Abstract
Fanconi anemia (FA) is an inherited chromosomal instability syndrome characterized by bone marrow failure, myelodysplasia (MDS), and acute myeloid leukemia (AML). Eight FA proteins associate in a nuclear core complex to monoubiquitinate FANCD2/FANCI in response to DNA damage. Additional functions have been described for some of the core complex proteins; however, in vivo genetic proof has been lacking. Here we show that double-mutant Fancc−/−;Fancg−/− mice develop spontaneous hematologic sequelae including bone marrow failure, AML, MDS and complex random chromosomal abnormalities that the single-mutant mice do not. This genetic model provides evidence for unique core complex protein function independent of their ability to monoubiquitinate FANCD2/FANCI. Importantly, this model closely recapitulates the phenotypes found in FA patients and may be useful as a preclinical platform to evaluate the molecular pathogenesis of spontaneous bone marrow failure, MDS and AML in FA.
Introduction
Characteristically, Fanconi anemia (FA) patients develop progressive bone marrow failure (BMF), ultimately requiring stem cell transplantation in the second or third decade of life.1 The progressive BMF can also lead to hematologic neoplasms, particularly myelodysplasia (MDS) and acute myeloid leukemia (AML),1-3 both of which are almost universally associated with aneuploidy and cytogenetic abnormalities.4,5 The interactions among the FA proteins are complex and incompletely understood. Thirteen complementation groups and their underlying gene defects (FANCA, FANCB, FANCC, FANCD1/BRCA2, FANCD2, FANCE, FANCF, FANCG/XRCC9, FANCI, FANCJ/BRIP1/BACH1, FANCL/PHF9/POG, FANCM, and FANCN/PALB2) have been identified to date.6-13 It is known that certain FA proteins (FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL and FANCM) associate in a core nuclear complex and function, at least in part, to catalyze the monoubiquitination of the target proteins, FANCD2 and FANCI, in response to DNA damage.7,8,14-18 The monoubiquitinated forms of FANCD2/FANCI translocate to chromatin where they co-localize with BRCA1, BRCA2, and RAD51 in nuclear foci thought to be sites of DNA repair.7,8,18-21 Inactivation of any of the FA core complex proteins results in complete failure to monoubiquitinate FANCD2/FANCI, leading to cellular hypersensitivity to DNA cross-linking agents.22,23
Additional functions have been ascribed to individual FA proteins suggesting a critical role in maintenance of normal hematopoiesis,24-31 yet these mechanisms have not been evaluated using a genetic model. Murine knockouts of the homologues of FANCA (Fanca), FANCC (Fancc), FANCG (Fancg), FANCD1 (FancD1), FANCD2 (Fancd2), and FANCM (Fancm) have been established.32,33 Although all strains of FA knockout mice are hypersensitive to mitomycin C (MMC), a DNA cross-linking agent, none of these single knockout mice display spontaneous aplastic anemia or myeloid malignancies characteristic of FA in patients.32,33 We reasoned that if FA proteins have divergent functions independent of FANCD2/FANCI monoubiquitination in hematopoietic cells, double knockout mice might display a more aggressive hematopoietic phenotype. We therefore generated Fancc−/−;Fancg−/− mice and report here that these mice more faithfully recapitulate the hematologic manifestations observed in FA patients.
Methods
Antibodies and chemicals
MMC, β-mercaptoethanol, benzidine and Ficoll-hypaque (density, 1.119) were purchased from Sigma-Aldrich. Recombinant stem cell factor (SCF), granulocyte-macrophage colony-stimulating factor (GM-CSF) and interleukin 3 (IL-3) were purchased from PeproTech and erythropoietin (EPO) from Amgen. Antibodies specific to CD45.1 and CD45.2 were purchased from BD PharMingen.
Mice
Mice containing a disruption in either the Fancc34 and Fancg35 genes were back-crossed 10 generations into the C57BL/6J strain. The mice were genotyped using PCR to detect wildtype (WT) and either mutant Fancc or Fancg alleles as described previously.34,35 Mice deficient in both Fancc and Fancg were generated by mating Fancc+/− with Fancg+/− to obtain double heterozygous mice (F1). These mice were then used to generate the homozygous mutant mice (F2). WT B6.SJL-PtrcaPep3b/BoyJ (B6.BoyJ) and WT C57BL/6J mice were purchased from The Jackson Laboratory. The institutional animal care and use committee of Indiana University approved all animal studies.
Hematopoietic progenitor growth
Bone marrow cells (BMCs) were isolated as described previously.36,37 Clonogenic methylcellulose assays were performed in triplicate (2 × 104 LDMNC per 35-mm plate). Cultures were established in 1% Iscove Modified Dulbecco Medium (IMDM) methylcellulose, StemCell Technologies, 30% fetal calf serum (FCS), 20mM glutamine, 200 U/mL penicillin and streptomycin, 80μM β-mercaptoethanol, 50 ng/mL recombinant murine SCF, 10 ng/mL recombinant murine GM-CSF, 2 U/mL recombinant human EPO, 10 ng/mL IL-3 and 5% vol/vol pokeweed mitogen spleen conditioned media (PWMSCM). In specified experiments, MMC was added at indicated concentrations. The cells were incubated at 37°C and 5% CO2, colonies were scored on day 7 after initiation of culture. To count the cells according to hematopoietic cell lineage, the colonies were stained with 1.5 mg/mL benzidine in 5% glacial and 25 μL of H2O2.
Bone marrow transplantation
BMCs were isolated from donor mice as described previously36,37 and resuspended in 500 μL IMDM containing 20% FCS at indicated cell concentrations for the specified experiments. The cells were injected into the tail vein of lethally irradiated recipient mice. The recipients were pre-conditioned by exposing them to a split dose of 1100 rads (700 + 400 rads, 4 hours apart) of ionizing radiation using a gammacell-4 exactor (Nordion) containing a 137Cs source.
Statistical and bioinformatics analysis
Statistical analysis on hematopoiesis assays was performed using GraphPad Prism 4.
Complete blood counts and chimerism
Peripheral blood was collected from the tail vein of mice and stored in microtainer brand tubes with ethylenediaminetetraacetic acid (Becton Dickinson). Complete blood counts were performed using a Hemavet950 (Drew Scientific Group). The remaining blood was then treated with red cell lysis buffer (Gentra Systems) for 10 minutes at room temperature. The cells were washed in phosphate-buffered saline (PBS) with 0.1% bovine serum albumin (BSA) and split into 2 aliquots. Each aliquot was stained with either CD45.2 FITC or CD45.1 FITC (BD PharMingen) at 4°C for 20 minutes. The cells were then washed in PBS with 0.1% BSA and analyzed by flow cytometry.
Histology
Spleens and humeri or tibiae from selected animals were fixed in 10% buffered formalin and embedded in paraffin. Sections were obtained and stained with hematoxylin and eosin (H&E). All histologic sections were analyzed in a blinded fashion.
Microscopy
Pictures were taken on a Zeiss Axioscope (Zeiss) with the Plan Neofluar 20×/.50 (Figures 2C, 3C, supplemental Figure 1B spleen [available on the Blood Web site; see the Supplemental Materials link at the top of the online article]) or the Ph3 Neofluar 40×/1.30 oil (Figure 3C pullout panel, supplemental Figure 1B BM; Zeiss). Images were captured using a SPOT RT Color camera, model 2.2.1 (Diagnostic Instruments) and edited using SPOT Advanced software Version 4.1.2 (Diagnostic Instruments).
Cytogenetic analysis
Cytogenetic analysis was performed on BMCs from non-competitive transplant recipients as described.38 Briefly, short-term, synchronized cultures were initiated and metaphase cells were prepared using standard cytogenetic techniques after mitotic arrest with 1% colcemid (10 μg/mL, Invitrogen Life Technologies). Between 5 and 15 G-banded metaphase cells were completely analyzed for each culture. Mouse chromosomes were classified using standardized mouse chromosome ideograms and following nomenclature standards of the International Committee on Standard Genetic Nomenclature for Mice (http://www.informatics.jax.org/mgihome/nomen/anomalies.shtml).
SKY analysis
Spectral karyotyping (SKY) was performed using mouse reagents (SkyPaint for mouse, Applied Spectral Imaging [ASI]) and following the manufacturer's protocol for mouse chromosome analysis with the exception of a longer hybridization time of 42 hours. Fluorochromes used included: rhodamine, Texas red, Cy5, FITC, and Cy5.5. An interferogram for each metaphase was generated using a SD200 Spectracube (ASI) mounted to a Leica DM 5000 fluorescence microscope. Three to 10 metaphase cells were captured and karyotyped for each culture.
RNA isolation and GeneChip Array
RNA was isolated using the RNeasy Mini Kit (QIAGEN) from whole BMCs obtained from 3-month-old syngeneic mice (WT, Fancc−/−, Fancg−/−, and Fancc−/−;Fancg−/−). Microarray assays were performed in the Affymetrix Microarray Core, a unit of the Gene Microarray Shared Resource located in the Knight Cancer Institute at Oregon Health & Science University. Hybridization targets were prepared from 2 μg total RNA using the 1-cycle cDNA, IVT amplification/labeling protocol procedure as described in the Affymetrix GeneChip Expression Analysis Technical Manual, rev.5 (Affymetrix). After fragmentation, each sample target was hybridized to an MOE 430 2.0 GeneChip array, a chip containing 45 000 probe sets that measure the expression level of > 39 000 transcripts and variants from > 34 000 well-characterized mouse genes (Affymetrix). Scanning was performed with the Affymetrix 3000 GeneArray Scanner and image processing and expression analysis were performed using Affymetrix GeneChip Operating Software (GCOS) v1.2 software. The quality of project hybridizations was assessed using: (1) performance metrics extracted from the GCOS report file for each array and (2) pair-wise scatter plots of the “absolute analysis” data from each array assay. All original microarray data have been deposited in the GEO public database under accession number GSE22094.
Microarray data management and analysis
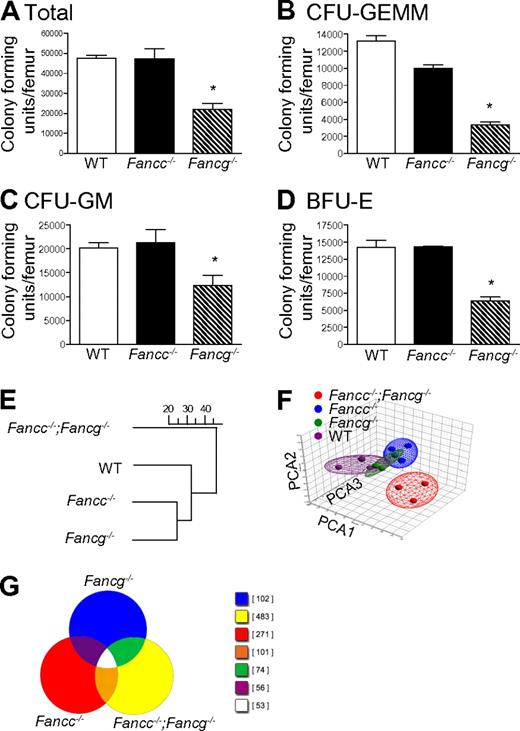
At the 1.5-fold threshold in double mutant cells, 3197 genes were differentially expressed in the pair-wise comparison with WT cells. Only 520 and 304 genes were differentially expressed (compared with WT) in Fancc and Fancg single mutant cells, respectively. Of the 520 (Fancc mutant), 263 were found in the set of 3197 genes. Of the 304 (Fancg mutant), 216 were found in the set of 3197. The processed image files (.CEL) were imported into GeneSifter and Partek Genomics Suite (Partek Inc) software programs. MAS was used for normalization. Hierarchical clustering was performed using GeneSifter for multiple datasets filtered by thresholds of 1.5-, 2-, 3-, 4-, and 5-fold using analysis of variance (ANOVA; P < .01) and the Benjamini-Hochberg correction for multiple comparisons. The dendrogams for each threshold were identical (Figure 1E shows the dendrogram for the 1.5-fold threshold). Principal component analysis was performed using both GeneSifter and Partek Genomics Suite (Figure 1F shows the results filtered at the 1.5-fold threshold, displayed using the Partek ellipsoid graphic tool).
Results
In initial experiments, multipotent, erythroid and myeloid progenitors from syngeneic 3-month-old WT, Fancc−/− and Fancg−/− mice were examined. Differences in clonogenic potential were observed in multiple hematopoietic compartments (Figure 1A-D). These functional results provided the first suggestion that FANCC and FANCG have non-overlapping roles in hematopoiesis. To further evaluate this possibility, we conducted GeneChip arrays to test the hypothesis that loss of both Fancc and Fancg will result in a significant change in expression profile. To accomplish this, unsupervised hierarchical clustering and principal component analyses were performed on expression microarray data generated from BMC RNA from 3-month-old syngeneic mice with 4 genotypes: WT, Fancc−/−, Fancg−/−, and mice with disruptions at both the Fancc and the Fancg loci (Fancc−/−;Fancg−/−). Interestingly, the results demonstrated that double mutant cells diverged more widely from the WT cells than did either of the single mutants. Hence, there were more commonalities found in the expression space between the 2 single mutants and also between them and the WT samples, but significantly greater dissimilarities between these 3 groups and the double mutant samples (Figure 1E-F). A Venn diagram provides a visual representation of the differential gene expression by each genotype (Figure 1G), providing evidence that each strain has uniquely expressed genes. The differences did not include variations in lineage specific gene expression (supplemental Table 1) assuring that the biologic differences observed (Figure 1E-F) did not derive from modest differences in the populations present in the bone marrow of the animals.
Loss of Fancg results in a more severe defect in multiple hematopoietic compartments than loss of Fancc. (A) Total number of progenitors per femur isolated from syngeneic 3-month-old mice with statistically similar weights (25 ± 0.6 g) and bone marrow cellularity (10 × 106 ± 0.7 × 106 cells per femur). *p = 0.0009 by 1-way ANOVA. (B) Number of multipotential colony-forming unit–granulocyte, erythroid, monocyte, megakaryocyte progenitors (CFU-GEMMs), *P < 0.0001 by 1-way ANOVA. (C) colony-forming unit–granulocyte-macrophage progenitors (CFU-GMs), *p = 0.0230 by 1-way ANOVA. (D) burst forming unit–erythroid progenitors (BFU-Es), *p = 0.0030 by 1-way ANOVA. Data represent mean ± SEM of 3 independent experiments each of which was plated in triplicate cultures. (E-F) Genome-wide transcriptomal analysis of marrow cells from 3-month-old mice from each of the 4 indicated FA genotypes was performed. A total of 12 BMC samples from 12 mice (Fancc−/−;Fancg−/−, Fancc−/−, Fancg−/− and WT) were analyzed using 12 AffyMetrix MOE 430 2.0 GeneChip arrays. (E) Unsupervised hierarchical clustering, the scale represents the level of correlation between the groups. (F) Principal component analysis. In this case the multidimensional data were reduced to a new coordinate system such that the greatest variance (20.8%) by any possible projec-tion of the data are plotted on the first coordinate (the first principal component, PCA1). The second greatest variance (14.5%) was plotted on the second coordi-nate (PCA2), and the third greatest (10.8%) plotted on the third coordinate (PCA3). Results from both analyses confirmed that double mutant cells diverged widely from both the WT samples and both of the single mutant samples.
Loss of Fancg results in a more severe defect in multiple hematopoietic compartments than loss of Fancc. (A) Total number of progenitors per femur isolated from syngeneic 3-month-old mice with statistically similar weights (25 ± 0.6 g) and bone marrow cellularity (10 × 106 ± 0.7 × 106 cells per femur). *p = 0.0009 by 1-way ANOVA. (B) Number of multipotential colony-forming unit–granulocyte, erythroid, monocyte, megakaryocyte progenitors (CFU-GEMMs), *P < 0.0001 by 1-way ANOVA. (C) colony-forming unit–granulocyte-macrophage progenitors (CFU-GMs), *p = 0.0230 by 1-way ANOVA. (D) burst forming unit–erythroid progenitors (BFU-Es), *p = 0.0030 by 1-way ANOVA. Data represent mean ± SEM of 3 independent experiments each of which was plated in triplicate cultures. (E-F) Genome-wide transcriptomal analysis of marrow cells from 3-month-old mice from each of the 4 indicated FA genotypes was performed. A total of 12 BMC samples from 12 mice (Fancc−/−;Fancg−/−, Fancc−/−, Fancg−/− and WT) were analyzed using 12 AffyMetrix MOE 430 2.0 GeneChip arrays. (E) Unsupervised hierarchical clustering, the scale represents the level of correlation between the groups. (F) Principal component analysis. In this case the multidimensional data were reduced to a new coordinate system such that the greatest variance (20.8%) by any possible projec-tion of the data are plotted on the first coordinate (the first principal component, PCA1). The second greatest variance (14.5%) was plotted on the second coordi-nate (PCA2), and the third greatest (10.8%) plotted on the third coordinate (PCA3). Results from both analyses confirmed that double mutant cells diverged widely from both the WT samples and both of the single mutant samples.
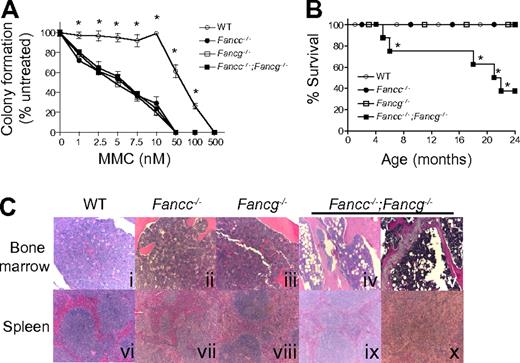
Because the hallmark of FA cells is hypersensitivity to DNA cross-linking agents, we evaluated the growth of hematopoietic progenitors from single and double mutant 3-month-old FA mice and syngeneic WT control mice in the presence of MMC (Figure 2A). The progenitors from Fancc−/−, Fancg−/− or Fancc−/−;Fancg−/− mice had comparable hypersensitivity to MMC (Figure 2A).
Fancc−/−;Fancg−/− mice have a shortened life span and an increased risk of BMF despite similar MMC sensitivity of Fancc−/−, Fancg−/− and Fancc−/−;Fancg−/− cells. (A) Sensitivity of WT, Fancc−/−, Fancg−/−, and Fancc−/−;Fancg−/− cells to MMC. Data represent mean ± SEM of 3 independent experiments each of which was plated in triplicate cultures isolated from the bone marrow of 3-month-old syngeneic mice. WT is significantly different from any of the other experimental groups, which had comparable sensitivity, *P < .001 by 2-way ANOVA. (B) Kaplan-Meier curve shows a significant decrease in survival of Fancc−/−;Fancg−/− mice compared with WT, Fancc−/− or Fancg−/− mice, n = 8/genotype. *P = 0.0004 for Fancc−/−;Fancg−/− compared with WT, Fancc−/− or Fancg−/− by the log-rank test for trend. (C) Fancc−/−;Fancg−/− mice, but not WT, Fancc−/− or Fancg−/− mice, develop aplastic anemia and myeloid malignancies. Subpanels iv and ix are from a 20-month-old Fancc−/−;Fancg−/− mouse and subpanels v and x are from a 23-month-old Fancc−/−;Fancg−/− mouse.
Fancc−/−;Fancg−/− mice have a shortened life span and an increased risk of BMF despite similar MMC sensitivity of Fancc−/−, Fancg−/− and Fancc−/−;Fancg−/− cells. (A) Sensitivity of WT, Fancc−/−, Fancg−/−, and Fancc−/−;Fancg−/− cells to MMC. Data represent mean ± SEM of 3 independent experiments each of which was plated in triplicate cultures isolated from the bone marrow of 3-month-old syngeneic mice. WT is significantly different from any of the other experimental groups, which had comparable sensitivity, *P < .001 by 2-way ANOVA. (B) Kaplan-Meier curve shows a significant decrease in survival of Fancc−/−;Fancg−/− mice compared with WT, Fancc−/− or Fancg−/− mice, n = 8/genotype. *P = 0.0004 for Fancc−/−;Fancg−/− compared with WT, Fancc−/− or Fancg−/− by the log-rank test for trend. (C) Fancc−/−;Fancg−/− mice, but not WT, Fancc−/− or Fancg−/− mice, develop aplastic anemia and myeloid malignancies. Subpanels iv and ix are from a 20-month-old Fancc−/−;Fancg−/− mouse and subpanels v and x are from a 23-month-old Fancc−/−;Fancg−/− mouse.
To further evaluate abnormalities in the hematopoietic compartment, cohorts of WT, Fancc−/−, Fancg−/− and Fancc−/−;Fancg−/− mice were observed up to 24 months. These mice were killed and evaluated when they developed signs of illness. A significant decrease in survival of Fancc−/−;Fancg−/− mice was observed compared with other genotypes (Figure 2B). To determine the cause of the increased mortality in these mice, serial evaluations of complete peripheral blood counts of primary Fancc−/−;Fancg−/− mice were performed. At the time of death or sacrifice due to illness, 50% of double knockout mice had a moderate leukocytosis (WBC > 15.9 K/μL), dysplastic megakaryocytes, a myeloid maturation arrest and splenomegaly (> 200 mg) with an invasion of myeloid cells leading to a disruption of normal splenic architecture (Figure 2C representative samples). Another 25% of the Fancc−/−;Fancg−/− mice had anemia (Hb < 7.0 g/dL), decreased bone marrow cellularity (< 50% of WT) and hypoplastic marrow containing large numbers of adipocytes (Figure 2C representative samples). The surviving mice did not have signs of illness at 24 months of age.
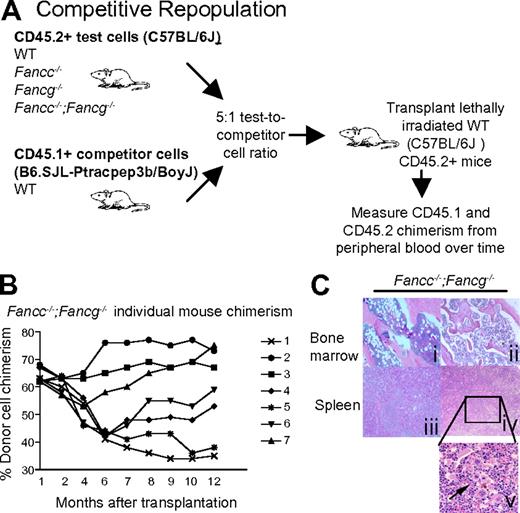
To examine the phenotype of the Fancc−/−;Fancg−/− hematopoietic stem cells in greater detail, we used a competitive repopulation assay. In this assay, stem cell activity is quantified by comparing the ability of mutant and WT BMCs to establish long-term hematopoiesis in competition with isogenic WT cells.39-41 Upon transplantation of mixed test and competitor cells (at a ratio of 5:1) into lethally irradiated recipients, peripheral blood chimerism was assessed monthly for 1 year by flow cytometry to determine test cell contribution to hematopoiesis (Figure 3A). The test cell repopulating activity was decreased in recipients reconstituted with either Fancc−/− or Fancg−/− BMCs compared with WT BMCs (supplemental Figure 1A). Interestingly, the peripheral blood chimerism of recipients reconstituted with Fancc−/−;Fancg−/− BMCs showed widely variable chimerism over time (Figure 3B) in contrast to stable chimerism in recipients reconstituted with all the other test cell genotypes (supplemental Figure 1A). In some Fancc−/−;Fancg−/− BMC recipients, the test cell population progressively out-competed the competitor cells while in other recipients the repopulating activity of the test cells was progressively reduced (Figure 3B). To further evaluate this unstable chimerism and the stem cell functionality, secondary transplants were performed for all genotypes and we found that only those transplanted with the Fancc−/−;Fancg−/− donor cells had a continued increase in chimerism variability (supplemental Figure 1B). The dramatic increase in test cell chimerism in some of the recipients suggests the development of clonal evolution of repopulating cells.
Fancc−/−;Fancg−/− hematopoietic stem cells undergo clonal evolution and malignant transformation in vivo. (A) Experimental design: Fancc−/−;Fancg−/− BMCs and isogenic WT competitor cells were co-transplanted into lethally irradiated recipient mice. Cells were transplanted at a 5:1 ratio (2.5 × 106 test cells and 0.5 × 106 competitor cells). Donor cells were collected from syngeneic 3-month-old mice. (B) Fluctuations of donor Fancc−/−;Fancg−/− cell chimerism were sequentially examined in peripheral blood of individual recipients, each symbol represents and individual recipient mouse (n = 7). (C) Mice reconstituted with BMCs from Fancc−/−;Fancg−/− mice showed abnormal bone marrow and spleen architecture 12 months after transplantation. Subpanels i and iii are from mouse 5 in panel B of this figure, with a chimerism of 38% at 12 months after transplantation. Subpanels ii, iv, and v are from mouse 2 in panel B of this figure.
Fancc−/−;Fancg−/− hematopoietic stem cells undergo clonal evolution and malignant transformation in vivo. (A) Experimental design: Fancc−/−;Fancg−/− BMCs and isogenic WT competitor cells were co-transplanted into lethally irradiated recipient mice. Cells were transplanted at a 5:1 ratio (2.5 × 106 test cells and 0.5 × 106 competitor cells). Donor cells were collected from syngeneic 3-month-old mice. (B) Fluctuations of donor Fancc−/−;Fancg−/− cell chimerism were sequentially examined in peripheral blood of individual recipients, each symbol represents and individual recipient mouse (n = 7). (C) Mice reconstituted with BMCs from Fancc−/−;Fancg−/− mice showed abnormal bone marrow and spleen architecture 12 months after transplantation. Subpanels i and iii are from mouse 5 in panel B of this figure, with a chimerism of 38% at 12 months after transplantation. Subpanels ii, iv, and v are from mouse 2 in panel B of this figure.
To evaluate whether the progressive increase in test cell chimerism in Fancc−/−;Fancg−/− BMC recipients was associated with the development of hematologic malignancies, spleen and bone marrow were harvested from the recipient mice. Recipients reconstituted with either WT, Fancc−/− or Fancg−/− cells all had normal splenic architecture and histology (supplemental Figure 1C). In contrast, recipient mice reconstituted with Fancc−/−;Fancg−/− BMCs showed a range of hematologic abnormalities including myeloid malignancies and aplastic anemia (Figure 3C, Table 1). The spleens from these recipients were enlarged and displayed complete disruption of normal splenic architecture and abnormal cell histology (Figure 3C). To confirm that these phenotypes were indeed due to the reconstitution of the hematopoietic system by donor cells and not the recipient's endogenous BMCs, CD45.2+ cells were sorted, cultured for growth of progenitors in semisolid medium, and individual colonies were genotyped. We found that less than 10% of the colonies were from the recipient's residual BMCs consistent with previous results (data not shown).36 Cytogenetic evaluation showed that only the transplanted Fancc−/−;Fancg−/− BMCs accumulated genetic abnormalities such as aneuploidy and multiple chromosomal translocations (Figure 4, supplemental Tables 2-3). Collectively, the spectrum of hematologic abnormalities is consistent with phenotypes observed in FA patients such as hypoplastic bone marrow and hematopoietic malignancies.42
Mice reconstituted with Fancc−/−;Fancg−/− donor cells have an increased risk of myeloid malignancy
| Donor cell genotype . | Malignancy . |
|---|---|
| WT | 0/16 |
| Fancc−/− | 0/15 |
| Fancg−/− | 0/16 |
| Fancc−/−;Fancg−/− | 9/17 |
| Donor cell genotype . | Malignancy . |
|---|---|
| WT | 0/16 |
| Fancc−/− | 0/15 |
| Fancg−/− | 0/16 |
| Fancc−/−;Fancg−/− | 9/17 |
This summary represents all of the data from 2 cohorts of competitive repopulation transplants and 1 cohort of non-competitive transplants.
WT indicates wild-type.
Spectral karyotyping shows aneuploidy and chromosomal translocations in Fancc−/−;Fancg−/− cells from the bone marrow of non-competitive recipients. WT shows a normal spectral karyogram. See supplemental Tables 2 and 3 for detailed descriptions of the chromosomal abnormalities found.
Spectral karyotyping shows aneuploidy and chromosomal translocations in Fancc−/−;Fancg−/− cells from the bone marrow of non-competitive recipients. WT shows a normal spectral karyogram. See supplemental Tables 2 and 3 for detailed descriptions of the chromosomal abnormalities found.
To rule out the possibility that the observed hematopoietic phenotypes occur after the loss of any 2 core complex proteins, we inter-crossed the Fanca−/− and Fancc−/− mice. The Fanca−/−;Fancc−/− mice did not develop bone marrow aplasia or myeloid malignancies although they failed to monoubiquitinate FANCD2 (Y.S. and D.W.C., unpublished data, June 28, 2005). These data are consistent with a previous study using an inter-cross of Fancc−/− mice with Fanca hypomorphic mice where Fancc−/− and Fanca−/− mice had similar numbers of myeloid progenitors and comparable MMC hypersensitivity to each other and Fanca−/−;Fancc−/− mice.43
Discussion
Numerous elegant studies have demonstrated the seminal role of FANCD2/FANCI monoubiquitination and phosphorylation in the activation of downstream effectors of DNA repair.12,22,23,44,45 It has also been shown that the FA core complex proteins are collectively required for activating FANCD2/FANCI and that the loss of any single FA core complex protein is sufficient to prevent FANCD2/FANCI monoubiquitination.22,23 However, recent studies have provided cellular and biochemical evidence that the FA core complex proteins have functions that are independent of their respective roles in activating FANCD2.24-31
Specifically, FANCC and FANCG, the human homologues of the genes inter-crossed here, have been shown to participate in molecular networks outside of the FA core complex. For example, FANCG has recently been shown to have a critical role in supporting the function of mitochondrial periredoxin-3 that is implicated in rendering FA cells a characteristic hypersensitivity to oxidative stress.28 Also, FANCG but not FANCC has been shown to form a complex with FANCD1, FANCD2 and XRCC3, and this complex is thought to function in homologous recombination repair.25 Similarly, FANCC is known to participate in the PKR signaling pathway, while the other FA core complex proteins are not.24,26 Whether mechanistically these interactions contribute to BMF and myeloid malignancies independently of FANCD2/FANCI is unclear. However, given the distinct hematopoietic phenotypes observed in syngeneic Fancc and Fancg deficient mice (Figure 1), we tested the hypothesis that Fancc and Fancg may cooperate in stem cell control by conducting a genetic inter-cross. We predicted that if these 2 FA core complex proteins are multifunctional and influence hematopoietic cell survival independent of their role in the FA nuclear core complex, then the Fancc/Fancg double knockout mice would display distinct pathologic phenotypes from those found in mice with inactivation of Fancc or Fancg only. Evidence that the phenotypes we observed in Fancc−/−;Fancg−/− bone marrow cells was specific to disruption of both the Fancc and Fancg genes was confirmed by the observation that Fancc−/−;Fancg−/− myeloid progenitors expressing transgenes that complement both Fancc and Fancg were resistant to MMC while Fancc−/−;Fancg−/− cells containing a transgene that complements Fancc or Fancg only were hypersensitive to MMC (supplemental Figure 2). Interestingly, the Fancc−/−;Fancg−/− mice developed BMF, myelodysplasia and complex cytogenetic abnormalities that are characteristic of bone marrow phenotypes observed in Fanconi anemia patients, but not present in the single knockout mice. Prior in vitro data supporting cooperation of FANCC and FANCG in the human system include a report in an adenocarcinoma cell line that genetic disruption of both FANCC and FANCG increased cytogenetic abnormalities.46 The model described here provides an in vivo platform that may be useful for questions regarding the pathogenesis of MDS and bone marrow failure.
Cross-breeding of FA mutant mice with other mice with inactivating gene mutations of non-FA genes has been reported and resulted in increased tumor formation and bone marrow hypocellularity.47-49 However, the majority of malignancies observed in those crosses were not representative of those commonly observed in FA patients. The development of the model outlined here provides interesting opportunities for modeling of therapeutic protocols. The failure of established single knockout mice to develop BMF has been a major limitation in translational preclinical applications to test questions surrounding stem cell engraftment, homing and gene transfer. For instance, it has been hypothesized that FA patients with hypoplasia or overt BMF will not require myelopreparation prior to transplantation of autologous genetically corrected stem/progenitor cells but the lack of a spontaneous BMF model has limited effectively testing this question prior to phase 1 trials. The Fancc−/−;Fancg−/− mice now provide a model that can more accurately allow testing of this experimental question.
Given the phenotypes observed in the Fancc−/−;Fancg−/− mice, we asked whether these phenotypes would be broadly applicable to any inter-cross of FA core complex proteins. Current experimental data suggest that this is not the case. While consistent pathologic phenotypes were observed in the Fancc−/−;Fancg−/− mice, using a similar experimental design as outlined in studies shown in this manuscript, inter-crossed mice that are deficient in both Fanca and Fancc have no observable cooperative defects in stem cell function. These data are consistent with studies by Noll et al43 who did not see biologic changes in hematopoietic progenitor cells exposed to alkylating agents in vitro. This suggests that there is something unique in the function of Fancg, because Fancc is common to both inter-crosses and studies to assess this function of Fancg are an important area of future investigation.
In summary, we report here the first genetic and in vivo evidence that the combined inactivation of Fancc and Fancg lead to a cooperative impairment of hematopoietic stem cell function, supporting the hypothesis that FANCC and FANCG function in divergent molecular pathways of relevance to hematopoiesis in addition to their non-divergent role in FANCD2/FANCI monoubiquitination. Our observations also reveal that the Fancc−/−;Fancg−/− mouse is a model that best recapitulates the spontaneous clinical hematopoietic phenotypes of human FA, including malignancies and bone marrow aplasia. Therefore, we predict that the Fancc−/−;Fancg−/− double knockout mouse will be useful in the studies of pathogenesis and experimental treatment of FA-related hematopoietic diseases.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Special thanks to Susan Stanley for help with manuscript submission.
This work was supported by NIH-NHLBI/P01 HL053586-14, NIH-NCI/R01 CA138287-01 to D.W.C., T32 HL07910-09 to A.C.P.-L. from the National Institutes of Health (Bethesda, MD) and DFG SPP 1230 to H.H.
National Institutes of Health
Authorship
Contribution: A.C.P.-L., S.L.C., X.L., Y.S., L.M., D.S., J.Y., J.L., and P.A. performed experiments; A.C.P.-L., S.L.C., G.N., X.L., Y.S., L.M., D.S., P.A., A.O., G.H.V., F.-C.Y., H.H., G.C.B., and D.W.C. analyzed data; and A.C.P.-L., S.L.C., G.N., H.H., G.C.B., and D.W.C. wrote the manuscript.
Conflict-of-interest disclosure: H.H. may receive royalties for the sales of CH-296 (RetroNectin) based on a license agreement between Indiana University and Takara Shuzo Inc. The remaining authors declare no competing financial interests.
Correspondence: D. Wade Clapp, Herman B Wells Center for Pediatric Research, Cancer Research Institute, 1044 W Walnut St, Rm 402A, Indianapolis, IN 46204-5254; e-mail: dclapp@iupui.edu.
REFERENCES
Author notes
A.C.P.-L. and S.L.C. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal