Abstract
Deregulation of Notch signaling has been linked to the development of T-cell leukemias and several solid malignancies. Yet, it is unknown whether Notch signaling is involved in the pathogenesis of mycosis fungoides and Sézary syndrome, the most common subtypes of cutaneous T-cell lymphoma. By immunohistochemistry of 40 biopsies taken from skin lesions of mycosis fungoides and Sézary syndrome, we demonstrated prominent expression of Notch1 on tumor cells, especially in the more advanced stages. The γ-secretase inhibitor I blocked Notch signaling and potently induced apoptosis in cell lines derived from mycosis fungoides (MyLa) and Sézary syndrome (SeAx, HuT-78) and in primary leukemic Sézary cells. Specific down-regulation of Notch1 (but not Notch2 and Notch3) by siRNA induced apoptosis in SeAx. The mechanism of apoptosis involved the inhibition of nuclear factor-κB, which is the most important prosurvival pathway in cutaneous T-cell lymphoma. Our data show that Notch is present in cutaneous T-cell lymphoma and that its inhibition may provide a new way to treat cutaneous T-cell lymphoma.
Introduction
Primary cutaneous T-cell lymphomas (CTCLs) compose a heterogeneous group of lymphoproliferative disorders of the skin. The most frequent entities are mycosis fungoides (MF) and Sézary syndrome (SS). The disease typically presents in the form of skin patches and/or plaques, which can progress to skin tumors, with subsequent involvement of lymph nodes, peripheral blood, and visceral organs. Large-cell transformation increases the likelihood of systemic dissemination and is associated with a worse prognosis.1
Notch is a transmembrane receptor family composing in humans 4 homologues (Notch1-4), which is initiated through direct cell contact with membrane-bound ligands of the Jagged or Delta family. Notch signaling is indispensable in the normal T-cell development.2-4 The receptor-ligand interaction induces 2 successive proteolytic cleavages by tumor necrosis factor-α converting enzyme and the γ-secretase/presenilin complex, resulting in the release and nuclear translocation of the intracellular domain (N-IC). In the nucleus, N-IC binds with transcriptional regulators such as CBF-1 (also termed CSL or RBP-Jκ), resulting in the expression of various effector genes, including HES (Hairy Enhancer of Split).5 Because these signaling pathways are crucially involved in the control of proliferation, differentiation, and apoptosis, deregulation of Notch may result in cancer. A t(7;9)(q34;q34.3) chromosomal translocation, which brings an activated form of the Notch1 receptor under the control of the T-cell receptor gene, is a cause of the acute lymphoblastic T-cell leukemia.6,7 Subsequently, Notch has been shown to be up-regulated and potentially involved in various solid tumors8-10 and hematologic malignancies, such as B-chronic lymphocytic leukemia, acute myeloid leukemia, multiple myeloma, Hodgkin lymphoma, and nodal anaplastic large cell lymphoma.11-14
The known role of Notch1 in normal T-cell development and its powerful influence on malignant T-cell growth and survival impelled us to investigate its potential oncogenic role in CTCL. We were encouraged by our earlier findings of a markedly increased expression of Notch1 in primary cutaneous CD30+ lymphoproliferative disorders, such as anaplastic large cell lymphoma and lymphomatoid papulosis.15 Here, we studied the expression and functional significance of Notch in CTCL. We found that Notch is expressed in a stage-dependent manner in MF and in SS. Notch inhibition, either with γ-secretase inhibitors (GSIs) or a specific Notch1 down-regulation, induced apoptosis in CTCL cell lines, paralleled by the inhibition of major prosurvival pathways mediated by nuclear factor-κB (NF-κB) and FoxO3a.
Methods
Tissue samples and patient data
Forty cases of MF (n = 35) and SS (n = 5) were selected from the archives of our institutions. The patients were 26 males and 13 females, with a median age of 65 years (range, 33-84 years) at diagnosis. One patient had biopsies taken on 2 occasions, and biopsies from stages with tumor MF and transformed MF were included from this patient. Histology and immunohistochemistry were confirmed by 2 pathologists (L.M.R.G. and E.R.) in accordance with the World Health Organization–Europeon Organization of Research and Treatment of Cancer1 classification. Early patch lesions were found in 6 patients with MF, 10 had infiltrated plaques, 9 cases had tumors, and 10 cases had MF with transformation to large T-cell lymphomas. Five of the latter were positive for CD30. Five patients had SS. A multiblock containing samples of tonsils, kidney, and liver was used as control. The study was approved by the Ethics Committee of Copenhagen and Frederiksberg and the Danish Data Protection Agency.
Cell culture and harvesting
Three CTCL cell lines have been used: MyLa2000 (derived from a plaque biopsy of a patient with MF),16 SeAx,17 and HuT-7818 (derived from peripheral blood of patients with SS). MyLa and SeAx were cultured in Dulbecco modified Eagle medium containing 4.5 g/L glucose, 25mM N-2-hydroxyethylpiperazine-N-2-ethanesulfonic, and at 37°C under 5% CO2 and HuT-78 in RPMI 1640 medium with l-glutamine, with 10% fetal calf serum, at 37°C and 5% CO2. P1051 are alloantigen-specific CD4+ T cells generated from a healthy donor (described in detail elsewhere19 ) cultured in RPMI 1640, 10% human serum, 2mM l-glutamine, 0.1 mg/mL penicillin, and 0.1 mg/mL streptomycin as well as 103 U/mL interleukin-2. Peripheral blood mononuclear cells were isolated from buffy coats provided by the blood bank at Rigshospitalet, Copenhagen, using density gradient centrifugation (Lymphoprep) and cultured under the same conditions as P1051. The cell lines were tested regularly to be negative for Mycoplasma.
Isolation of CD7−CD4+ Sézary cells
Peripheral blood mononuclear cells were isolated from blood of a 53-year-old male patient diagnosed with SS according to the World Health Organization–Organization of Research and Treatment of Cancer classification. This patient had peripheral lymphocytosis of 9.5 × 109/L with a CD4/CD8 ratio of 19. T-cell receptor gene rearrangement showed a monoclonal population containing T-cell receptor-γ-V2/V4-J1/J2 in the peripheral blood and in the bone marrow. CD7−CD4+ cells were isolated using Anti-Biotin Multisort Kit (Miltenyi Biotec) together with an anti-CD7 biotin-conjugated antibody (eBioscience) and anti-CD4 microbeads (Miltenyi Biotec) as described previously.20
Reagents and antibodies
GSI I (Z-Leu-Leu-Nle-CHO), GSI IX (DAPT, N-[N-(3,5-difluorophenacetyl-L-alanyl)]-S-phenylglycine t-butyl ester), GSI XX (dibenzazepine, (S,S)-2-[2-(3,5-difluorophenyl)acetylamino]-N-(5-methyl-6-oxo-6,7-dihydro-5H-dibenzo[b,d]azepin-7-yl)propionamide), GSI XXI (compound E, (S,S)-2-[2-(3,5-difluorophenyl)-acetylamino]-N-(1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-propionamide), MG132 (Z-Leu-Leu-Leu-CHO), LY294002 (2-(4-morpholino)-8-phenyl-4H-1-benzopyran-4-one), and Akt inhibitor X (10-(4′-(N-diethylamino)butyl)-2-chlorophenoxazine, HCl) were from Merck Calbiochem. Rapamycin (23,27-epoxy-3H-pyrido[2,1-c][1,4]oxaazacyclohentriacontine) and helenalin were from Sigma-Aldrich. GSIs, LY294002, rapamycin, MG132, and helenalin were dissolved in dimethyl sulfoxide (DMSO). Mock-treated cells were cultured with DMSO at final concentrations of 0.04% to 0.2%. Antibodies specific for Notch1, Notch3, Notch4, Delta, and NF-κB p65 were from Santa Cruz Biotechnology. The Notch2 antibody developed by Artavanis-Tsakonas was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa, Department of Biology (Iowa City, IA). Antibodies against poly(ADP-ribose) polymerase (PARP), Akt total, phosphorylated Akt (Ser473; Thr 308), total FoxO3a, and phosphorylated FoxO3a (Ser318/321) were from Cell Signaling. Anti-HES1 was from Abcam. To detect total protein in Western blotting, anti-actin antibody (Sigma-Aldrich) was used.
Immunohistochemistry
Affinity-purified monoclonal rabbit antihuman Notch1 c-20 was purchased from Santa Cruz Biotechnology. The sections were deparaffinized, rehydrated, and incubated in 3% H2O2 for 10 minutes. After microwave heat-induced epitope retrieval, sections were incubated for 60 minutes at room temperature with a 1:200 dilution of Notch1 in 1% bovine serum albumin/Tris-buffered saline and then stained using the DAKO EnVision+ Detection System-HRP for use with rabbit primary antibodies (Dako Denmark). After diaminobenzidine treatment, the slides were incubated in 0.5% copper sulfate and counterstained in Mayer hematoxylin. The procedure was performed manually.
Scoring of Notch1 labeling in tumor cells was performed by trained pathologists (L.M.R.G. and E.R.). The malignant cells were scored as negative (no visible staining, or positive staining in < 10%), moderately positive (positive staining in 10%-50% of the tumor cells), and positive in a majority (> 50%) of the tumor cells.
Western blot
Whole-cell extracts were prepared by lysis in sample buffer (0.5M Tris-HCl, pH 6.8; 5% glycerol; 10% sodium dodecyl sulfate; 0.2M dithiothreitol) supplemented with protease inhibitor cocktail from Roche Diagnostics. Equal amounts of protein were separated by a 12% Bis-Tris gel electrophoresis at 200 V followed by electrophoretic transfer to a nitrocellulose membrane (Bio-Rad). Membranes were blocked for 1 hour at 4°C with Li-Cor blocking agent before incubation with primary mouse, rabbit, or rat antibody overnight at 4°C. Subsequently, they were incubated for 1 hour with the appropriate secondary antibodies labeled with 800IR dye (anti–rabbit; Li-Cor), Alexa Fluor 680 (anti–mouse or anti–rat; both from Invitrogen). Protein bands were detected and quantified with the infrared Odyssey imaging System (Li-Cor). Preparation of cytoplasmic and nuclear protein extractions was made using the NucBuster protein extraction kit from Novagen, following the manufacturer's protocol (Novagen, Merck KgaA).
Cell viability, proliferation, and apoptosis
Viability was assessed with the fluorescent dye, calcein-AM (Invitrogen), according to the manufacturer's protocol. Briefly, cell cultures were treated with GSI, washed twice with phosphate-buffered saline (PBS), and incubated for 40 minutes at 37°C with calcein-AM in PBS. In 6-day cultures, the media was changed at day 3. Cellular fluorescence was measured using a Wallac 1420 Victor3 II microplate reader (PerkinElmer Life and Analytical Sciences).
For assessment of cell proliferation, cell lines were grown in 96-well round-bottom tissue culture plates in a final volume of 200 μL and treated with GSI I for 24 hours. Sixteen hours before harvest, 3H-thymidine (1 μCi [0.037 MBq]/well) was added, the cells harvested onto glass fiber filters, and 3H-thymidine incorporation was measured in a TopCount scintillation counter (PerkinElmer Life and Analytical Sciences). The proliferation was expressed as mean counts per minute of triplicate cultures. To determine the effects on the cell cycle, cells were pelleted by centrifugation, washed in PBS, and resuspended in Nuclear Isolation and Staining Solution (NPE Systems). DNA content was analyzed by flow cytometry using a Cell Lab Quanta SC MPL (Beckman Coulter) and Cell Lab Quanta SC MPL Analysis Software, Version 1.0.
The Caspase-Glo 3/7 (Promega) detecting the activity of apoptotic terminal caspases 3 and 7 was performed according to the manufacturer's protocol. The luminescence was measured using a Wallac 1420 Victor3 II microplate-based luminometer (PerkinElmer Life and Analytical Sciences). In addition, apoptosis was determined by flow cytometry using fluorescein isothiocyanate-annexin V/propidium iodide (PI) protocol of the manufacturer (Beckman Coulter) and analyzed by flow cytometry in a Cell Lab Quanta SC MPL flow cytometer.
Confocal microscopy
Cytospin preparations of cells were fixed at 4°C in acetone for 20 minutes followed by permeabilization with 0.5% Triton X-100 for 10 minutes and rehydration with 0.5% PBS/bovine serum albumin for 15 minutes. The cells were incubated with primary antibodies, washed twice in PBS, and incubated with Alexa 568–conjugated anti–rabbit antibody (Invitrogen). Cells were imaged with an Olympus IX70 laser scanning microscope (Olympus, FluoView Confocal System). Fluorescence was determined in randomly selected 10 cells using Fluoview Version 4.3 software.
siRNA
One day before transfection, the cells were transferred to Petri dishes 2 cm in diameter at 300 000 cells/mL. Transfections were done by lipofection using Notch1, Notch2, Notch3, or nontargeting ON-TARGETplus SMART-pool siRNAs (Dharmacon RNA Technologies) at 20nM and Lipofectamine RNAiMax (Invitrogen) according to the manufacturer's protocol. One day after transfection, the media was changed and the cells were assayed for viability and apoptosis. The extent of protein knockdown was quantified by Western blotting.
Statistics
Notch1 immunostaining data were analyzed with Kruskal-Wallis test for nonparametric data. Continuous data are reported as mean plus or minus SD, and the differences were evaluated by the Student t test. Because all experiments were performed at a minimum in triplicate and repeated 2 or 3 times, all data were pooled for statistical analysis. P values less than .05 were considered to be statistically significant. Statistical analysis was performed by SPSS Version 17.0, GraphPad Prism Version 4.03 software, or Excel (Microsoft).
Results
CTCL cell lines (SeAx, MyLa, and Hut-78), but not normal human lymphocytes, express Notch
Using antibodies directed against the intracellular domains of the Notch1 to Notch4 receptors, we detected as the fragments of expected length approximately 110 kDa (Notch1 and Notch2), approximately 90 kDa (Notch3), and approximately 52 kDa (Notch4) in all 3 tested cell lines (Figure 1A). They represent the active, intracellular forms of the receptors. The full-length receptors were difficult to visualize by Western blot, and their expression was not possible to evaluate reproducibly. Further evidence for the functional activation of Notch was provided by demonstrating the presence of its natural ligand Delta in CTCL cell lines as well as the expression of HES1, which is stimulated almost exclusively by Notch. In contrast, normal proliferating T cells (P1051) and nonproliferating leukocytes from 2 healthy donors showed either very weak or undetectable protein levels of the Notch family members, with an exception of Notch2 in P1051 (Figure 1B). However, the P1051 did not express Hes1, so it was probable that the Notch signaling pathway was inactive.
Expression of Notch family proteins in CTCL. (A) Western blot analysis of whole-cell lysates from CTCL cell lines: MyLa (lane 1), SeAx (lane 2), and Hut78 (lane 3). The blots were probed with the antibodies against Notch1 to Notch4; the intracellular domains are visualized as bands at approximately 110 kDa (Notch1 and Notch2), approximately 90 kDa (Notch3), and approximately 52 kDa (Notch4), Notch ligand, Delta, and Hes, the transcriptional factor induced by Notch signaling. (B) Western blot analysis of normal proliferating T cells (P1051) and nonproliferating leukocytes from 2 healthy donors probed for Notch1 to Notch4 and Hes1. (C) Immunostaining for Notch1 in skin biopsies from a patient with tumor-stage MF (top images, UplanSApo 40×/0.90; bottom images, UplanSApo 60×/0.90 using Olympus BX51 microscope with camera Olympus DP70) showing high Notch1 expression in tumor cells. The staining was mainly cytoplasmic and membranous. Arrowhead (top right image) indicates epidermiotropic Notch1-positive tumor cells as displayed in bottom right image at higher magnification. The arrow indicates Notch1-negative small reactive T lymphocytes. (D) Quantification of Notch1 expression in biopsies from lesional skin from patients with MF in stage I (patch and plaque), stage II (tumors), transformed MF, and SS. The histograms represent the percentage of patients classified into one of the 3 categories (negative, moderately positive, and positive), as described in “Immunohistochemistry.” Comparison of the expression profiles shows statistical significant increase in Notch expression with stage (P = .017).
Expression of Notch family proteins in CTCL. (A) Western blot analysis of whole-cell lysates from CTCL cell lines: MyLa (lane 1), SeAx (lane 2), and Hut78 (lane 3). The blots were probed with the antibodies against Notch1 to Notch4; the intracellular domains are visualized as bands at approximately 110 kDa (Notch1 and Notch2), approximately 90 kDa (Notch3), and approximately 52 kDa (Notch4), Notch ligand, Delta, and Hes, the transcriptional factor induced by Notch signaling. (B) Western blot analysis of normal proliferating T cells (P1051) and nonproliferating leukocytes from 2 healthy donors probed for Notch1 to Notch4 and Hes1. (C) Immunostaining for Notch1 in skin biopsies from a patient with tumor-stage MF (top images, UplanSApo 40×/0.90; bottom images, UplanSApo 60×/0.90 using Olympus BX51 microscope with camera Olympus DP70) showing high Notch1 expression in tumor cells. The staining was mainly cytoplasmic and membranous. Arrowhead (top right image) indicates epidermiotropic Notch1-positive tumor cells as displayed in bottom right image at higher magnification. The arrow indicates Notch1-negative small reactive T lymphocytes. (D) Quantification of Notch1 expression in biopsies from lesional skin from patients with MF in stage I (patch and plaque), stage II (tumors), transformed MF, and SS. The histograms represent the percentage of patients classified into one of the 3 categories (negative, moderately positive, and positive), as described in “Immunohistochemistry.” Comparison of the expression profiles shows statistical significant increase in Notch expression with stage (P = .017).
Notch1 is expressed in advanced MF and SS in tumor cells
To address whether lymphoma cells in vivo resemble CTCL cell lines with respect to the expression of Notch, we performed immunohistochemical stainings of skin specimens from MF patients in all stages (I-IV) with an anti-Notch1 antibody. All biopsies were suitable for immunohistochemical analyses. Of 40 tumor specimens, 21 cases displayed positivity for Notch1 in tumor cells (Table 1), as recognized by the irregular and/or pleomorphic nuclei (Figure 1C). The stain was mainly cytoplasmic and occasionally nuclear. Expression of Notch increased with the more advanced stage; 8 of 9 cases of tumor-stage MF and 7 of 10 cases of transformed MF displayed positivity, whereas positive Notch staining was seen in only 1 of 6 patch-stage MF and 2 of 10 cases of plaque-stage MF (Figure 1C-D). A Kruskal-Wallis test confirmed the statistical significant dependence of Notch expression on the stage (P = .017). In SS, one-half of the cases displayed positivity for Notch1 in tumor cells. In reactive tonsils, scattered large lymphoid cells in the paracortical and interfollicular areas as well as in the germinal centers were positive for Notch1, displaying cytoplasmic and occasionally nuclear staining. In the skin biopsies, endothelium, fibroblasts, and basal keratinocytes showed cytoplasmatic positivity to a various degree as reported by others.21 Small reactive-appearing lymphocytes were mainly negative (Figure 1C).
Patient characteristics and Notch1 immunohistochemical stain scores
| Patient no. . | Age, y . | Sex . | Diagnosis . | Stage . | Notch status in tumor cells . |
|---|---|---|---|---|---|
| 1 | 75 | Female | MF patch | IB | Negative |
| 2 | 44 | Female | MF patch | IB | Negative |
| 3 | 67 | Male | MF patch | IB | Negative |
| 4 | 59 | Male | MF patch | IB | Negative |
| 5 | 59 | Male | MF patch | IB | Negative |
| 6 | 55 | Female | MF patch | IB | > 50% |
| 7 | 44 | Female | MF plaque | I | Negative |
| 8 | 57 | Male | MF plaque | IB | Negative |
| 9 | 80 | Female | MF plaque | I | Negative |
| 10 | 77 | Male | MF erythrodermic | IIIA | Negative |
| 11 | 80 | Male | MF plaque | IB | Negative |
| 12 | 49 | Female | MF plaque | IB | Negative |
| 13 | 33 | Male | MF plaque | IA | Negative |
| 14 | 56 | Male | MF plaque | I | > 10% - < 50% |
| 15 | 44 | Male | MF folliculotropic | IB | > 10% - < 50% |
| 16 | 33 | Male | MF plaque | IA | > 50% |
| 17 | 74 | Female | MF tumor | IIB | Negative |
| 18 | 84 | Female | MF tumor | IIB | > 50% |
| 19 | 50 | Female | MF tumor | IVB | > 50% |
| 20 | 70 | Female | MF tumor | IIB | > 50% |
| 21 | 82 | Male | MF tumor | IIB | > 50% |
| 22 | 66 | Male | MF tumor | IIB | > 50% |
| 23 | 57 | Female | MF tumor | IIB | > 50% |
| 24 | 79 | Male | MF tumor | IIB | > 50% |
| 25 | 58 | Male | MF tumor | IIB | > 50% |
| 26 | 84 | Female | MF transformed | IIB | Negative |
| 27 | 83 | Male | MF transformed | IIB | Negative |
| 28 | 71 | Male | MF transformed | IIB | Negative |
| 29 | 77 | Male | MF transformed | II | > 10% - < 50% |
| 30 | 63 | Male | MF transformed | IIB | > 10% - < 50% |
| 31 | 57 | Male | MF transformed | IVB | > 10% - < 50% |
| 32 | 51 | Female | MF transformed | IVB | > 10% - < 50% (same patient as patient 19) |
| 33 | 61 | Female | MF transformed | IVA | > 50% |
| 34 | 67 | Male | MF transformed | IIA | > 50% |
| 35 | 77 | Male | MF transformed | IVA | > 50% |
| 36 | 68 | Male | SS | IVA | Negative |
| 37 | 71 | Male | SS | IVB | Negative |
| 38 | 72 | Male | SS | IVA | Negative |
| 39 | 73 | Male | SS | IVB | > 50% |
| 40 | 65 | Male | SS | IV | > 50% |
| Patient no. . | Age, y . | Sex . | Diagnosis . | Stage . | Notch status in tumor cells . |
|---|---|---|---|---|---|
| 1 | 75 | Female | MF patch | IB | Negative |
| 2 | 44 | Female | MF patch | IB | Negative |
| 3 | 67 | Male | MF patch | IB | Negative |
| 4 | 59 | Male | MF patch | IB | Negative |
| 5 | 59 | Male | MF patch | IB | Negative |
| 6 | 55 | Female | MF patch | IB | > 50% |
| 7 | 44 | Female | MF plaque | I | Negative |
| 8 | 57 | Male | MF plaque | IB | Negative |
| 9 | 80 | Female | MF plaque | I | Negative |
| 10 | 77 | Male | MF erythrodermic | IIIA | Negative |
| 11 | 80 | Male | MF plaque | IB | Negative |
| 12 | 49 | Female | MF plaque | IB | Negative |
| 13 | 33 | Male | MF plaque | IA | Negative |
| 14 | 56 | Male | MF plaque | I | > 10% - < 50% |
| 15 | 44 | Male | MF folliculotropic | IB | > 10% - < 50% |
| 16 | 33 | Male | MF plaque | IA | > 50% |
| 17 | 74 | Female | MF tumor | IIB | Negative |
| 18 | 84 | Female | MF tumor | IIB | > 50% |
| 19 | 50 | Female | MF tumor | IVB | > 50% |
| 20 | 70 | Female | MF tumor | IIB | > 50% |
| 21 | 82 | Male | MF tumor | IIB | > 50% |
| 22 | 66 | Male | MF tumor | IIB | > 50% |
| 23 | 57 | Female | MF tumor | IIB | > 50% |
| 24 | 79 | Male | MF tumor | IIB | > 50% |
| 25 | 58 | Male | MF tumor | IIB | > 50% |
| 26 | 84 | Female | MF transformed | IIB | Negative |
| 27 | 83 | Male | MF transformed | IIB | Negative |
| 28 | 71 | Male | MF transformed | IIB | Negative |
| 29 | 77 | Male | MF transformed | II | > 10% - < 50% |
| 30 | 63 | Male | MF transformed | IIB | > 10% - < 50% |
| 31 | 57 | Male | MF transformed | IVB | > 10% - < 50% |
| 32 | 51 | Female | MF transformed | IVB | > 10% - < 50% (same patient as patient 19) |
| 33 | 61 | Female | MF transformed | IVA | > 50% |
| 34 | 67 | Male | MF transformed | IIA | > 50% |
| 35 | 77 | Male | MF transformed | IVA | > 50% |
| 36 | 68 | Male | SS | IVA | Negative |
| 37 | 71 | Male | SS | IVB | Negative |
| 38 | 72 | Male | SS | IVA | Negative |
| 39 | 73 | Male | SS | IVB | > 50% |
| 40 | 65 | Male | SS | IV | > 50% |
Negative indicates that < 10% of tumor cells are labeled in Notch1.
Notch targeting by GSIs induces apoptosis and decreases cell viability
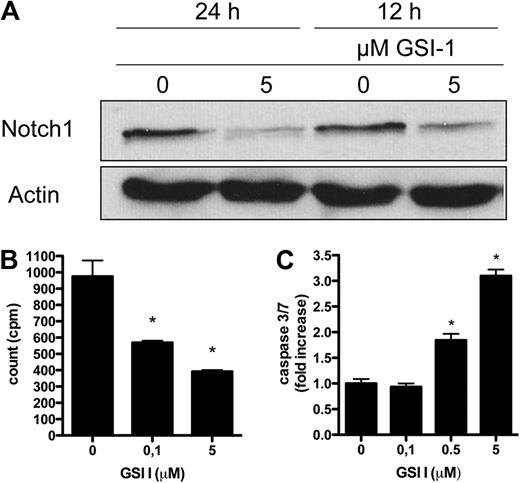
GSIs block the proteolytic cleavage of all Notch receptors and hereby prevent release of N-IC.22 The expression of Notch in malignant T cells suggests that GSIs could have a therapeutic effect in CTCL. First, we confirmed that inhibition of γ-secretase with GSI I treatment for 18 hours blocked Notch processing, resulting in concentration-dependent decreased levels of active, intracellular Notch1 to Notch4 (Figure 2A). To exclude that the observed effect was the result of a general cytotoxicity and protein degradation, the expression of STAT3 was also examined, showing no decrease in total protein level (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). MyLa, SeAx, and Hut78 were incubated for 48 hours with increasing GSI concentrations or a vehicle, and apoptosis was assessed by caspase 3/7 activity. Results showed that GSI I induced apoptosis in a concentration-dependent manner with the greatest effect observed at a concentration of 1μM or greater (P < .001). GSI IX, XX, and XXI had a much less pronounced effect (Figure 2B; supplemental Figure 2A-B). To evaluate the time-response of GSIs on apoptosis, caspase 3/7 activity was measured 0, 3, 12, 48, and 72 hours after treatment with GSI I at 5μM and GSI IX, XX, and XXI at 20μM. For GSI I treatment, the apoptotic effect peaked after 24 hours in MyLa and Hut78, whereas the peak was seen as rapidly as after 3 hours for SeAx (P < .001; Figure 2C; supplemental Figure 2C-D). The finding of a superior potency of GSI I (a tripeptide GSI, z-Leu-Leu-Nle-CHO) was expected in view of the earlier data reported for other neoplasms.23,24
Inhibition of Notch by GSIs stimulates apoptosis and blocks cell proliferation in CTCL cell lines. (A) SeAx cells were treated with GSI I (0.05-5μM) or the vehicle (DMSO) and analyzed with Western blot for activated Notch1 to Notch4 and Hes, as in Figure 1. (B-C) Induction of proapoptotic caspases 3 and 7 after treatment of MyLa by different GSIs: I, IX, XX, and XXI. The cells were treated by the increasing concentrations of GSIs for 48 hours (B) or with 5μM GSI I and 20μM of GSI IX, XX, and XXI for 0 to 72 hours (C). Data are fold increase over the residual caspase 3/7 activity in the control, vehicle-treated cells. P < .05 in both graphs for GSI I treatment compared with vehicle treatment. P < .05 for all tested GSI XX concentrations and for GSI XXI 20μM. P < .05 for GSI IX and XXI after 24 hours, for GSI XXI after 48 hours, and GSI XXI after 72 hours in MyLa. Data are mean ± SD. (D) Sub-G1 population in MyLa, SeAx, and Hut78 cells, which were treated with 5μM GSI I (0, 3, 24, and 48 hours). DNA was stained with NiM-DAPi, and the cells were analyzed by flow cytometry. Bars represent mean values. The experiment was repeated 3 times with similar results. *P < .05. (E) MyLa was treated with GSI I (5μM; 0, 3, and 24 hours) and stained with annexin V and PI for flow cytometry, as described in “Cell viability, proliferation and apoptosis.” Cells were gated according to the annexin V (green FL1 channel, x-axis) and PI-specific (red, FL3 channel, y-axis) fluorescence. Values in the quadrants represent the percentages of cells. The experiment was repeated twice with similar results. (F) PARP cleavage in MyLa, SeAx, and Hut78 cells treated with 5μM GSI I for 24 hours. Whole-cell extracts were prepared for Western blot as in Figure 1, and the blots were probed with the antibody against PARP detecting its intact form (116 kDa) and the caspase-cleaved 89-kDa fragment. (G) Cell proliferation of MyLa, SeAx, and Hut78 cells treated with increasing concentrations of GSI I for 24 hours. Sixteen hours before harvest, 3H-thymidine (1 μCi [0.037 MBq]/well) was added, and the results are expressed as mean counts per minute of triplicate experiments ± SD. *P < .05.
Inhibition of Notch by GSIs stimulates apoptosis and blocks cell proliferation in CTCL cell lines. (A) SeAx cells were treated with GSI I (0.05-5μM) or the vehicle (DMSO) and analyzed with Western blot for activated Notch1 to Notch4 and Hes, as in Figure 1. (B-C) Induction of proapoptotic caspases 3 and 7 after treatment of MyLa by different GSIs: I, IX, XX, and XXI. The cells were treated by the increasing concentrations of GSIs for 48 hours (B) or with 5μM GSI I and 20μM of GSI IX, XX, and XXI for 0 to 72 hours (C). Data are fold increase over the residual caspase 3/7 activity in the control, vehicle-treated cells. P < .05 in both graphs for GSI I treatment compared with vehicle treatment. P < .05 for all tested GSI XX concentrations and for GSI XXI 20μM. P < .05 for GSI IX and XXI after 24 hours, for GSI XXI after 48 hours, and GSI XXI after 72 hours in MyLa. Data are mean ± SD. (D) Sub-G1 population in MyLa, SeAx, and Hut78 cells, which were treated with 5μM GSI I (0, 3, 24, and 48 hours). DNA was stained with NiM-DAPi, and the cells were analyzed by flow cytometry. Bars represent mean values. The experiment was repeated 3 times with similar results. *P < .05. (E) MyLa was treated with GSI I (5μM; 0, 3, and 24 hours) and stained with annexin V and PI for flow cytometry, as described in “Cell viability, proliferation and apoptosis.” Cells were gated according to the annexin V (green FL1 channel, x-axis) and PI-specific (red, FL3 channel, y-axis) fluorescence. Values in the quadrants represent the percentages of cells. The experiment was repeated twice with similar results. (F) PARP cleavage in MyLa, SeAx, and Hut78 cells treated with 5μM GSI I for 24 hours. Whole-cell extracts were prepared for Western blot as in Figure 1, and the blots were probed with the antibody against PARP detecting its intact form (116 kDa) and the caspase-cleaved 89-kDa fragment. (G) Cell proliferation of MyLa, SeAx, and Hut78 cells treated with increasing concentrations of GSI I for 24 hours. Sixteen hours before harvest, 3H-thymidine (1 μCi [0.037 MBq]/well) was added, and the results are expressed as mean counts per minute of triplicate experiments ± SD. *P < .05.
To further document the effects of GSIs on apoptosis and cell cycle, nucleos isolation medium-4,6-diamidino-2-phenylindole dihydrocholride (NiM-DAPi) stainings for total DNA were used to evaluate cell cycle phase in the 3 cell lines treated with GSI I 5μM over a time course of 0 to 48 hours. The percentage of cells in the sub-G0/G1 phase increased significantly in all 3 cell lines after 24 hours of treatment, indicative of apoptosis (Figure 2D). Correspondingly, analysis by annexin V/PI staining after treatment with GSI I 5μM showed an increase in apoptotic cells in all cell lines most pronounced after 24 hours (the early and late apoptosis rate after 24 hours treatment was, respectively, 34.3% and 38.6% for MyLa, 13.4% and 42.9% for SeAx, and 32.0% and 52.0% for Hut78, compared with, respectively, 10.1% and 15.1% for MyLa, 7.1% and 8.9% for SeAx, and 6.8% and 11.4% in the vehicle-treated cells; Figure 2E; supplemental Figure 2E). The effect of GSI I on apoptosis was also confirmed by Western blot detection of PARP cleavage (Figure 2F; supplemental Figure 2F).
Calcein assays after GSI I treatment for 0, 1, 3, and 24 hours showed a significant decrease in cell viability compared with control cultures after 24 hours (mean percentage decrease in calcein activity for MyLa, SeAx, and Hut78 was 84.6% ± 5.5%, 90.0% ± 4.1% and 78.8% ± 5.4%, respectively). Proliferation, measured by 3H-thymidine incorporation, demonstrated a significant dose-dependent decrease in proliferation of the malignant T cells after treatment with GSI I for 24 hours (Figure 2G). The influence on cell viability induced by GSI IX, XX, and XXI was concordant with the lower effects seen on apoptosis data. The mean percentage decrease in viable cells reached significant values for GSI IX only in SeAx at days 3 and 6 (18.7% ± 7.1% and 35.5% ± 10.6%, respectively), for GSI XX in MyLa at days 3 and 6 (22.4% ± 8.9% and 35.8% ± 7.8%, respectively), and in SeAx at days 2, 3, and 6 (17.6% ± 6.7%, 25.1% ± 5.7%, and 48.4% ± 7.4%, respectively) and for GSI XXI in MyLa at 24 hours (28.9% ± 6.8%) and in SeAx at day 6 (35.6% ± 7.5%).
Cell line results were validated in the short-term culture of leukemic cells (CD7− CD4+) obtained from peripheral blood of a patient with SS. Figure 3A shows a decrease in N-IC expression after treatment with 5μM GSI I for 12 and 24 hours, respectively. As expected of a primary culture, leukemic Sézary cells had a very low rate of proliferation, and a further, significant decrease in proliferation in response to GSI I treatment was detected (Figure 3B). Apoptosis was induced by treatment with increasing concentrations of GSI I as observed in the tumor cell lines (Figure 3C).
GSI I treatment in primary leukemic Sézary cells inhibits Notch and induces apoptosis. (A) CD7− CD4+ cells isolated from peripheral blood of a patient diagnosed with SS were treated with GSI I 5μM for 12 and 24 hours and analyzed with Western blot for Notch1. (B) Primary leukemic Sézary cells treated with increasing concentrations of GSI I for 24 hours and analyzed for thymidine incorporation as in Figure 1G. *P < .05. (C) Induction of proapoptotic caspases 3 and 7 after treatment of primary leukemic Sézary cells with increasing concentrations of GSI I for 24 hours. Data are mean ± SD. *P < .05.
GSI I treatment in primary leukemic Sézary cells inhibits Notch and induces apoptosis. (A) CD7− CD4+ cells isolated from peripheral blood of a patient diagnosed with SS were treated with GSI I 5μM for 12 and 24 hours and analyzed with Western blot for Notch1. (B) Primary leukemic Sézary cells treated with increasing concentrations of GSI I for 24 hours and analyzed for thymidine incorporation as in Figure 1G. *P < .05. (C) Induction of proapoptotic caspases 3 and 7 after treatment of primary leukemic Sézary cells with increasing concentrations of GSI I for 24 hours. Data are mean ± SD. *P < .05.
Specific down-regulation of Notch1 by RNA interference technology induces apoptosis in vitro
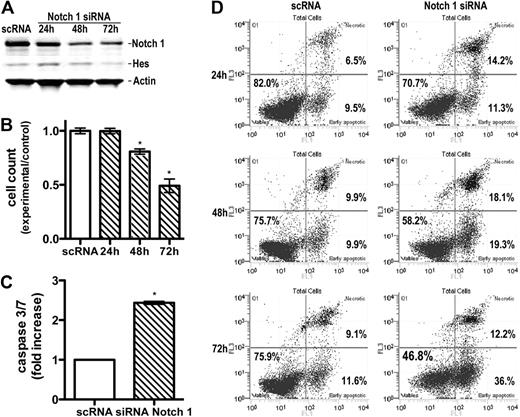
Because GSIs may affect signaling pathways other than γ-secretase,25 we specifically down-regulated Notch by siRNA nucleofection. SeAx line was successfully transfected with control nontargeting or Notch1, Notch2, or Notch3 siRNA. At 24, 48, and 72 hours after transfection, cells were examined for expression of Notch by Western blot to demonstrate a knockdown of protein levels. Data are shown for Notch1 in Figure 4A. The protein level of intracellular Notch1 was down-regulated in Notch1 siRNA-transfected cells compared with the control siRNA-transfected groups. To verify that the reduction of Notch1 by siRNA was sufficient to block the Notch1 pathway, HES1 expression was analyzed. Down-regulation of Notch1 expression by siRNA led to a decrease in HES1 protein expression.
Specific down-regulation of Notch1 by siRNA induces apoptosis. (A) Down-regulation of Notch1 and Hes expression by Notch1 siRNA. Whole-cell extracts were prepared from Seax cells 24 to 72 hours after transfection with siRNA or scRNA (control) and probed with antibodies against intracellular Notch1 and Hes1, as in Figure 1. Band intensity quantification showed a decrease in Notch1 and Hes expression by 73% and 45%, respectively, 72 hours after transfection. (B) Influence of Notch1 siRNA on cell proliferation. Cells were transfected as in panel A and enumerated by flow cytometry 24, 48, and 72 hours after transfection. Bars represent mean values (n = 3) ± SD. *P < .05 compared with the scRNA-treated, control group. (C) Increase in caspase 3/7 activity in SeAx cells transfected with Notch1 siRNA. Caspases were measured, as described in Figure 2, 24 hours after transfection. Data are mean ± SD. *P < .001. (D) Induction of annexin V-positive, apoptotic cells 24 to 72 hours after transfection with Notch1 siRNA compared with the scRNA-transfected cells. Flow cytometry and enumeration of cells were done as described in Figure 2.
Specific down-regulation of Notch1 by siRNA induces apoptosis. (A) Down-regulation of Notch1 and Hes expression by Notch1 siRNA. Whole-cell extracts were prepared from Seax cells 24 to 72 hours after transfection with siRNA or scRNA (control) and probed with antibodies against intracellular Notch1 and Hes1, as in Figure 1. Band intensity quantification showed a decrease in Notch1 and Hes expression by 73% and 45%, respectively, 72 hours after transfection. (B) Influence of Notch1 siRNA on cell proliferation. Cells were transfected as in panel A and enumerated by flow cytometry 24, 48, and 72 hours after transfection. Bars represent mean values (n = 3) ± SD. *P < .05 compared with the scRNA-treated, control group. (C) Increase in caspase 3/7 activity in SeAx cells transfected with Notch1 siRNA. Caspases were measured, as described in Figure 2, 24 hours after transfection. Data are mean ± SD. *P < .001. (D) Induction of annexin V-positive, apoptotic cells 24 to 72 hours after transfection with Notch1 siRNA compared with the scRNA-transfected cells. Flow cytometry and enumeration of cells were done as described in Figure 2.
Results showed that down-regulation of Notch1 influenced cell proliferation (Figure 4B) and induced apoptosis. The caspase 3/7 activity increased significantly in the Notch1 siRNA-transfected group compared with the control siRNA-transfected group 24 hours after transfection (P < .001; Figure 4C). In contrast, transfection with Notch2 and Notch3 did not exhibit any significant increase in apoptosis (supplemental Figure 3). The early and late apoptosis rate in the Notch1 siRNA-transfected group was, respectively, 11.3% and 14.2% after 24 hours, 19.3% and 18.1% after 48 hours, and 36.0% and 12.2% after 72 hours, compared with, respectively, 9.5% and 6.5% after 24 hours, 9.9% and 9.9% after 48 hours, and 11.6% and 9.1% after 72 hours in the control siRNA-transfected group (Figure 4D). These values were lower than obtained for GSI I, and there are 2 possible explanations. One is that some residual Notch1 was detectable in the cells despite the successful siRNA transfection. Second, unlike GSIs, which prevent activation of all 4 Notch receptors, RNA interference technology allows a specific down-regulation of each receptor. Our data suggest that tumor cells are mainly dependent on Notch1 for the protection against apoptosis. This was not surprising given the major role Notch1 plays in both normal T-cell development2-4 and other hematologic malignancies.7,11-14
GSI I treatment is accompanied by decreased activity in Akt and NF-κB signaling
To explore the molecular mechanisms underlying Notch inhibition, we focused on the phosphoinositide 3-kinase-Akt signaling, which is important for development and progression of cancers.26 Previous studies have indicated that Notch is able to modulate the activity of Akt.27-30 We examined the phosphorylation status of Akt using anti-phospho-Akt Ser473 and -Thr308 antibodies 4 hours after GSI I treatment. Treatment resulted in reductions in phosphorylated Akt (supplemental Figure 4A). However, the effects of 2 different blockers of Akt (Ly294002 and Akt Inhibitor X) and mTOR (rapamycin) were very modest and not comparable with the effect detected by GSI I (mean fold increase in caspase activity for MyLa, SeAx, and Hut78 was in the range of 1.1-5.1; supplemental Figure 4B). Neither did we find any additive effects of Akt blockers on GSI I-induced apoptosis, implying that GSI I-induced Akt inhibition is only a minor contributor to apoptosis (data not shown).
NF-κB is constitutively activated in CTCL cells, and its inhibition leads to apoptosis.31 In preliminary experiments, we confirmed that 2 NF-κB inhibitors (MG-132 and helenalin) potently induced apoptosis in MyLa and SeAx cell lines. Because Notch can control NF-κB activity,13,32,33 we studied the effect of Notch inhibition by GSI I on the activation status of NF-κB in MyLa and SeAx cells. As shown in Figure 5A, the nuclear fraction of NF-κB (representing the active form) decreased after GSI I treatment in a concentration-dependent manner, suggesting that the GSI I mechanism of apoptosis involves the inhibition of NF-κB. Results from Western blot were confirmed by studies of nuclear NF-κB localization using confocal microscopy, which revealed a decrease in p65 nuclear staining of the NF-κB subunit p65 after GSI I treatment of CTCL cell lines (data not shown).
Inhibition of NF-κB signaling and nuclear translocation of FoxO3a by GSI I. (A) Western blot of cytoplasmic (C) and nuclear (N) extracts of MyLa cells treated with GSI I (0-5μM, 6 hours) showing a concentration-dependent decrease in the nuclear NF-κB content. The positive control was treated with the NF-κB inhibitor, MG132 (10μM). Immunoblotting with anti-PARP documented the purity of the cytoplasmic and nuclear extracts. (B) The cytoplasmic (C) and nuclear (N) fractions of MyLa cells were obtained as in Figure 4A, and the blots were probed with the FoxO3a-specific antibody. The blot shows a translocation of FoxO3a from the cytoplasm to the nucleus. PARP is a marker of fraction purity, as in Figure 4A. (C) MyLa cells were treated with 5μM GSI I or the vehicle for 4 hours or 12 hours, fixed and stained with the anti-FoxO3a antibody followed by the AlexaFluor568-conjugated secondary antibody. The cells were observed in confocal microscope (Olympus, FluoView Confocal System). Original scale of the images is 50 μm × 50 μm. (D) Nuclear FoxO3a fluorescence was quantified from the confocal microscopy images of CTCL lines treated with GSI I as in panel A (n = 10 cells per group). *P < .05 compared with the control (t test). Bars represent mean ± SD. (E) Phosphorylation of FoxO3a in CTCL cells treated with the vehicle or 5μM GSI I for 4 hours. The cellular content of the whole FoxO3a and FoxO3a phosphorylated at Ser318/321 was determined by Western blot. Compared with MyLa and HuT-78, SeAx had low basal levels of phosohorylated FoxO3a, and band intensity quantification exhibited no decrease in phosphorylation after treatment.
Inhibition of NF-κB signaling and nuclear translocation of FoxO3a by GSI I. (A) Western blot of cytoplasmic (C) and nuclear (N) extracts of MyLa cells treated with GSI I (0-5μM, 6 hours) showing a concentration-dependent decrease in the nuclear NF-κB content. The positive control was treated with the NF-κB inhibitor, MG132 (10μM). Immunoblotting with anti-PARP documented the purity of the cytoplasmic and nuclear extracts. (B) The cytoplasmic (C) and nuclear (N) fractions of MyLa cells were obtained as in Figure 4A, and the blots were probed with the FoxO3a-specific antibody. The blot shows a translocation of FoxO3a from the cytoplasm to the nucleus. PARP is a marker of fraction purity, as in Figure 4A. (C) MyLa cells were treated with 5μM GSI I or the vehicle for 4 hours or 12 hours, fixed and stained with the anti-FoxO3a antibody followed by the AlexaFluor568-conjugated secondary antibody. The cells were observed in confocal microscope (Olympus, FluoView Confocal System). Original scale of the images is 50 μm × 50 μm. (D) Nuclear FoxO3a fluorescence was quantified from the confocal microscopy images of CTCL lines treated with GSI I as in panel A (n = 10 cells per group). *P < .05 compared with the control (t test). Bars represent mean ± SD. (E) Phosphorylation of FoxO3a in CTCL cells treated with the vehicle or 5μM GSI I for 4 hours. The cellular content of the whole FoxO3a and FoxO3a phosphorylated at Ser318/321 was determined by Western blot. Compared with MyLa and HuT-78, SeAx had low basal levels of phosohorylated FoxO3a, and band intensity quantification exhibited no decrease in phosphorylation after treatment.
A major downstream target of the NF-κB pathway is the Forkhead FoxO3a transcription factor (FXHRL1).26,34,35 Active, nuclear FXHRL1 induces cell cycle arrest or apoptosis, whereas inactivation through phosphorylation and nuclear exclusion promotes cell growth and tumorigenesis. As predicted, Western blot showed that GSI I treatment induced FoxO3A dephosphorylation (not observed for SeAx, which has very low, barely detectable levels of phosphorylated FoxO3a before treatment) and nuclear translocation. Confocal microscopy confirmed the nuclear translocation of FoxO3a after GSI I treatment (Figure 5B-E). Taken together, these data suggest that inhibition of the NF-κB is involved in the proapoptotic effect of GSI.
Discussion
Aberrant Notch signaling has been linked to a variety of neoplastic malignancies, such as breast cancer,8,10 malignant melanoma,9,36 acute lymphoblastic T-cell leukemia,6,7 and B-chronic lymphocytic leukemia.13 In contrast to its oncogenic role in these cancers, Notch acts as a tumor suppressor in other neoplasms, such as murine basal cell carcinoma,37 demonstrating the complexity of this pathway. In primary cutaneous CD30+ lymphoproliferative disorders, we have recently reported a more intense expression of Notch1 in anaplastic large cell lymphoma compared with the clinically more benign variant lymphomatoid papulosis.15 To further explore a potential oncogenic role for Notch signaling in CTCL, we here expanded our studies to the most common subtypes of CTCL: MF and SS.
We showed that cell lines derived from MF and SS patients express activated Notch1 to Notch4. Immunohistochemical stainings of skin biopsies from 40 patients with MF stage I-IV and SS showed the presence of Notch. Importantly, Notch expression was positively correlated with the stage of the disease, which supports the hypothesis on the involvement of Notch in the progression of the lymphoma. A similar stage-specific expression of Notch has been described for other cancers, and the level of expression has been found to correlate inversely to the clinical outcomes,10,36 probably via increased apoptotic resistance conferred by the high Notch activity.38 Although oncogenic Notch signaling in acute lymphoblastic T-cell leukemia has been ascribed to activating chromosomal translocations,6,7 no translocations or mutations of Notch have been found in CTCL.39 It is thus probable that the activation of Notch in CTCL occurs because of continuous ligand stimulation, as previously observed in Hodgkin lymphoma and nodal anaplastic large cell lymphoma.11
Therapeutic targeting of Notch by small molecules has been achieved by GSIs, which inhibit the proteolytic cleavage and release of the biologically active intracellular fragment of Notch to the cytoplasm. Treatment of CTCL cell lines with different GSIs resulted in apoptosis. There were marked differences between different compounds tested; GSI IX, XX, and XXI exhibited only a modest effect, whereas GSI I in the micromolar range was a very potent inducer of cell death. The superiority of GSI I has also been reported in other malignancies,23,24 and GSI plasma concentration in the micromolar range has been shown to be physiologically achievable in clinical trials of human leukemias.40 Furthermore, we reproduced the proapoptotic activity of GSI I in a short-term culture model of leukemic cells obtained from a patient with SS, which further underscores the potential of Notch blockade for the therapy of cutaneous lymphoma.
We have considered the possibility that the effect of GSI I was not specific to the inhibition of Notch. GSIs are known to exert pleiotropic effects, including inhibition of proteolytic cleavage of amyloid precursor protein, the epidermal growth factor receptor, and the cell-adhesion molecules E-cadherin and CD44.25 However, this possibility was considered unlikely because the specific blocking of individual Notch receptors by RNA interference resulted in apoptosis. Down-regulation of Notch1 had a potent proapoptotic effect, but Notch2 or Notch3 did not. Thus, in cutaneous lymphoma, Notch1 is primarily responsible for cell survival, similar to what has been observed for normal T-cell development2-4 and in other hematologic malignancies.7,11-14
The molecular mechanisms through which Notch activation affects apoptotic programs are not understood, but a complex cross-talk with the phosphoinositide 3-kinase/Akt survival pathway has been suggested in different models, including T cells.27,41 Indeed, we were able to show that GSI I treatment resulted in dephosphorylation of Akt. However, this contribution of Akt inactivation to the apoptotic effect of GSI I seems to be small because the specific inhibition of Akt by Ly294002, rapamycin, and Akt inhibitor X resulted in a weak apoptotic response. Moreover, we did not see any additive effect between GSI I and Akt inhibitors on apoptosis in CTCL cell lines.
Previous reports have suggested that Notch can display both stimulatory and inhibitory control of NF-κB activity.32,33,42,43 Regulation occurs through several potential mechanisms, including direct interactions of Notch with NF-κB subunits, indirect effects on IκB phosphorylation, and transcriptional effects. It has been hypothesized that activated Notch in T cells may result in constitutive NF-κB activation, leading to T-cell leukemia/lymphoma.32,33 It is also known that NF-κB inhibition produces apoptosis in cutaneous lymphoma lines, including SeAx and MyLa.31,44 We observed that GSI I had a potent blocking effect on NF-κB inhibiting its translocation from the cytoplasm to the nucleus. Moreover, FoxO3a, which is an effector of both NF-κB and Akt inhibition, was shuffled to the nucleus. It is thus probable that the blockade of the NF-κB pathway significantly contributes to apoptosis after inhibition of Notch1.
Treatment of advanced cutaneous lymphoma is an area of high medical need. Besides radiotherapy, no treatments are available. Several chemotherapy regimens, including pegylated, liposomal doxorubicin, gemcitabine, fludarabine, and CHOP (cytoxan, hydroxyrubicin, oncovin, prednisone), produce transient responses, but the effect is short-lived and relapses are usually observed. Our data open the possibility of using inhibitors of Notch in the therapy of CTCL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ms Vibeke Pless, Ms Eva Hoffmann, Ms Ingelise Pedersen, and Ms Anne Jørgensen for excellent technical assistance as well as Gunhild L. Skovgaard for general support and critical comments.
This work was supported in part by research funding from the Danish Cancer Society, Aage Bang Foundation, Minister Erna Hamilton Foundation, Jens and Maren Thestrup Foundation, and Soeren and Helene Hempel Foundation.
Authorship
Contribution: M.R.K. and R.G. designed the research; M.R.K., E.B., and B.T.L. performed the research; M.R.K., L.M.R.G., E.R., and R.G. analyzed the data; M.R.K., L.M.R.G., and R.G. wrote the paper; and A.W. and N.Ø. advised in research design.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Maria Rørbæk Kamstrup, Department of Dermatology D92, Bispebjerg Hospital, Bispebjerg Bakke 23, Copenhagen DK-2400, Denmark; e-mail: mk43@bbh.regionh.dk.


![Figure 2. Inhibition of Notch by GSIs stimulates apoptosis and blocks cell proliferation in CTCL cell lines. (A) SeAx cells were treated with GSI I (0.05-5μM) or the vehicle (DMSO) and analyzed with Western blot for activated Notch1 to Notch4 and Hes, as in Figure 1. (B-C) Induction of proapoptotic caspases 3 and 7 after treatment of MyLa by different GSIs: I, IX, XX, and XXI. The cells were treated by the increasing concentrations of GSIs for 48 hours (B) or with 5μM GSI I and 20μM of GSI IX, XX, and XXI for 0 to 72 hours (C). Data are fold increase over the residual caspase 3/7 activity in the control, vehicle-treated cells. P < .05 in both graphs for GSI I treatment compared with vehicle treatment. P < .05 for all tested GSI XX concentrations and for GSI XXI 20μM. P < .05 for GSI IX and XXI after 24 hours, for GSI XXI after 48 hours, and GSI XXI after 72 hours in MyLa. Data are mean ± SD. (D) Sub-G1 population in MyLa, SeAx, and Hut78 cells, which were treated with 5μM GSI I (0, 3, 24, and 48 hours). DNA was stained with NiM-DAPi, and the cells were analyzed by flow cytometry. Bars represent mean values. The experiment was repeated 3 times with similar results. *P < .05. (E) MyLa was treated with GSI I (5μM; 0, 3, and 24 hours) and stained with annexin V and PI for flow cytometry, as described in “Cell viability, proliferation and apoptosis.” Cells were gated according to the annexin V (green FL1 channel, x-axis) and PI-specific (red, FL3 channel, y-axis) fluorescence. Values in the quadrants represent the percentages of cells. The experiment was repeated twice with similar results. (F) PARP cleavage in MyLa, SeAx, and Hut78 cells treated with 5μM GSI I for 24 hours. Whole-cell extracts were prepared for Western blot as in Figure 1, and the blots were probed with the antibody against PARP detecting its intact form (116 kDa) and the caspase-cleaved 89-kDa fragment. (G) Cell proliferation of MyLa, SeAx, and Hut78 cells treated with increasing concentrations of GSI I for 24 hours. Sixteen hours before harvest, 3H-thymidine (1 μCi [0.037 MBq]/well) was added, and the results are expressed as mean counts per minute of triplicate experiments ± SD. *P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/14/10.1182_blood-2009-12-260216/4/m_zh89991057210002.jpeg?Expires=1765895224&Signature=f5g3vELAEY8MvkVRGrzVR5eKGKLkd0Lx64totxjY~lrFMY-K2RFWjJp7n-va2cuyYGBvlTFmZ0KJUxitEFoiiwLdnh7jFq4bvWf-PElHV30L1rhOolSMZBvYO-RlFFRAg8L0xtPJDiJttwyXjWjh0T134NkvtpW7kRlDet8KTtZt-w2yk8lRUqV1VRCYlV00f9lloJicqIljg6FgFDX5erA7fSgEFA~3wKKkhx7OscYjLU0W6uke9nzHfEt28ajdf~o~LqZyNzWbKu1KWH~Pjisx9vzVsrZ99DykHnHMVQUj4FD4XbamHPG71Q~bYzx123Ko9GQgZ3UnhS3KzEz4UA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal