60/+19 region has been shown to be a critical area within the VWF gene promoter, possessing 2 Ets-binding motifs, with the 5′-most motif being more critical to transcriptional activation.6 In transgenic mice, a larger VWF fragment (2182 bp) containing the 5′-flanking sequences and including exon 1 and intron 1 of the gene has been shown to activate transgene expression in endothelial cells of the heart, skeletal muscles, and brain,8 suggesting that additional distant VWF gene sequences are required for expression in other vascular endothelial cells in vivo. Specific interaction between positive and negative regulatory sequences together with their transcription factors have been identified within this region and have contributed to the endothelial-specific VWF mRNA expression levels.6,9-11 The local vascular microenvironment also appears to contribute to the regulation of endothelial VWF expression,8 and it is well recognized that there is considerable heterogeneity in the distribution and nature of endothelial VWF expression throughout the vasculature.12
Despite some progress in the past 2 decades with respect to the mechanisms that regulate endothelial expression of VWF, many details of this process remain unresolved. Natural transcriptional mutants resulting in the quantitative trait, type 1 VWD, provide an opportunity to add new knowledge in this area. In this study, we provide details of the functional characterization of a 13-bp deletion within the basal promoter of the VWF gene. We document evidence using both in vitro and in vivo experimental approaches that this deletion reduces transcriptional activation of the VWF regulatory region. Overall, these results provide strong evidence that this promoter deletion plays a key pathogenic role in the type 1 VWD phenotype in this patient.
Methods
Confirmation of the c.-1522_-1510del13 mutant, patient phenotype, and screening of normal population
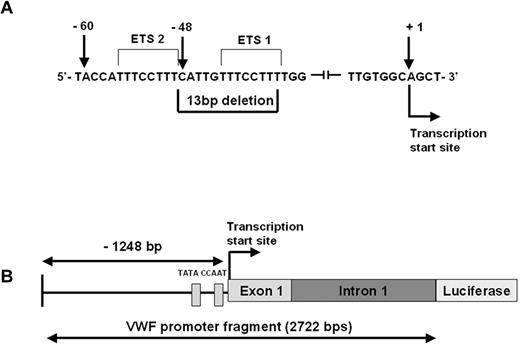
This mutation was identified as a part of the Canadian type 1 VWD study3 following the direct DNA sequencing of the 52 exons of the VWF gene and approximately 50 bp around the exon–intron boundaries. The mutation is a heterozygous deletion of 13 bp (c.-1522_-1510del13: 5′-CATTGTTTCCTTT-3′) within the promoter of the VWF gene, 48 bp upstream of the transcription start site (Figure 1A). The index case is a 40-year-old male who has a clinical history of mild bleeding symptoms (easy bruising and bleeding after procedures). The VWF:Ag was 0.49 IU/mL, VWF:RCo 0.35 IU/mL, and FVIII:C 0.92 IU/mL (single determination). ABO blood group is AB. The mutation was also identified in the son of the index case. We confirmed the mutation in the index case by sequencing of cloned DNA from the patient. The VWF promoter deletion was not found in 100 normal individuals.
The c.-1522_-1510del13 VWF promoter mutation. (A) VWF sequence showing the location of the 13-bp deletion within the promoter of the VWF gene, 48 bp upstream of the transcription start site. The 2 Ets binding sites are indicated. (B) Schematic representation of the VWF promoter fragment including 1248 bp of 5′ flanking sequence, exon 1 and intron 1 (total length 2722 bp). The CAAT and TATA box locations relative to the mutation site are shown.
The c.-1522_-1510del13 VWF promoter mutation. (A) VWF sequence showing the location of the 13-bp deletion within the promoter of the VWF gene, 48 bp upstream of the transcription start site. The 2 Ets binding sites are indicated. (B) Schematic representation of the VWF promoter fragment including 1248 bp of 5′ flanking sequence, exon 1 and intron 1 (total length 2722 bp). The CAAT and TATA box locations relative to the mutation site are shown.
Construction of the HD-Ad vector
A 2722-bp region of the VWF promoter including exon 1 and intron 1 was amplified from the patient's DNA into 2 fragments and cloned into the PCR2.1 plasmid (Figure 1B). Wild-type (WT) and mutant (Mut) clones were confirmed by sequencing. WT and Mut vectors were constructed by ligating the 2 fragments into the shuttle plasmid pSC7 containing the LacZ gene and subsequently cloned into the helper-dependent adenovirus (HD-Ad) precursor plasmid pSC9C containing the luciferase cDNA sequence. All plasmids were kindly provided by Dr Mary Hitt (University of Alberta, Edmonton, AB).13 Construction of the pSC9C vector was performed so that the VWF promoter fragment regulates the transcription of the luciferase gene whereas the LacZ gene acts as an internal transduction control regulated by the ubiquitous β-actin promoter. An HD-Ad vector lacking the VWF promoter fragment was constructed as an additional control. Constructs made were as follows: HD-Ad −Luc WT, HD-Ad −Luc Mut, and HD-Ad −Luc control.
Growing and purification of HD-Ad vector carrying the wild-type and mutant VWF promoter
Growing and purification of HD-Ad vector was performed according to Palmer and Ng.14 Briefly, all HD-Ad plasmids were digested with PmeI to release the ITR-flanked viral genome and used to transfect 116 cells by a standard calcium phosphate method. The transfected cells were infected 16 hours later with Ad2 helper virus (AdNg163; kindly provided by Dr Robin Parks, University of Ottawa, Ottawa, ON). Infected cells were harvested when the complete cytopathic effect was reached 48 hours after transduction. Vectors were further amplified in 4-6 rounds of propagation and the large scale HD-Ad vector was then prepared by coinfecting 3 L of 116 cells at 3-4 × 105 cells/mL with both the AdNg163 and the crude lysate saved from the small scale virus isolation. HD-Ad vectors were purified by 1 step, and 2 continuous CsCl gradients, and then dialyzed in tris(hydroxymethyl)aminomethane (Tris)–HCl buffer (pH 8.0). Ad-vector concentration was determined by spectrophotometric analysis of DNA using NanoDrop 2000 (ThermoScientific). An average of 1 × 1012 virus particles/mL of the total virus yield was obtained using this protocol. The dialyzed virus was used for all in vitro endothelial and nonendothelial cell experiments and the in vivo mouse studies.
Cell culture
In vitro studies were performed by transduction of WT and Mut HD-Ad-VWF vectors into passage 4-6 human blood outgrowth endothelial cells (BOECs). BOECs were isolated from 50 mL of blood from healthy volunteers according to Matsui et al.15 HepG2 cells were used as the nonendothelial cell type for the transduction studies. Early passage human BOECs were also used as the source of nuclear extracts for the electrophoretic mobility shift assays (EMSAs) and chromatin immunoprecipitation (ChIP) analysis.
In vitro expression studies in BOECs using luciferase and β-galactosidase assays
To investigate whether or not there is a difference in expression between the WT and Mut HD-Ad-VWF vectors, BOECs were grown in 6-well culture plates (seeded 1.6 × 105 cells/well) and transduced at 80% confluence with WT and Mut HD-Ad vectors at a multiplicity of infection (MOI) of 25 000. Forty-eight hours later, the cells were harvested, washed with phosphate- buffered saline (PBS), then lysed in 100 μL of lysis buffer. Fifty microliters of the lysate was assayed for luciferase (Luciferase assay kit; Promega) and 10 μL used for the β-gal assays (Galacto-Light TM; Applied Biosystems). HD-Ad vector without the VWF promoter fragment was included as an additional control. All assays were performed in duplicates in 16 independent experiments. Some plates were secured for X-Gal staining to demonstrate the percentage of transduced cells.
EMSAs
To investigate the binding of endothelial cell transcription factors to the region of the VWF promoter involved in this deletion, in silico analysis was performed with TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html) and MatInspector (http://www.genomatrix.de/online_help/help_matinspector/matinspector_help.html). Two endothelial transcription factors, GATA-3 and Ets, showed differential binding between the WT and Mut sequences. Accordingly, nuclear extracts from BOECs were produced using NE-PER Nuclear and Cytoplasmic Extraction Reagent Kit (Thermo Scientific Pierce). Mobility shift assays were performed with a WT-VWF oligonucleotide spanning nucleotides −61 to −32 and a Mut oligonucleotide spanning −70 to −32 (numbers are relative to the VWF transcription start site). Oligonucleotides were labeled with 2 μL of 5mM [α-P32] dATP and annealed. Three microliters of nuclear extract was preincubated for 5 minutes at 4°C in the absence or presence of competitors (50× of double stranded unlabeled oligonucleotides) in binding buffer consisting of 0.8 μL of poly dI-dC, 1mM dithiothreitol (DTT), 10mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) pH 7.8, 50mM KCl,1mM MgCl2, 0.1mM EDTA (ethylenediaminetetraacetic acid), 0.1% NP-40, 1mM spermidine, and 10% glycerol. The probes were added and the reaction mixture was then incubated for 15 minutes at 24°C. Samples were analyzed by non denaturating 6% PAGE in 0.25× TBE. In supershift assays, antibodies to Ets and GATA-3 (Santa Cruz Biotechnology) were incubated with the BOEC nuclear extracts for 30 minutes at room temperature before the addition of the probes. Cross competition assays were performed where we tested the WT and Mut DNA probes with unlabeled competitor oligos (WT and Mut) at 10-, 50-, and 100-fold excess. An NFY probe was used as a control.16
Mice biophotonic studies and analysis of organ-specific reporter gene expression
Animal studies were approved by the Queen's University Animal Care Committee. Nine adult WT Balb/c mice were injected via the tail vein with 3.5 × 1010 viral particles of purified WT or Mut HD-Ad-VWF 24 hours after they had received an intravenous infusion of 250 μL of clodronate liposomes. Mice were imaged 48 hours later. Mice were anesthetized with isoflurane (Abbott Laboratories), injected intraperitoneally with 300 μL of 15 mg/mL D-luciferin (Gold Biotechnology) and placed in a light-tight box. Photon emission was detected 5 minutes later with a cooled charge-coupled device (CCCD) camera (IVIS; Caliper Life Sciences), and digital images were recorded. Mice were killed 4 days after vector injection. The mice were terminally perfused with PBS and organs—liver, kidney, spleen, lungs and heart—were isolated for β-gal transgene expression. β-Gal ELISA kit (Roche) was used according to the manufacturer's instructions. In addition, liver tissues from WT and Mut vector-treated mice were subjected to quantitative real-time PCR for assessment of viral genome copies.
Chromatin immunoprecipitation
DNA/nuclear protein interactions were studied using ChIP-IT Express (Active Motif). Briefly, BOECs were grown to 90% confluency and fixed using 1% paraformaldehyde. The cells were resuspended in sodium dodecyl sulfate lysis buffer containing protease inhibitors, and the cell pellets were treated enzymatically to shear the chromatin into 200-bp fragments. Chromatin complexes were collected by centrifugation at 4°C and diluted 5-fold in ChIP dilution buffer containing protease inhibitor. We wanted to investigate the binding to the DNA sequence adjacent to the deletion to specific Ets and GATA endothelial transcription factors as revealed in the in silico analyses. The sheared samples were immunoprecipitated with antibodies against endothelial transcription factors: Ets-1 and Ets-2 as well as GATA-2, GATA-3, and GATA-6 (Santa Cruz Biotechnology). The liver-specific transcription factor HNF-1 was included as a negative control in the assay. Positive (RNA pol II) and negative (immunoglobulin G [IgG]) controls were also included. Immunoprecipitated chromatin complexes were washed and eluted with elution buffer (50mM Tris-HCl pH 8.0, 10mM ethylenediaminetetraacetic acid, 1% sodium dodecyl sulfate). PCR reaction was designed to amplify the promoter region from −111 to +28, giving an amplicon of 139 bp containing the deletion site. Another PCR was designed to amplify a 164-bp amplicon from +129 to +293 known to contain a GATA binding site at +220. Input DNA representing the sheared chromatin before immunoprecipitation was also included and served as a reference for each PCR.
Allele-specific qRT-PCR analysis of VWF-RNA from BOECs
Total RNA was isolated from BOECs derived from the index case and 6 healthy volunteers using QIAamp RNA blood mini kit according to the manufacturer's instructions. The analysis was performed using a 2-step quantitative real-time polymerase chain reaction (qRT-PCR; RT reaction, then PCR to amplify the cDNA). The cDNA specific fluorogenic probes were obtained from Applied Biosystems, and all real-time experiments were carried out in an ABI Prism 7000 sequence detection system.
Standard curves for VWF and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) control transcripts were generated. The fluorescence intensity was obtained using the standard curves generated from 10-fold dilutions of the cloned gene-specific regions. The VWF cDNA was analyzed using a TaqMan MGB FAM labeled probe, spanning exons 39/40 and normalized using FAM labeled GAPDH endogenous control.
To determine the relative allelic contributions for VWF, a TaqMan SNP genotyping assay, specific for VWF c.2365A/G, p.Thr789Ala (rs1063856), was utilized. The index case is heterozygous (G/A) for this SNP. Healthy controls consisted of 2 homozygotes for allele 1 (A), 1 homozygote for allele 2 (G), and 3 heterozygotes (G/A). Fluorescence values were obtained using standard curves specific for each SNP allele. The transcript ratios of the “A” allele to the “G” allele 2 were compared between the index and healthy individuals.
VWF nomenclature
The c.-1522_-1510del13 designation refers to the new nomenclature based on the Human Genome Variation Society17 and as reported by Goodeve and coworkers,18 where A of ATG initiator codon in exon 2 is +1. According to this nomenclature, genomic DNA nt should be numbered from the initiator ATG as +1. Numbering of the cDNA nucleotides should be sequential throughout the coding regions of the gene, from 1 to 8439, with the A of the initiator ATG site as +1.
Analysis
Statistical comparison of the WT and mutant patterns of expression in the transduction, in vivo mouse biophotonic, and qRT-PCR studies was performed using unpaired Student t tests. Statistical significance was defined as P < .05.
Results
Confirmation of the 13-bp VWF promoter deletion
The mutation c.-1522_-1510del13: 5′-CATTGTTTCCTTT-3′ was previously reported.3 In this study, confirmation of the mutation and the precise site and number of nucleotides deleted was achieved through sequencing of cloned DNA from the index case. This analysis confirmed that the patient is heterozygous for a 13-bp deletion located 48 bp upstream of the transcription start site of the VWF gene (Figure 1A). The pathogenic nature of the deletion is supported by its absence from 100 normal subjects.
In this index case, we also identified 13 additional sequence variations, all of which have been previously reported as polymorphisms in the VWF mutation database. Three SNPs were found in the distal promoter, 4 intronic polymorphisms were identified, and 6 exonic variants were documented. Although we have previously reported an influence from the promoter SNP haplotype, we have no reason to believe that any of these changes has additional significant effects on VWF expression.
The promoter deletion reduces transcriptional activity in transduced endoethelial (BOECs) and nonendothelial (HepG2) cells
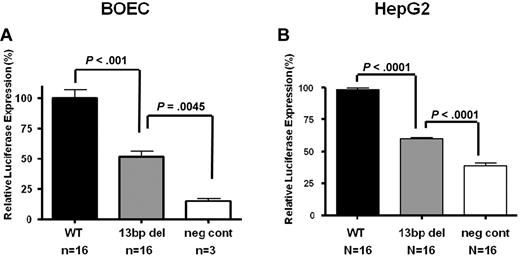
A HD-Ad system was used to quantify transcriptional activity in vitro in BOECs and HepG2 cells. Using this protocol, transduction efficiencies of > 80% were achieved in BOECs as determined by X-gal staining. Because the reporter vector contains both Luc and LacZ coding regions, we were able to measure both test Luciferase and control β-galactosidase activity in transduced cell lysates. Our results showed a consistent 50.5% reduction in the normalized luciferase:β-gal expression levels with the Mut vector compared with the WT vector (n = 16, P < .001; Figure 2A). However, reporter gene transactivation is not abolished by the promoter deletion, as can be seen comparing expression results achieved with the vector backbone alone (n = 3, P = .004). Furthermore, the effect of the mutation is not endothelial cell–specific since a similar 40% reduction in reporter gene expression was documented with the transduction of HepG2 cells (n = 16, P < .0001; Figure 2B).
In vitro transduction studies of VWF WT and Mut vectors in endothelial and HepG2 cells. Transduction of HD-Ad-VWF WT-Luc, HD-Ad–VWF Mut-Luc, Mut and HD-Ad–VWF WT-neg in (A) BOECs and (B) HepG2 cells. In both cell types, there is a significant reduction in normalized luciferase:β-gal expression from the mutant vector compared with the WT vector in BOECs (n = 16, 50.5% reduction, P < .001) and in HepG2 cells (n = 16, 40% reduction, P < .0001). All values shown are means ± SEM.
In vitro transduction studies of VWF WT and Mut vectors in endothelial and HepG2 cells. Transduction of HD-Ad-VWF WT-Luc, HD-Ad–VWF Mut-Luc, Mut and HD-Ad–VWF WT-neg in (A) BOECs and (B) HepG2 cells. In both cell types, there is a significant reduction in normalized luciferase:β-gal expression from the mutant vector compared with the WT vector in BOECs (n = 16, 50.5% reduction, P < .001) and in HepG2 cells (n = 16, 40% reduction, P < .0001). All values shown are means ± SEM.
In vivo biophotonic studies confirm reporter transgene expression with the WT but not mutant vector
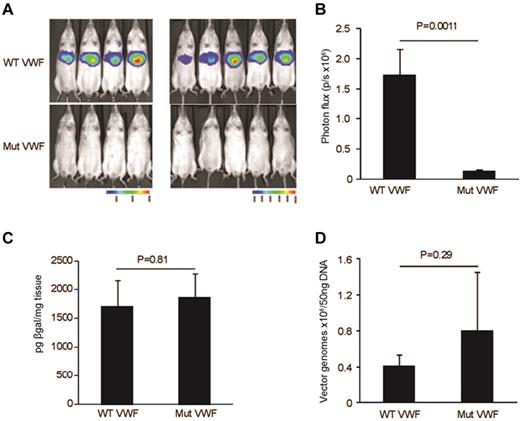
To determine the magnitude and localization of luciferase reporter transgene expression in vivo, bioluminescent imaging was performed in living animals 48 hours after vector administration. Imaging was monitored 10 minutes after administration of luciferin. Data presented in Figure 3A-B show a significant reduction in bioluminescence expression (n = 9, P = .001). From the anatomical localization of the bioluminescent signal, the transgene expression is likely originating from the liver. Previous studies have shown the liver to be the major site of Ad5 mediated sequestration and transduction after intravenous delivery.19-21
In vivo mice biophotonic studies quantifying expression from the VWF-WT and Mut vectors. (A) Bioluminescent imaging was performed in 9 Balb/c mice 48 hours after tail vein injection of 3.5 × 1010 viral particles of HD-Ad. Imaging was monitored 5 minutes after administration of luciferin. (B) Significant reduction in bioluminescence expression (photon flux/second; n = 9, P = .001). (C) β-galactosidase assays on liver isolated from mice injected with WT and Mut vectors. No significant difference in β-gal expression was documented indicating equivalent liver cell transduction (n = 5, P = .8). (D) Quantitative real-time PCR to test the differential HD-Ad liver tropism in mice injected with WT and Mut vectors. The graph shows no significant changes in vector copy number between WT and Mut vector (n = 5, P = .29) complementing results of the β-gal assay. All values shown are means ± SEM.
In vivo mice biophotonic studies quantifying expression from the VWF-WT and Mut vectors. (A) Bioluminescent imaging was performed in 9 Balb/c mice 48 hours after tail vein injection of 3.5 × 1010 viral particles of HD-Ad. Imaging was monitored 5 minutes after administration of luciferin. (B) Significant reduction in bioluminescence expression (photon flux/second; n = 9, P = .001). (C) β-galactosidase assays on liver isolated from mice injected with WT and Mut vectors. No significant difference in β-gal expression was documented indicating equivalent liver cell transduction (n = 5, P = .8). (D) Quantitative real-time PCR to test the differential HD-Ad liver tropism in mice injected with WT and Mut vectors. The graph shows no significant changes in vector copy number between WT and Mut vector (n = 5, P = .29) complementing results of the β-gal assay. All values shown are means ± SEM.
To determine whether the reduction in luciferase expression seen with the Mut vector in the biophotonic studies was influenced by a difference in vector amounts in the liver, we quantified β-gal expression and vector copy numbers in liver tissue isolated from mice injected with the WT and Mut vectors. In these studies, neither β-gal expression (n = 5, P = .81; Figure 3C) nor vector copy numbers (n = 4, P = .29; Figure 3D) showed differences between the WT and Mut vector studies. These results confirm that the reduction in reporter transgene expression is due to the promoter deletion mutation rather than to differences in vector distribution. These results also support the proposal that the liver is the site of reporter transgene expression as predicted by the biophotonic studies.
Endothelial transcription factors bind aberrantly to the mutant VWF region as shown in EMSAs
The in silico analyses, using 2 complementary programs, revealed that Ets transcription factors are predicted to bind to both the WT and mutant sequence. These analyses showed 2 strong Ets binding sites (Figure 1A) and 4 much weaker candidate GATA sites adjacent to and overlapping the promoter deletion. With the MatInspector program, the core and matrix similarity scores for the 2 Ets sites ranged from 0.79 to 1.0 and 0.74 to 0.92, respectively, whereas the putative GATA binding sites showed much lower scores of 0.75 and 0.70-0.73, respectively. EMSAs using endothelial nuclear proteins derived from BOECs confirmed that the pattern of DNA-protein complexes was distinct with probes representing the WT and Mut VWF sequences. Although the patterns of bound complexes appear different in Figure 4A-D, we believe that this is due to differences in electrophoretic separation for independent experiments. However, most importantly, there is always a difference between the patterns obtained with the WT and Mut probes. In addition, in each instance, the complexes were specific, because they disappeared in competition studies using an excess of the corresponding unlabeled probe. Both sets of complexes were “super-shifted” using anti-Ets and anti-GATA3 antibodies (Figure 4A-C). These results suggest that the promoter deletion is altering the binding of these 2 transcription factors at this site, but clearly both transcription factors still bind the mutant element. To verify the quantitative difference in binding between Wt and Mut sequences, we performed cross-competition EMSAs. Our results, shown in Figure 4D, indicate that, whereas the unlabeled WT probe successfully competed the labeled WT at 50- and 100-fold excess, thus confirming the same specificity shown in our previous experiments, the unlabeled Mut probe was only able to compete with binding of the labeled WT probe at 100-fold excess. These results verify that the WT probe binds to endothelial cell nuclear proteins in a specific and stronger fashion than the Mut sequence.
Electrophoretic Mobility Shift Assays using endothelial nuclear extracts derived from BOECs. Nuclear extracts from BOECs were tested with a 32P- labeled VWF WT probe spanning nucleotides −61 to −32 and a Mut probe spanning −70 to −32. (A) The pattern of DNA-protein complexes is distinct for the WT and Mut probes. Complexes are specific, because they disappeared in competition studies using 50× unlabeled probe. DNA-protein complexes with both the WT and mutant probes were “super-shifted” using anti–GATA-3 antibodies. (B) WT-DNA protein complexes from a repeated and independent experiment. The DNA-protein complex is “super-shifted” using anti-Ets antibodies (C) Mut DNA-protein complexes. The complex with the mutant probe is also “super-shifted” using anti-Ets antibodies. The images shown in panels B and C are from the same electrophoresis gel run. Repeated lanes have been removed to avoid confusion. (D) Cross-competition EMSAs. WT and Mut probes consistently show distinct banding patterns. Cross competition using cold WT and cold Mut oligos (10-, 50-, and 100-fold excess) shows that the cold WT probe competes both WT and Mut probe binding, whereas cold Mut competes only the Mut probe binding. Successful competition with WT probe binding is seen with the cold Mut oligo only at 100-fold excess. NE = nuclear extract. The vertical lines inserted between lanes in panels B and C indicate places where lanes from the same gel and experiment have been juxtaposed to clarify the data.
Electrophoretic Mobility Shift Assays using endothelial nuclear extracts derived from BOECs. Nuclear extracts from BOECs were tested with a 32P- labeled VWF WT probe spanning nucleotides −61 to −32 and a Mut probe spanning −70 to −32. (A) The pattern of DNA-protein complexes is distinct for the WT and Mut probes. Complexes are specific, because they disappeared in competition studies using 50× unlabeled probe. DNA-protein complexes with both the WT and mutant probes were “super-shifted” using anti–GATA-3 antibodies. (B) WT-DNA protein complexes from a repeated and independent experiment. The DNA-protein complex is “super-shifted” using anti-Ets antibodies (C) Mut DNA-protein complexes. The complex with the mutant probe is also “super-shifted” using anti-Ets antibodies. The images shown in panels B and C are from the same electrophoresis gel run. Repeated lanes have been removed to avoid confusion. (D) Cross-competition EMSAs. WT and Mut probes consistently show distinct banding patterns. Cross competition using cold WT and cold Mut oligos (10-, 50-, and 100-fold excess) shows that the cold WT probe competes both WT and Mut probe binding, whereas cold Mut competes only the Mut probe binding. Successful competition with WT probe binding is seen with the cold Mut oligo only at 100-fold excess. NE = nuclear extract. The vertical lines inserted between lanes in panels B and C indicate places where lanes from the same gel and experiment have been juxtaposed to clarify the data.
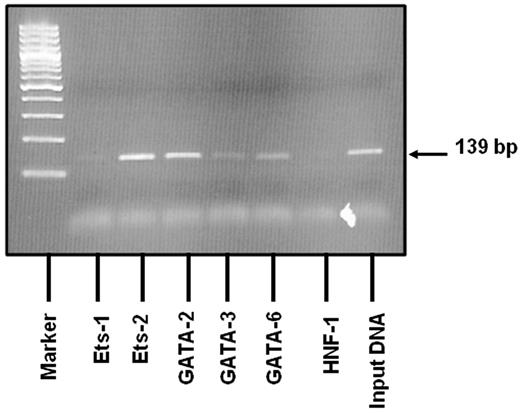
ChIP analysis shows that Ets and GATA proteins bind to the site of the promoter deletion in endothelial cell nuclei
To verify the role of the deletion mutation in the aberrant transcription factor binding seen in EMSA studies, we performed ChIP assays in BOECs. Native chromatin was immunoprecipitated with antibodies against Ets and GATA transcription factors or control IgG, and the immunoprecipitated fractions were subjected to PCR using VWF-specific primers spanning nucleotides −111 to +28 (the region containing the deletion). We have tested the following transcription factors for binding to this region: Ets-1, Ets-2, GATA-2, GATA-3, and GATA-6. As shown in Figure 5, amplified bands matched the predicted PCR fragment size, and the DNA region of the promoter involved in the deletion mutation clearly binds to Ets-2 and GATA-2. Weaker binding is seen for GATA-3 and GATA-6. Our data show evidence to support the binding of these endothelial-specific transcription factors to the region of interest. Liu and coworkers recently validated the functional significance of GATA-3 binding at +220 as a basal promoter element in the context of a similarly large VWF promoter fragment.22 Accordingly, we have included this region as an additional positive control for GATA binding and indeed showed binding of GATA-3 to this fragment (data not shown). These in vivo transcription factor binding data complement the EMSA results and indicate that the deletion mutation has occurred in a region of the VWF promoter to which both Ets and GATA transcription factors bind.
Results of the ChIP assay. Native chromatin derived from BOECs was immunoprecipitated with antibodies against Ets-1, Ets-2, GATA-2, GATA-3 and GATA-6. Binding of the liver-specific transcription factor, HNF-1, known not to bind to this region was also evaluated. PCR was designed to amplify a 139-bp region (−111 to +28; a region containing the deletion mutation). The 2% agarose gel shows that amplified bands match the predicted PCR fragment size in relation to the DNA ladder. The VWF sequence in which the deletion is located binds clearly to Ets-2 and GATA-2. Weaker binding is seen with GATA-3 and GATA-6. Positive (RNA pol II) and negative (mouse IgG) controls produced the expected results (data not shown).
Results of the ChIP assay. Native chromatin derived from BOECs was immunoprecipitated with antibodies against Ets-1, Ets-2, GATA-2, GATA-3 and GATA-6. Binding of the liver-specific transcription factor, HNF-1, known not to bind to this region was also evaluated. PCR was designed to amplify a 139-bp region (−111 to +28; a region containing the deletion mutation). The 2% agarose gel shows that amplified bands match the predicted PCR fragment size in relation to the DNA ladder. The VWF sequence in which the deletion is located binds clearly to Ets-2 and GATA-2. Weaker binding is seen with GATA-3 and GATA-6. Positive (RNA pol II) and negative (mouse IgG) controls produced the expected results (data not shown).
Reduced endothelial cell expression of the VWF mRNA transcript associated with the promoter deletion
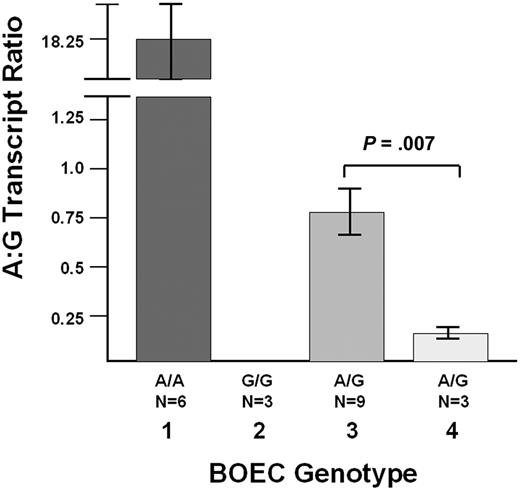
Finally, we have evaluated the effect of the promoter deletion on the allelic expression of VWF from BOECs derived from the index case. Quantitative RT-PCR studies were performed with BOEC-derived mRNA using the SNP c.2365G/A (rs1063856) to differentiate the 2 VWF transcripts. Studies were performed in 2 normal homozygous A/A subjects (n = 6 experiments), 1 normal G/G subject (n = 3 experiments), 3 G/A heterozygotes (n = 9 experiments), and the index case (n = 3 experiments), who is a G/A heterozygote. Determination of SNP phase in the index case's family shows that the promoter deletion is linked (in cis) to the “A” SNP allele. The results in Figure 6 show that the ratio of “A” to “G” transcripts in the index case BOECs (n = 3 experiments) is 4.6-fold lower that the mean “A” to “G” ratio in the 3 normal heterozygous subjects (P = .007, n = 3 and 9 experiments for index case and healthy controls, respectively). This finding confirms a significant association of the promoter deletion with reduced VWF transcript production.
Quantitative RT-PCR studies with BOEC-derived mRNA. Using the SNP c.2365G/A (rs1063856) to differentiate the 2 VWF transcripts, the index case who is heterozygous G/A for this SNP (n = 3 experiments) was studied and compared with 6 healthy control individuals: 2 A/A subjects (lane 1, n = 6 experiments), 1 G/G subject (lane 2, n = 3 experiments), and 3 G/A heterozygotes (lane 3, n = 9 experiments). The graph shows the average A:G transcript ratio (± SEM). In lane 1, The A:G ratio in the A/A homozygotes is not infinite because there is some variable “background” binding of the G probe to the A transcripts. In lane 2, the A:G ratio was always “0,” because there were never any A transcripts detected in the G/G homozygote. In the index case (lane 4, n = 3 experiments), the A:G ratio is 4.6-fold lower that the mean A:G ratio in the 3 normal G/A heterozygous subjects (P = .007). The VWF mutation is in cis with the c.2365A/G (rs1063856) SNP “A” allele, and thus, the promoter deletion is associated with reduced VWF transcript production.
Quantitative RT-PCR studies with BOEC-derived mRNA. Using the SNP c.2365G/A (rs1063856) to differentiate the 2 VWF transcripts, the index case who is heterozygous G/A for this SNP (n = 3 experiments) was studied and compared with 6 healthy control individuals: 2 A/A subjects (lane 1, n = 6 experiments), 1 G/G subject (lane 2, n = 3 experiments), and 3 G/A heterozygotes (lane 3, n = 9 experiments). The graph shows the average A:G transcript ratio (± SEM). In lane 1, The A:G ratio in the A/A homozygotes is not infinite because there is some variable “background” binding of the G probe to the A transcripts. In lane 2, the A:G ratio was always “0,” because there were never any A transcripts detected in the G/G homozygote. In the index case (lane 4, n = 3 experiments), the A:G ratio is 4.6-fold lower that the mean A:G ratio in the 3 normal G/A heterozygous subjects (P = .007). The VWF mutation is in cis with the c.2365A/G (rs1063856) SNP “A” allele, and thus, the promoter deletion is associated with reduced VWF transcript production.
Discussion
The aim of this study was to characterize the functional significance of a VWF promoter deletion with reference to the type 1 VWD phenotype present in the index case. The results of the study show that the promoter deletion significantly reduces transcriptional activity in vitro in endothelial and liver cells and in vivo in a mouse model. Furthermore, evaluation of allele-specific VWF expression from endothelial cells derived from the index case shows a marked reduction of the transcript regulated by the deleted promoter. In summary, these studies represent the first functional characterization of a transcriptional mutant resulting in type 1 VWD.
Type 1 VWD is the most common inherited bleeding disorder in humans. Three recent studies have provided the first details of the molecular genetic pathology associated with this highly variable mild/moderate quantitative trait.3-5 The conclusion of these studies is that approximately 65% of type 1 VWD patients have candidate mutations in the coding region of VWF, the consensus splice sequences and the proximal promoter. The location of the genetic changes contributing to the type 1 phenotype in the other 35% of cases remains unresolved, but variation within VWF introns and at more distant regulatory sequences are possibilities that need evaluation. In the Canadian type 1 VWD study, 16% of index cases had candidate mutations documented in the promoter region. All but 1 of these 8 putative transcriptional mutants represented point mutations whereas the other variant was a 13-bp deletion that forms the basis of this study.
Transcriptional mutations have been described as the cause of genetic disease in many conditions23 but the paradigmatic conditions for understanding the influence of transcriptional mutants are the quantitative hemoglobin pathologies. Molecular genetic analysis of the thalassemias24 and hereditary persistence of fetal hemoglobin25 has documented a spectrum of mutations that alter normal transcriptional processes. Furthermore, analysis of the distant locus control regions (LCRs) of the globin genes has confirmed that mutations at locations many kilobases from the transcribed locus can have a deleterious effect.26 The potential of an LCR for VWF and its possible contribution to quantitative trait variability remains to be explored.
Previous investigation of the elements responsible for regulating VWF expression has revealed a complex array of both positive and negative mediators.7,10,27-30 These studies have been complemented by in vivo assessment of VWF regulatory sequence potential in transgenic mice, which has shown that even with 2.1 kb of 5′ flanking sequence (plus exon 1 and intron 1 sequence) only a subset of endothelial cells in brain, heart, and skeletal muscle demonstrate transgene expression.8 There is also evidence that polymorphic sequence variation in the VWF promoter plays a role in regulating VWF expression and that some of these sequences may mediate responses to biomechanical forces.11,30
The 13-bp deletion documented in the index case involves 10 nucleotides at the 5′ end of the deleted sequence (nucleotides −48 to −39) that are conserved in the murine and bovine VWF promoters.31 These evolutionarily conserved sequences contain 2 strong Ets binding sites and several weaker candidate GATA binding sites. The promoter deletion is also just 4 bp upstream of the TAATAA TATA-box equivalent at the VWF locus. Both of these factors highlight the likely functional significance of the deleted sequence. In addition, in this study we have shown that the binding of 2 endothelial transcription factors, GATA and Ets is altered by the mutation. Interestingly, although the patterns of DNA-protein and antibody super-shifted complexes are distinct for the WT and mutant probe sequences, it appears that both transcription factors still bind the mutant element, although less effectively, as verified by our cross-competition EMSA results. In addition, the results of the ChIP assay have provided evidence that Ets-2 and GATA-2 (and to a lesser extent GATA-3 and GATA-6) bind to the region of the VWF promoter containing the deletion mutation. In aggregate, these results suggest that Ets and, likely to a lesser extent, GATA-mediated transactivation through this region of the promoter will be adversely influenced by the deletion mutation. Details of the specific contribution of these 2 transcription factors to transactivation of the locus through their binding to this region will require further study. Finally, given the proximity of the VWF TATA sequence, this deletion may also destabilize the preinitiation complex on this allele.
The speculations in the previous paragraph are supported by the results of the allele-specific expression studies performed on mRNA derived from the index case's endothelial cells. In these studies, we have shown that the mutation-linked “A” transcripts are significantly reduced relative to expression of the “G” allele. While our results have documented a 4.6-fold relative reduction of the mutant “A” transcript, the absolute level of disruption of expression from this allele cannot be determined with confidence from this study due to the small size of the study population and limitations of the qRT-PCR protocol.
While there are several complementary pieces of data in this study that support an important transcriptional regulatory role for the 13 nucleotides deleted from the proximal VWF promoter in this index case, our results raise further questions concerning the cell type-specificity of gene expression mediated by the proximal 5′ flanking sequence. In this study, transactivation was down-regulated to a similar extent in transduced endothelial and liver cells. Furthermore, our in vivo mouse studies show transgene expression in the liver with the WT sequence but absent transgene expression with the deletion mutant. All of these studies have been performed with a reporter gene construct that incorporates 1.3 kb of 5′ flanking sequence, exon 1 and intron 1 of the VWF gene. Previous in vivo analysis of a VWF reporter transgene regulated by 2.1 kb of 5′ flanking sequence (plus exon 1 and intron 1 sequence) showed transgene expression in endothelial cells in the brain, heart, and skeletal muscle8 There are at least 2 other key differences between these 2 studies. First, in vivo, our transgene constructs will have been targeted to the liver because of the route of administration (tail vein injection) and because they were delivered by an adenoviral vector, a known hepatotropic delivery system. Second, in contrast to the integrated regulatory constructs of the earlier study, our transgenes will have remained extrachromosomal in location and may have undergone an aberrant form of chromatin remodeling. Overall, the results of these studies highlight the complexity of the cell type–specificity of expression mediated by the VWF 5′ flanking sequence. Additional studies exploring more distant upstream and downstream elements are required to better understand the cellular heterogeneity of VWF expression. These considerations are further emphasized by the recent demonstration of a transcriptional activation sequence in intron 51 of VWF that is functional in a subset of pulmonary endothelial cells.32
In summary, this investigation has confirmed an important functional role for the 13-bp sequence deleted from the promoter of one of the VWF genes in this index case of type 1 VWD. The coagulation phenotype in this patient (VWF:Ag 0.49 IU/mL) is consistent with a haploinsufficient mechanism of action, with a marked reduction of transcript production from the mutant allele. We speculate that the haploinsufficient effect of this deletion mutation results from its location in the proximal VWF promoter, adjacent to the VWF TATA sequence, and the fact that it disrupts the binding of both Ets and GATA transcription factors. We also propose that this mutation would result in a severe phenotype (either severe type 1 or type 3 VWD) if coinherited along with a second, VWF null allele. These results represent the first functional characterization of a transcriptional mutation resulting in this quantitative trait. Future studies are required to determine the contribution of other regulatory sequence variants on the type 1 VWD phenotype. The magnitude of their effect on the VWF phenotype will presumably depend upon their location, and the extent to which they disrupt the normal transcriptional regulatory machinery.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This project was funded through the Zimmerman Program for Molecular and Cellular Biology of von Willebrand Disease by National Institutes of Health Program Project Grant HL081588 and by Canadian Institutes of Health Research operating grant no. PO-97 849. D.L. holds a Canada Research Chair in Molecular Hemostasis.
National Institutes of Health
Authorship
Contribution: M.O. designed and performed research, analyzed data, and wrote the paper; C.B., Y.C., C.N., N.H., D.H., S.B., A.L.P., A.B., and P.J. performed research; S.W. performed research and analyzed data; and D.L. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr David Lillicrap, Richardson Laboratory, Department of Pathology and Molecular Medicine, Queen's University, Kingston, ON, Canada K7L 3N6; e-mail: lillicrap@cliff.path.queensu.ca.