Abstract
Mutations in the isocitrate dehydrogenase gene (IDH1) were recently described in patients with acute myeloid leukemia (AML). To investigate their prognostic significance we determined IDH1 status in 1333 young adult patients, excluding acute promyelocytic leukemia, treated in the United Kingdom MRC AML10 and 12 trials. A mutation was detected in 107 patients (8%). Most IDH1+ patients (91%) had intermediate-risk cytogenetics. Mutations correlated significantly with an NPM1 mutation (P < .0001) but not a FLT3/ITD (P = .9). No difference in outcome between IDH1+ and IDH1− patients was found in univariate or multivariate analysis, or if the results were stratified by NPM1 mutation status. However, when stratified by FLT3/ITD status, an IDH1 mutation was an independent adverse factor for relapse in FLT3/ITD− patients (P = .008) and a favorable factor in FLT3/ITD+ patients (P = .02). These results suggest that metabolic changes induced by an IDH1 mutation may influence chemoresistance in a manner that is context-dependent.
Introduction
Although considerable progress has been made in identifying acquired biomarkers that impact on prognosis in patients with acute myeloid leukemia (AML), a significant proportion of patients lack presence of a specific cytogenetic or molecular abnormality, and new markers are still required.1 Recently, whole genome sequencing of an AML case revealed a p.Arg132Cys mutation in the isocitrate dehydrogenase (IDH1) gene.2 Mutations of the same codon were detected in a further 15 of 187 (8%) patients. These mutations were first described in gliomas,3 although no mutated cases had been detected in earlier studies of 145 AML patients.4,5 In gliomas, IDH1-mutated patients have a better outcome than nonmutated cases,4 whereas in AML there are reports of a possible adverse effect of an IDH1 mutation on survival among normal karyotype (NK) patients either without2,6 or with7 a nucleophosmin (NPM1) mutation. Others, however, found no significant impact of an IDH1 mutation on outcome.8-11 To investigate this further, we analyzed mutation status and clinical outcome in a large series of younger adult AML patients with known fms-like tyrosine kinase/internal tandem duplication (FLT3/ITD), FLT3/tyrosine kinase domain (TKD), NPM1, and CCAAT/enhancer binding protein-α (CEBPA) mutant status who were uniformly treated in the UK Medical Research Council (MRC) AML10 and 12 trials.
Methods
Patients and mutation analysis
Genomic DNA was available from diagnostic samples of 1333 patients, excluding acute promyelocytic leukemia. Details comparing the characteristics of patients with and without IDH1 data are given in supplemental data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Ethical approval for the trials and tissue collection for research was obtained from the Multi-Center Research Committee of Wales; informed consent was obtained in accordance with the Declaration of Helsinki. Amplicons of IDH1 exon 4 were screened by denaturing high-performance liquid chromatography (see supplemental data). Abnormal chromatograms were mutation-specific and were confirmed by sequencing or restriction enzyme digestion. FLT3/ITD, FLT3/TKD, NPM1, and CEBPA mutation status were determined as described previously.12-14
Therapy, clinical endpoints, and statistical methods
Details of trial protocols, clinical endpoints, and statistical methods are in the supplemental data.
Results and discussion
Of 1333 patients screened, 107 (8%) had an IDH1 codon 132 mutation; 54 (50%) p.Arg132His, 35 (33%) p.Arg132Cys, 12 (11%) p.Arg132Gly, 4 (4%) p.Arg132Ser, 2 (2%) p.Arg132Leu. Patient details are in supplemental Table 1. IDH1 mutant-positive patients (IDH1+) were significantly older than IDH1− patients (median 49 vs 42 years; P < .0001) and more likely to be female (64% vs 36%; P = .007). There was no difference in type of leukemia (de novo/secondary) or presenting white blood cell count (WBC). Cytogenetics were available in 1077 patients. Using the MRC classification,15 most IDH1+ patients (91%) were in the intermediate-risk group; 74% of IDH1+ patients had an NK, 11% of all NK patients were IDH1+. Mutations were also detected in 7% with adverse-risk and 1% favorable-risk cytogenetics (supplemental Table 1). The presence of IDH1 mutations significantly correlated with NPM1 mutations, 65% of IDH1+ patients were NPM1+ (P < .0001), 77% in the NK only group, but not with FLT3/ITDs, FLT3/TKD, or CEBPA mutations.
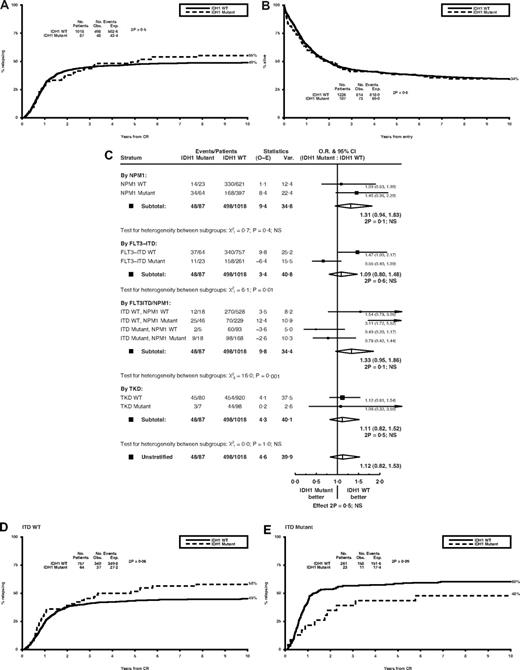
There was no difference between IDH1+ and IDH1− patients in response to therapy (remission rates of 81% and 83%, respectively), nor incidence of resistant disease or induction death (Table 1). Surviving patients were censored at July 1, 2009, with follow-up complete for more than 97% of patients, median length 12.0 years (range, 2.9-21.2 years). Neither relapse risk nor overall survival (OS) differed according to mutation status (cumulative incidence of relapse [CIR] at 10 years: 55% with vs 49% without a mutation, P = .5; OS: 34% for both, P = .6; Figure 1A-B and Table 1). This did not change in multivariate analysis adjusting for age, WBC, sex, type of leukemia, cytogenetic risk group, performance status, FLT3/ITD and NPM1 mutant status (Table 1). Similar results were obtained considering only NK patients (supplemental Figure 1). There was also no significant difference in the impact of an IDH1 mutation if the results were stratified according to NPM1 mutation status. CIR at 10 years for IDH1+ and IDH1− patients was 62% versus 53% for NPM1− patients (P = .7), 53% versus 42% for NPM1+ patients (P = .09; supplemental Figure 2A-B), OS 27% versus 28%, respectively, for NPM1− patients (P = .3), 39% versus 46% for NPM1+ patients (P = .2). There was no significant heterogeneity between the subgroups, P = .4 for relapse (Figure 1C), P = .9 for OS. Similar results were obtained if only NK patients were considered (supplemental Figure 3).
Response to therapy and outcome data according to IDH1 mutant status in total cohort of 1333 patients
| Endpoint . | IDH1 WT (n = 1226) . | IDH1 mutant (n = 107) . | Unadjusted . | Adjusted . | ||
|---|---|---|---|---|---|---|
| OR/HR (95% CI) . | P . | OR/HR (95% CI) . | P . | |||
| Response to therapy | ||||||
| CR/CRi | 83% | 81% | 1.15 (0.68-1.95) | .6 | 0.60 (0.26-1.38) | .2 |
| RD | 10% | 10% | 1.01 (0.52-1.93) | > .999 | 0.72 (0.25-2.09) | .5 |
| ID | 6% | 8% | 1.37 (0.62-3.03) | .4 | 0.53 (0.17-1.65) | .3 |
| Outcome at 10 years | ||||||
| OS | 34% | 34% | 1.07 (0.83-1.36) | .6 | 1.06 (0.79-1.40) | .7 |
| RFS | 34% | 30% | 1.07 (0.82-1.40) | .6 | 1.14 (0.85-1.53) | .4 |
| CIR | 49% | 55% | 1.12 (0.82-1.53) | .5 | 1.13 (0.80-1.58) | .5 |
| CIDCR | 17% | 15% | 0.94 (0.55-1.59) | .8 | 1.14 (0.64-2.05) | .7 |
| Endpoint . | IDH1 WT (n = 1226) . | IDH1 mutant (n = 107) . | Unadjusted . | Adjusted . | ||
|---|---|---|---|---|---|---|
| OR/HR (95% CI) . | P . | OR/HR (95% CI) . | P . | |||
| Response to therapy | ||||||
| CR/CRi | 83% | 81% | 1.15 (0.68-1.95) | .6 | 0.60 (0.26-1.38) | .2 |
| RD | 10% | 10% | 1.01 (0.52-1.93) | > .999 | 0.72 (0.25-2.09) | .5 |
| ID | 6% | 8% | 1.37 (0.62-3.03) | .4 | 0.53 (0.17-1.65) | .3 |
| Outcome at 10 years | ||||||
| OS | 34% | 34% | 1.07 (0.83-1.36) | .6 | 1.06 (0.79-1.40) | .7 |
| RFS | 34% | 30% | 1.07 (0.82-1.40) | .6 | 1.14 (0.85-1.53) | .4 |
| CIR | 49% | 55% | 1.12 (0.82-1.53) | .5 | 1.13 (0.80-1.58) | .5 |
| CIDCR | 17% | 15% | 0.94 (0.55-1.59) | .8 | 1.14 (0.64-2.05) | .7 |
Adjusted analyses were performed using logistic/Cox regression methods adjusted for age, sex, cytogenetic risk group, performance status, de novo/secondary disease, presenting white blood cell count, and FLT3/ITD and NPM1 mutation status.
WT indicates wild-type; OR, odds ratio; HR, hazard ratio; CI, confidence interval; CR, complete remission; CRi, CR with incomplete hematologic recovery; RD, resistant disease; ID, induction death; OS, overall survival; RFS, relapse-free survival; CIR, cumulative incidence of relapse; and CIDCR, cumulative incidence of death in CR
Clinical outcome stratified by IDH1 mutant status. Kaplan-Meier curves for (A) cumulative incidence of relapse and (B) overall survival in the total cohort. (C) Mantel-Byar analysis for the effect of IDH1 mutations on relapse risk stratified by NPM1, FLT3/ITD, and FLT3/TKD mutant status. (D) Cumulative incidence of relapse by IDH1 status in FLT3/ITD-WT and (E) FLT3/ITD-mutant patients. WT indicates wild-type; CR, complete remission; O-E, observed minus expected; Var., variance; O.R., odds ratio; CI, confidence interval; and NS, not significant.
Clinical outcome stratified by IDH1 mutant status. Kaplan-Meier curves for (A) cumulative incidence of relapse and (B) overall survival in the total cohort. (C) Mantel-Byar analysis for the effect of IDH1 mutations on relapse risk stratified by NPM1, FLT3/ITD, and FLT3/TKD mutant status. (D) Cumulative incidence of relapse by IDH1 status in FLT3/ITD-WT and (E) FLT3/ITD-mutant patients. WT indicates wild-type; CR, complete remission; O-E, observed minus expected; Var., variance; O.R., odds ratio; CI, confidence interval; and NS, not significant.
However, the impact of an IDH1 mutation did differ if the results were stratified according to FLT3/ITD status, with a trend for a higher relapse rate in IDH1+FLT3/ITD− cases (CIR 58% with vs 45% without an IDH1 mutation, P = .06) but not in FLT3/ITD+ patients (48% vs 60% respectively, P = .09; Figure 1D-E), P = .01 for heterogeneity between the subgroups (Figure 1C). In multivariate analysis, an IDH1 mutation was an independent adverse factor for relapse in FLT3/ITD− patients (P = .008) and a favorable factor in FLT3/ITD+ patients (P = .02) (supplemental Table 2), with significant heterogeneity between the groups (P = .002). Data for OS were compatible with those for relapse (supplemental Figure 2C-D; supplemental Table 2). Comparable results were observed in the subgroup of NPM1+FLT3/ITD− patients; 17% were IDH1+ and were associated with a significantly higher relapse risk and lower OS (Figure 1C). There was no evidence that an IDH1 mutation influenced outcome in patients with a FLT3/TKD mutation, although only 9 patients had both mutations (Figure 1C). There was also no evidence that the treatment given influenced the impact of an IDH1 mutation, with similar results if the data were reanalyzed censoring at the time of allogeneic transplantation. Overall, these results suggest that metabolic changes induced by an IDH1 mutation may influence chemoresistance in a context-dependent manner.
Of note, IDH2 gene mutations have now been detected in AML patients at a similar or increased frequency to IDH1 mutations and, likewise, are associated with NPM1 mutations and predominate in NK patients.6,7,11,16 At present, their impact on outcome remains controversial, and further analysis is required to clarify whether they differ from IDH1 mutations.
The mechanism by which IDH1 mutations contribute to cancer is not clear and may be multifaceted. IDH1 is an important source of NADPH, which protects the cell from redox stress, and has been linked to the suppression of apoptosis and enhanced cell survival and growth.17 Arginine132 plays a key role in regulating IDH1 activity, and functional studies have shown that the mutations dramatically reduce enzyme activity by impairing affinity for its substrate, isocitrate, with a consequent loss in production of NADPH and α-ketoglutarate.4,18 This may both limit resistance to apoptosis and promote glycolysis through accumulation of hypoxia-inducible factor 1, which requires α-ketoglutarate for its degradation. However, recent studies have also suggested that the mutations may lead to a gain-of-function by shifting enzyme substrate specificity and enabling conversion of α-ketoglutarate to 2-hydroxyglutarate, accumulation of which leads to increased oxidative stress.19 It is unusual for a specific mutation to be associated with opposing prognostic associations in different subgroups of patients and this may relate to the dual metabolic impact of an IDH1 mutation, with increased IDH1 mutant-associated chemosensitivity only being apparent in highly activated or proliferative blast cells such as those carrying a FLT3/ITD. In less activated cells an antiapoptotic effect might predominate. This will be an important issue in AML as metabolic pathways are being actively pursued as future chemotherapeutic targets.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to the clinical investigators who entered and managed patients in these 2 trials.
This work was supported by Leukemia & Lymphoma Research UK and the UK Medical Research Council. The work was undertaken at UCLH/UCL, who received a proportion of funding from the Department of Health's NIHR Biomedical Research Centres funding scheme.
Authorship
Contribution: R.E.G. and D.C.L. designed the study; C.L.G. and C.M.E. performed assays; R.K.H. analyzed the data; A.K.B. is principal trial coordinator; and C.L.G., R.E.G., R.K.H. and D.C.L. wrote the manuscript, which was reviewed by all authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rosemary E. Gale, Department of Haematology, UCL Cancer Institute, Paul O'Gorman Bldg, 72 Huntley St, London, WC1E 6DD United Kingdom; e-mail: rosemary.gale@ucl.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal