Abstract
The nucleoside analog 2′-C-cyano-2′-deoxy-1-β-D-arabino-pentofuranosyl-cytosine (CNDAC), currently in clinical trials for hematologic malignancies, has a novel action mechanism of causing a single-strand break after its incorporation into DNA. Double-strand breaks (DSBs) are generated thereafter in vivo and, if not repaired, pose lethal impact on cell survival. This study sought to define the mechanisms by which CNDAC-induced DSBs are formed and repaired. We demonstrated that single-strand breaks induced by CNDAC incorporation into DNA were converted to DSBs when cells progressed into the subsequent S-phase. CNDAC-induced DSBs were products of replication, rather than a consequence of apoptosis. ATM, the activator of homologous recombination (HR), was essential for cell survival after CNDAC treatment in cell lines and in primary acute myeloid leukemia samples, as were the HR components, Rad51, Xrcc3, and Brca2. Furthermore, formation of sister chromatid exchanges, a hallmark of HR, increased significantly after CNDAC-treated cells had progressed into a second replication cycle. In contrast, neither the replication stress sensor ATR nor DNA-PK, the initiator of nonhomologous end-joining of DSB, was involved in repair of CNDAC-induced damage. Together, these results indicate that HR, but not nonhomologous end-joining, is the major repair or survival mechanism for DNA damage caused by CNDAC.
Introduction
A prodrug of 2′-C-cyano-2′-deoxy-1-β-D-arabino-pentofuranosyl-cytosine (CNDAC), sapacitabine, is an orally bioavailable cytosine nucleoside analog that is currently in clinical trials in relapsed acute leukemias and myelodysplastic syndromes.1 TAS-109, a formulation of the parent nucleoside, is being studied in solid tumors.2 Investigations of CNDAC have demonstrated a novel action mechanism of causing a single-strand nick after its incorporation into DNA.3-6 Ligation of the 3′-hydroxyl of the analog by incorporation of subsequent deoxynucleotides initiates β-elimination. In DNA, this process cleaves the phoshophodiester linkage 3′ to the analog as the CNDAC nucleotide is rearranged to form 2′-C-cyano-2′,3′-didehydro-2′,3′-dideoxycytidine (CNddC). Because CNddC lacks a 3′-hydroxyl group, it is a de facto chain terminator at a single-strand DNA break that represents the signature of this novel DNA self-strand breaking process.4,7
Repair of single-strand break (SSB) and double-strand break (DSB) is critical for genetic integrity and cell survival. Much attention has been focused on the repair of 2-ended DSBs, which arise from cleavage of both strands of DNA caused by ionizing radiation, cross-linked alkylation, and radiomimetic drugs, such as bleomycin.8 However, a one-ended DSB can be created when a replication fork encounters a DNA SSB or after collapse of a stalled replication fork.9,10 Two major mechanisms repair DSB in mammalian cells: nonhomologous end-joining (NHEJ) and homologous recombination (HR). HR requires substantial sequence homologies to function in an error-free manner.11 As such, the activity of the HR pathway is limited to the S or G2 phase of the cell cycle when a sister chromatid is present. In contrast, NHEJ ligates the 2 free ends of DNA after exonucleolytically generating single-stranded regions that are attracted by a few bases of microhomology. NHEJ is active in all phases of the cell cycle, although it is an intrinsically error-prone process.12
Central to the DNA damage and repair responses are sensors, particularly the phosphatidylinositol 3-kinase-related protein kinase family, including DNA-dependent protein kinase (DNA-PK), ataxia telangiectasia mutated (ATM) and ATM- and Rad3-related protein (ATR). Composed of a large catalytic subunit of 460 kDa, DNA-PKcs, and a dimeric DNA binding subunit, Ku70 and Ku80, DNA-PK plays an important role in mammalian NHEJ.13 Deficiency in either DNA-PKcs or Ku80 result in DNA DSB repair defects manifested as X-ray sensitivity and impaired V(D)J recombination.14,15 The ATM kinase is activated by autophosphorylation16 and is responsible for the phosphorylation of a wide range of downstream substrates in response to genotoxic stress, such as ionizing radiation, which generates DSBs.17,18 ATM signals for repair of DSBs, to activate and cause the association of HR repair components, such as Rad51, Xrcc3, Brca2, and H2AX.18 ATR initiates checkpoint responses to various agents that stall replication forks and damage DNA in multiple ways.19
Repair mechanisms of CNDAC-induced DNA damage are largely unknown. Lack of components of the transcription-coupled nucleotide excision repair (TC-NER) pathway sensitized clonogenic survival of CNDAC-treated cells by 3- to 5-fold.20 Presumably, this was directed at the chain-terminating analog at the nick. However, unrepaired SSBs may be converted to one-ended DSBs on subsequent replication, a possibility consistent with γ-H2AX formation and chromosomal breaks and gaps that are evident after CNDAC treatment.21 The objective of the present study was to determine how CNDAC-induced SSBs are converted to DSBs and whether DSB repair mechanisms contribute to viability. The results demonstrated that SSBs were converted to DSBs after replication and that HR repair has a major impact on survival in cell lines and in primary acute myeloid leukemia (AML) blasts. The substantial loss of clonogenicity in cells lacking HR function is probably the result of a synthetically lethal convergence of such genetic lesions and the drug mechanism.
Methods
Patient samples
Informed consent was obtained in accordance with the Declaration of Helsinki under the aegis of the University of Texas M. D. Anderson Cancer Center Institutional Review Board-approved protocols. Patient characteristics are shown in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). AML blasts were resuspended in RPMI 1640 supplemented with 10% fetal bovine serum for in vitro studies. Analysis of CNDAC metabolites and clonogenic assay were conducted as described in the next paragraph.
Quantitation of CNDAC and its metabolites in cells and DNA
To determine intracellular accumulation of CNDAC triphosphate (CNDAC-TP), AML primary samples were incubated with [3H]CNDAC (15 Ci/mmol, Sankyo Co, Ltd; final concentrations, 0.5-5μM) for the indicated times. To measure cellular elimination of CNDAC-TP, AML samples were incubated with 10μM CNDAC (containing [3H]CNDAC) for 4 hours, washed, and resuspended in drug-free media for indicated times. Cellular nucleotides were extracted with perchloric acid (PCA), and extracts were neutralized with potassium hydroxide (KOH) and stored at −20°C until analysis by high performance liquid chromatography (HPLC).3
To quantitate the incorporation of CNDAC into DNA, ML-1 cells were incubated with 0.3μM [3H]CNDAC for 18 hours, cells were processed, and nucleosides were separated by HPLC.7 The equation 1 pmol = 6.02 × 1011 molecules was used to calculate the number of molecules of total CNddC, CNDAC, and CNDC per cell. The percentage of the radioactivity in CNddC peak to total radioactivity in all peaks was calculated for each sample and used to calculate the number of molecules of CNddC per cell.
γ-H2AX staining and flow cytometric analysis
Cells were harvested at indicated times after drug treatment and prepared for flow cytometric analysis of H2AX phosphorylation.21 Fluorescence of at least 20 000 cells was determined on a BD Biosciences FACSCalibur flow cytometer.
Double-labeling with IdU and CldU and flow cytometric analysis
Cells were pulse-labeled with 5μM 5-iododeoxyuridine (IdUrd) and CNDAC for 10 minutes and supplemented with 0.2μM 5-chlorodeoxyuridine (CldUrd) 12 hours after wash. Cells were harvested after drug treatment, washed twice with ice-cold phosphate-buffered saline (PBS), fixed in 70% ethanol overnight at 4°C, washed in PBS, and resuspended in 0.04% pepsin for 12 minutes followed by denaturing in 2 N HCl 12 minutes at 37°C. The acid was neutralized with Na2B4O7.10 H2O, pH 8.5, and cells were washed once with 0.5% Tween 20/1% bovine serum albumin (BAS)/PBS, and rat–anti-CldU (1:40 dilution) was added for 1 hour at room temperature. After centrifugation, the pellet was suspended in goat anti–rat IgG, F(ab′)2-allophycocyanin-conjugated secondary antibody (SC-3832, Santa Cruz Biotechnology, 1:100 dilution) for 1 hour in the dark. After washing twice with 0.5% Tween 20/1% BSA/PBS, the pellet was stained with mouse–anti-IdUrd followed by fluorescein isothiocyanate (FITC)–conjugated secondary antibody. The pellet was washed twice and counterstained with propidium iodide (Sigma-Aldrich) containing RNase A (Roche Diagnostics) for 30 minutes at 4°C. Fluorescence of at least 20 000 cells was determined by flow cytometry.
Neutral comet assay
DNA DSBs were determined using single cell gel electrophoresis (comet) under neutral conditions (R&D Systems). Individual nuclei or comets were viewed using a Zeiss epifluorescence microscope attached to an imaging system (Kinetic Imaging Komet system Version 4.0); 100 cells per treatment condition were counted. Double-strand damage was quantified as an increase in Olive tail moment, the product of the amount of DNA (fluorescence) in the tail and the distance between the means of the head and tail fluorescence distributions.22
PFGE
To detect high molecular weight DNA fragmentation, 1 × 106 ML-1 cells per sample were washed with PBS and resuspended in PBS, mixed with of 1.2% agarose, and processed.23 Comparison of DNA concentrations among different samples was carried out by ethidium bromide staining. The intensity of each band was quantitated, and the percentage of DSB fragments versus total input in each lane was expressed.
Measurements of apoptosis
At various times after treatment, 1 × 106 ML-1 cells were taken for apoptosis analysis with FITC-conjugated annexin V antibody and propidium iodide, and analyzed for apoptotic cells.24
Immunoblotting of subcellular fractions
Subcellular fractionation was performed,25 fractions (S1, cytoplasmic; S2, nuclear soluble; P2, chromatin-bound) were loaded on 4% to12% Bis-Tris gradient gels (Bio-Rad) or 10% sodium dodecyl sulfate–polyacrylamide gels and transferred onto nitrocellulose membranes.21 Immunoblots were quantitated by Li-Cor Odyssey Version 3.0 software.
Depletion of ATM by small interfering RNA
ATM ON-TARGETplus SMARTpool (siATM) and 2 negative control siRNAs, siGENOME RISC-Free Control siRNA (siC1) and ON-TARGETplus Nontargeting Pool (siC2), were purchased from Dharmacon RNA Technologies. siRNA transfections were done with Opti-MEM I (Invitrogen) containing DharmaFECT II and 50nM final concentrations of oligos, after experimental optimization of the manufacturer's instructions. Cells were replated into 6-well plates 24 hours after transfection and incubated for additional 24 hours before introduction of CNDAC. Cells were washed 24 hours after drug treatment and incubated with complete medium to allow colony formation.
Clonogenic cell survival assay
Suspension culture cells were exposed to drugs for indicated times, washed with PBS (37°C), and resuspended in fresh medium. Clonogenicity was determined as described after 9 to 10 days.23 AML primary samples freshly purified or revived from liquid nitrogen were plated in methylcellulose medium in triplicates, exposed to drugs, and incubated at 37°C for 5 to 6 days.26 Colonies of more than or equal to 50 cells were counted under a dissecting microscope; results were expressed as a percentage of control colony formation.
Attached cells were seeded in 60-mm dishes or 6-well plates 1 day before cells were exposed to a range of concentrations of drugs for 24 hours. After washing into drug-free medium, cells were incubated for 6 to 10 days, and colonies in triplicate plates or wells were counted under a dissecting microscope (≥ 50 cells) or scored electronically (GelCount; Oxford Optronix).
Measurement of SCE
Exponentially growing AA8 cells were continuously incubated with 10μM bromodeoxyuridine (BrdU) for 2 population doubling times (∼ 30 hours), with 100 ng/mL colcemid being added for the final 1.5 hours. CNDAC (1μM) was added at 0 hour or after 15 hours. Mitomycin C, a positive control, was introduced after 15 hours. Cultures were harvested with Accutase, and metaphase spreads were prepared. Microscope slides were stained with Hoechst 33258 (Sigma-Aldrich), exposed to 366-nm ultraviolet light, and extracted in heated 2× saline sodium citrate buffer. Slides were then dried and stained with Giemsa. Images of at least 20 metaphase spreads were captured under bright-field microscopy, and sister chromatid exchange (SCE) was scored where a color switch occurred.
Statistical analysis
Fifty percent inhibitory concentration (IC50) values were calculated using the GraphPad Prism5 Version 5 software (GraphPad Software) based on at least 2 independent experiments in triplicate. Intergroup comparisons of clonogenic survival in response to drugs were made using a 2-tailed, paired t test.
Materials
Cell lines, chemicals and antibodies are described in supplemental Methods.
Results
Generation rate of SSBs and the cytotoxicity of CNDAC
As the clinical formulation of CNDAC, sapacitabine has activity in AML, we used human myelogenous leukemia ML-1 cells to model cellular responses to this drug. To elaborate the time course of SSB formation, ML-1 cells were incubated with 0.3μM [3H]CNDAC, washed into fresh medium, and samples were extracted and hydrolyzed into nucleosides. HPLC7 separated 3 radioactive peaks,7 which were identified as CNDAC, CNddC, and CNDC (an epimer of CNDAC generated under mild alkaline conditions in the procedure27 ), respectively, based on retention times that matched authentic standards (Figure 1A). Results demonstrated that CNddC constituted approximately 56% of the total analogs in DNA, after an 18-hour treatment, which increased to 65% over 48 hours after CNDAC washout (data not shown). The number of molecules in DNA of total CNddC, CNDAC, and CNDC per cell after 18-hour treatment was 1.38 × 105 molecules/cell. As the presence of CNddC indicates the formation of an SSB by β-elimination, the number of SSBs per cell was calculated to be 70 400 after 4 hours, which increased to 77 400 at 18 hours and 89 100 after 48 hours (Figure 1B). Although no indications of cell death were observed at the time of these determinations, it is clear from clonogenic assays that the repopulation capacity was greatly diminished by this treatment (Figure 1C left), indicating that subsequent events were critical to viability. Using clinically relevant CNDAC levels,1 clonogenic assays on AML primary cells also revealed a concentration-dependent cell killing effect (Figure 1C right). Although there was heterogeneity in CNDAC sensitivity among patient samples, no relationship with disease status was evident. AML primary cells exposed to CNDAC in vitro were capable of accumulating CNDAC-TP that was both concentration- and time-dependent (Figure 1D left and middle). Intracellular CNDAC-TP was eliminated rapidly, with a mean half-life of 2.8 hours (Figure 1D right).
Identification of CNDAC and its metabolites in cells and DNA. (A) ML-1 cells were incubated with 0.3μM [3H]CNDAC for 18 hours and washed into fresh RPMI medium, and DNA was extracted and enzymatically hydrolyzed. Components of DNA hydrolysates were separated by reversed-phase HPLC, and radioactivity was quantitated. (Top panel) An authentic standard of CNDAC, CNddC, and CNDC; the ordinate unit represents ultraviolet absorption at 270 nm (AU). (Bottom panel) An HPLC analysis of DNA hydrolysis products degraded with DNase I, phosphodiesterase, and alkaline phosphatase; the ordinate represents radioactivity (CPM). (B) Number of CNddC molecules per cell, calculated as described in “Quantitation of CNDAC and its metabolites in cells and DNA.” (C) Cytotoxicity of CNDAC in ML-1 cells (left) and purified primary samples from AML patients (right) determined by clonogenic assay. Exponentially growing ML-1 cells were exposed to a range of concentrations of CNDAC for 18 hours, washed into fresh medium, and allowed to form colonies. The survival curve is mean ± SD values from triplicates. AML primary samples (patients 11-17) were incubated with CNDAC at indicated concentrations for 5 to 6 days without washing out of drug. The curves are mean values from duplicates or triplicates. (D) HPLC analysis of intracellular accumulation of CNDAC-TP in AML primary samples exposed to CNDAC at indicated concentrations for 2 hours (left, patients 1, 2, 4, 6, 7, 8, 9, and 10) or 5μM CNDAC for indicated times (middle, patients 3-10), and elimination of CNDAC-TP from AML primary cells over 10 hours after a 4-hour incubation with 10μM CNDAC (right, patients 6, 7, and 10).
Identification of CNDAC and its metabolites in cells and DNA. (A) ML-1 cells were incubated with 0.3μM [3H]CNDAC for 18 hours and washed into fresh RPMI medium, and DNA was extracted and enzymatically hydrolyzed. Components of DNA hydrolysates were separated by reversed-phase HPLC, and radioactivity was quantitated. (Top panel) An authentic standard of CNDAC, CNddC, and CNDC; the ordinate unit represents ultraviolet absorption at 270 nm (AU). (Bottom panel) An HPLC analysis of DNA hydrolysis products degraded with DNase I, phosphodiesterase, and alkaline phosphatase; the ordinate represents radioactivity (CPM). (B) Number of CNddC molecules per cell, calculated as described in “Quantitation of CNDAC and its metabolites in cells and DNA.” (C) Cytotoxicity of CNDAC in ML-1 cells (left) and purified primary samples from AML patients (right) determined by clonogenic assay. Exponentially growing ML-1 cells were exposed to a range of concentrations of CNDAC for 18 hours, washed into fresh medium, and allowed to form colonies. The survival curve is mean ± SD values from triplicates. AML primary samples (patients 11-17) were incubated with CNDAC at indicated concentrations for 5 to 6 days without washing out of drug. The curves are mean values from duplicates or triplicates. (D) HPLC analysis of intracellular accumulation of CNDAC-TP in AML primary samples exposed to CNDAC at indicated concentrations for 2 hours (left, patients 1, 2, 4, 6, 7, 8, 9, and 10) or 5μM CNDAC for indicated times (middle, patients 3-10), and elimination of CNDAC-TP from AML primary cells over 10 hours after a 4-hour incubation with 10μM CNDAC (right, patients 6, 7, and 10).
SSBs induced by CNDAC incorporation are converted to DSBs at the subsequent S-phase
The single-strand nick caused by CNDAC is recognized and, in part, repaired by the TC-NER pathway.20 However, the overall removal rate of the analog from DNA is a relatively slow process (Figure 1B). Single-strand nicks are potential substrates for DSB formation by either the action of endonucleases or by torsional effects, particularly during mitotic processes.21 In addition, a second round of replication across a nick would potentially generate a one-ended DSB if this lesion is not bypassed.8 To test this hypothesis, ML-1 cells were incubated with aphidicolin to enrich the S-phase population, followed by a 4-hour pulse with CNDAC, and chase into fresh media. Samples were taken at different times and subjected to either flow cytometry for detecting H2AX phosphorylation (γ-H2AX), a DSB marker,28,29 or to neutral comet assay to evaluate double-strand DNA fragmentation. After release of the aphidicolin block, cells progressed through the cell cycle and entered the subsequent S-phase 20 hours later (Figure 2A). The pulse treatments with 0.8μM and 2.0μM CNDAC appeared to delay this progression by several hours. Flow cytometric analysis of γ-H2AX, concurrent with analysis of cellular DNA content, showed that a robust increase in H2AX phosphorylation was detectable simultaneously with the onset of the second S-phase in drug-treated cells (Figure 2B). In contrast, no change in γ-H2AX was evident in controls. The generation of DNA DSBs was also assessed by the single-cell gel electrophoresis assay under neutral conditions (the comet assay). The increase of Olive tail moment values was also detected approximately 24 hours after cells were washed free of CNDAC (Figure 2C). The increase γ-H2AX and tail moment values immediately after CNDAC washout may be attributed to transient delay of S-phase progression21 and inhibited DNA synthesis.3 After the time of entrance into the second S-phase, the growth rate of CNDAC-treated cells became clearly less than that of control cells (Figure 2D), concurrent with the appearance of DNA damage.
SSBs induced after CNDAC incorporation during the first S-phase were converted into DSBs in the second S-phase. ML-1 cells were synchronized in S phase using 1μM aphidicolin for 24 hours, followed by a 4-hour pulse with CNDAC and chase in to fresh media. Samples were taken at indicated times and subjected to flow cytometric analysis, comet assay, or electronic counting. (A-B) Samples were fixed by 4% paraformaldehyde solution and stained with FITC-conjugated γ-H2AX antibody and PI. Flow cytometry showed both cell-cycle progression (A) and γ-H2AX–positive profile (B) that was CNDAC dose-dependent (●, 0μM; ▾, 0.8μM; ♦, 2μM). (C) Generation of DNA DSBs was assessed by single-cell gel electrophoresis (comet assay) under neutral conditions. The level of DSBs was quantitated by determination of Olive tail moment before and after washing out of CNDAC (●, 0μM; ▾, 0.8μM; ♦, 2μM). (D) Absolute cell number was scored by an electronic particle counter at indicated time points, and cell growth curve was plotted for each drug concentration (●, 0μM; ▾, 0.8μM; ♦, 2μM). Dashed lines indicate time of drug washing out. Data are 1 representative of 2 independent experiments.
SSBs induced after CNDAC incorporation during the first S-phase were converted into DSBs in the second S-phase. ML-1 cells were synchronized in S phase using 1μM aphidicolin for 24 hours, followed by a 4-hour pulse with CNDAC and chase in to fresh media. Samples were taken at indicated times and subjected to flow cytometric analysis, comet assay, or electronic counting. (A-B) Samples were fixed by 4% paraformaldehyde solution and stained with FITC-conjugated γ-H2AX antibody and PI. Flow cytometry showed both cell-cycle progression (A) and γ-H2AX–positive profile (B) that was CNDAC dose-dependent (●, 0μM; ▾, 0.8μM; ♦, 2μM). (C) Generation of DNA DSBs was assessed by single-cell gel electrophoresis (comet assay) under neutral conditions. The level of DSBs was quantitated by determination of Olive tail moment before and after washing out of CNDAC (●, 0μM; ▾, 0.8μM; ♦, 2μM). (D) Absolute cell number was scored by an electronic particle counter at indicated time points, and cell growth curve was plotted for each drug concentration (●, 0μM; ▾, 0.8μM; ♦, 2μM). Dashed lines indicate time of drug washing out. Data are 1 representative of 2 independent experiments.
A second approach to evaluating the passage of cells with CNDAC-induced SSB into the subsequent S-phase was to label the S-phase fraction of exponentially growing cells simultaneously with a pulse of 5μM IdU and CNDAC (0.8μM or 2.0μM). After 10 minutes, cells were washed into fresh medium and CldU was added 12 hours later, when all cells labeled with IdU (and exposed to CNDAC) had passed out of S-phase. Using this timed sequence, only cells that progressed into the second S-phase would be positive for both IdU and CldU. Flow cytometric analysis demonstrated that double-labeled cells appeared after 18 hours (Figure 3A), indicating that cells labeled with both IdU and CNDAC had entered the second round of DNA replication. Consistent with this, cell growth was not inhibited 24 hours after 10-minute treatment by either concentration of CNDAC (Figure 3B). Pulsed-field gel electrophoresis indicated DSBs in control cells at 18 to 24 hours that were labeled with IdU and CldU in the absence of CNDAC (Figure 3C), probably because the incorporation of halogenated analogs contributed to strand breaks.30 This serves as a positive control for the generation of DSBs in cells that replicate across an SSB. Cells pulsed with 2μM CNDAC exhibited substantially greater DSB formation between 18 and 24 hours than did controls (Figure 3C), consistent with the postulate that attempted replication in cells that have CNDAC-induced strand breaks results in DSBs in DNA. Pulsing cells with a lesser CNDAC concentration had no significant effect.
DSBs induced by CNDAC are products of replication, rather than a consequence of induction of apoptosis. (A-C) ML-1 cells were pulse-labeled with various concentrations of CNDAC in the presence of IdU for 10 minutes and chased in to fresh media; CldU was added 12 hours after washing. Samples were taken at different times and subjected to flow cytometry and PFGE. (A) Samples were fixed and stained with FITC-IdU antibody and allophycocyanin-CldU antibody. Flow cytometric analysis determined the time at which double-labeled cells appeared, indicating cells in the second S-phase. (B) Absolute cell number was quantitated by electronic particle counter at different times. (C) The generation of DNA DSBs was assessed by PFGE. The intensity of each band was quantitated, and the percentage of DSB fragment versus total input in each lane was expressed in the graph. Note that 0.8μM CNDAC did not cause DSBs beyond the background level. (D-F) ML1 cells were pulse-labeled with 2μM CNDAC in the presence of IdU for 10 minutes and chased into fresh media, CldU was supplemented 12 hours after wash with or without aphidicolin. Samples were taken at different time points and subjected to flow cytometry and PFGE. (D) Absolute cell number was counted by Coulter counter at different time points, and cell growth was inhibited in the presence of aphidicolin. (E) The generation of DNA DSBs was assessed by PFGE. The intensity of each band was quantitated, and the percentage of DSB fragment versus total input in each lane was expressed in the graph. Note that aphidicolin prevented the induction of DSBs at 18 to 24 hours after treatment. Data are 1 representative of 2 independent experiments. (F) Time of onset of apoptosis in ML-1 cells after 2μM of CNDAC treatment for 10 minutes. Apoptosis was detectable after 31 hours after treatment, as assessed by annexin V staining.
DSBs induced by CNDAC are products of replication, rather than a consequence of induction of apoptosis. (A-C) ML-1 cells were pulse-labeled with various concentrations of CNDAC in the presence of IdU for 10 minutes and chased in to fresh media; CldU was added 12 hours after washing. Samples were taken at different times and subjected to flow cytometry and PFGE. (A) Samples were fixed and stained with FITC-IdU antibody and allophycocyanin-CldU antibody. Flow cytometric analysis determined the time at which double-labeled cells appeared, indicating cells in the second S-phase. (B) Absolute cell number was quantitated by electronic particle counter at different times. (C) The generation of DNA DSBs was assessed by PFGE. The intensity of each band was quantitated, and the percentage of DSB fragment versus total input in each lane was expressed in the graph. Note that 0.8μM CNDAC did not cause DSBs beyond the background level. (D-F) ML1 cells were pulse-labeled with 2μM CNDAC in the presence of IdU for 10 minutes and chased into fresh media, CldU was supplemented 12 hours after wash with or without aphidicolin. Samples were taken at different time points and subjected to flow cytometry and PFGE. (D) Absolute cell number was counted by Coulter counter at different time points, and cell growth was inhibited in the presence of aphidicolin. (E) The generation of DNA DSBs was assessed by PFGE. The intensity of each band was quantitated, and the percentage of DSB fragment versus total input in each lane was expressed in the graph. Note that aphidicolin prevented the induction of DSBs at 18 to 24 hours after treatment. Data are 1 representative of 2 independent experiments. (F) Time of onset of apoptosis in ML-1 cells after 2μM of CNDAC treatment for 10 minutes. Apoptosis was detectable after 31 hours after treatment, as assessed by annexin V staining.
To evaluate the possibility that the DSBs were caused by apoptosis, exponentially growing ML-1 cells were pulse-labeled with CNDAC in the presence of IdU for 10 minutes and chased into fresh media, CldU was supplemented 12 hours after washing, and aphidicolin was added to a portion of the culture to block progression into the second S-phase. Although cell growth was blocked by aphidicolin (Figure 3D), this inhibition of DNA replication prevented the generation of DNA DSBs at 18 to 24 hours after treatment compared with those cells in the absence of aphidicolin (Figure 3E). Evidence of apoptosis was not detected by annexin V assay until 31 hours (Figure 3F). Thus, the DSBs seen by pulsed-field gel electrophoresis (PFGE) 18 to 24 hours after treatment with 2μM CNDAC (Figure 3C,E) were products of replication, not the result of the induction of apoptosis.
DNA-PKcs and ATR are dispensable for cell survival after CNDAC
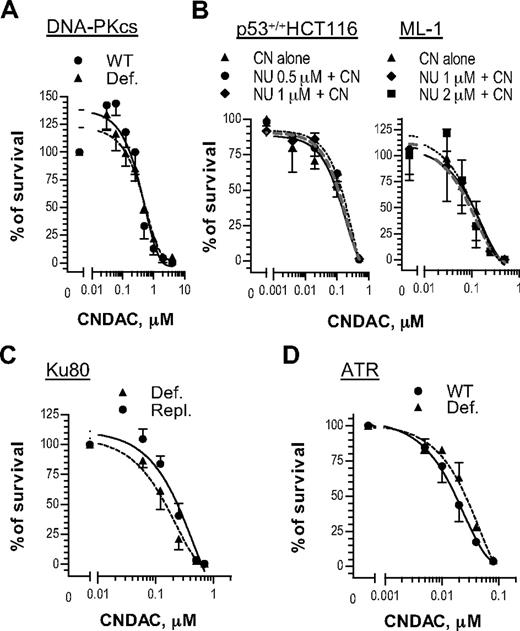
Phosphatidylinositol 3-kinase-related protein kinases sense DNA damage and signal for responses, including repair mechanisms that may be associated with drug resistance. For instance, it is possible that both NHEJ and HR may repair DSBs induced by CNDAC. To investigate this possibility, we evaluated the cellular response to CNDAC of cells deficient in proteins of the NHEJ pathway. The human cell line M059-J, isolated from a malignant glioma, lacks DNA-PKcs activity because of defective mRNA turnover associated with a frameshift mutation in the PRKDC gene.31 M059-J cells exhibited similar sensitivity to CNDAC compared with M059-K cells, which express the wild-type enzyme (Figure 4A). Consistent with this result, a specific DNA-PKcs inhibitor NU7441 did not increase sensitivity to CNDAC in the p53 wild-type lines, HCT116 and ML-1 (Figure 4B). Furthermore, the rodent xrs6 cell line, which lacks Ku80, appears to be related to the DNA-PK defect. Repletion of a CHO Ku80 gene in this line (xrs6-hamKu80) rescues its properties of NHEJ.32 When exposed to CNDAC, both xrs6 and xrs6-hamKu80 cells showed similar sensitivity to CNDAC (Figure 4A). Taken together, these data indicate that the NHEJ pathway is not the major pathway for repairing of CNDAC-induced DSBs.
DNA-PK and ATR are dispensable for cell survival after DNA damage by CNDAC. (A) Glioma-derived cell lines M059-K (●, wild-type) and M059-J (▴, DNA-PKcs-deficient) were incubated with a range of concentrations of CNDAC for 24 hours, and colonies were stained and quantitated after 10 days. (B) HCT116 cells and ML-1 cells, both p53 wild-type, were preincubated with NU7441 at indicated concentrations for 1 hour, followed by various concentrations of CNDAC for additional 24 hours. After washing, HCT116 cells were incubated for 7 to 8 days, and colonies were scored by GelCount; ML-1 cells were counted and distributed into methylcellulose to form colonies in 9 days. (C) CHO lines xrs6 (▴, Ku80-deficient) and xrs6-hamKu80 (●, Ku80-repleted) were incubated with a range of concentrations of CNDAC for 24 hours, and colonies were stained and counted after 6 days. (D) Human fibroblast lines 1BRhTERT (●, wild-type control) and F02-98hTERT (▴, ATR-Seckel) were exposed to different concentrations of CNDAC for 24 hours, and colonies were scored in 10 days. Representative data of 3 independent experiments are shown. All points are mean ± SD of triplicate samples.
DNA-PK and ATR are dispensable for cell survival after DNA damage by CNDAC. (A) Glioma-derived cell lines M059-K (●, wild-type) and M059-J (▴, DNA-PKcs-deficient) were incubated with a range of concentrations of CNDAC for 24 hours, and colonies were stained and quantitated after 10 days. (B) HCT116 cells and ML-1 cells, both p53 wild-type, were preincubated with NU7441 at indicated concentrations for 1 hour, followed by various concentrations of CNDAC for additional 24 hours. After washing, HCT116 cells were incubated for 7 to 8 days, and colonies were scored by GelCount; ML-1 cells were counted and distributed into methylcellulose to form colonies in 9 days. (C) CHO lines xrs6 (▴, Ku80-deficient) and xrs6-hamKu80 (●, Ku80-repleted) were incubated with a range of concentrations of CNDAC for 24 hours, and colonies were stained and counted after 6 days. (D) Human fibroblast lines 1BRhTERT (●, wild-type control) and F02-98hTERT (▴, ATR-Seckel) were exposed to different concentrations of CNDAC for 24 hours, and colonies were scored in 10 days. Representative data of 3 independent experiments are shown. All points are mean ± SD of triplicate samples.
ATR is thought to function mainly in response to replicative stress, although evidence exists for cooperation with ATM.33,34 The human fibroblast line F02-98hTERT, derived from a patient with Seckel syndrome and lacking ATR, was similarly sensitive to CNDAC as 1BRhTERT, an ATR-proficient line derived from a healthy person (Figure 4C). Thus, neither ATR nor DNA-PKcs appears to be required for cell survival after CNDAC exposure.
Homologous recombination is a major survival mechanism in response to CNDAC-induced DNA damage
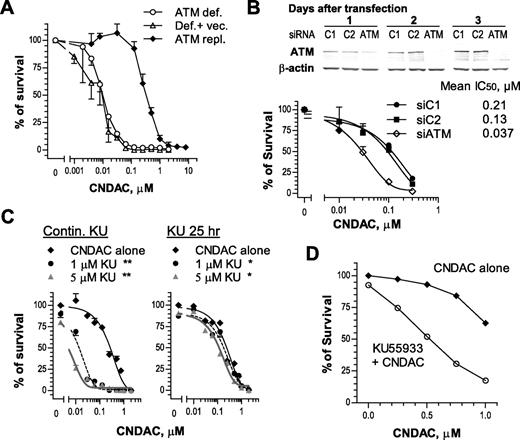
There is extensive evidence that ATM is a sensor of DSBs that is associated with the homologous recombination pathway.17,18 Human fibroblasts deficient in ATM or transfected with an empty vector were approximately 30-fold more sensitive to CNDAC (IC50 ≅ 0.01μM) than cells repleted with full-length ATM cDNA (IC50 ≅ 0.3μM; Figure 5A). Consistent with this, HCT116 cells with ATM knocked down by siRNA (Figure 5B top) were sensitized 3- to 5-fold in clonogenic assays after transfection (Figure 5B bottom). It is noteworthy that depletion of ATM by siRNA was transient and the ATM level was recovered to that of controls by the time colonies were fixed for quantitation (data not shown). Incubation with a specific ATM inhibitor, KU55933, during exposure to CNDAC and throughout colony formation sensitized OCI-AML3 cell clonogenicity by 20-fold (Figure 5C left). In contrast, removal of both compounds after exposure resulted in diminished potentiation of CNDAC-induced killing (Figure 5C right), indicating the need for sustained inhibition of ATM during colony formation to achieve maximum loss of viability. The strategy of small molecule inhibitor was applied to AML primary cells to further evaluate the dependence of ATM after CNDAC. The clonogenicity of AML blasts coincubated with a marginally cytotoxic concentration of KU55933 and CNDAC was significantly less (P = .015) compared with those exposed to CNDAC alone (Figure 5D), consistent with results from cell lines. In contrast, treatment of primary cells with NU7441 did not significantly alter CNDAC toxicity in 2 of 3 samples (data not shown). In addition, there was a significant increase in the phosphorylation of ATM on Ser 1981 and of the ATM downstream substrate, SMC1 (structural maintenance of chromosomes protein-1) on Ser 966 in the T-ALL line CCRF-CEM when exposed to CNDAC, but not in the phosphorylation of either DNA-PKcs on Ser 2056 or ATR on Ser 428 (data not shown). Therefore, ATM appears to be integral for repair of DNA damage caused by CNDAC.
ATM is essential for cell survival in response to CNDAC. (A) AT-C (○, ATM-deficient), AT-V (Δ, AT-C transfected with vector), and AT-AT (♦, AT-C repleted with full-length ATM cDNA) cells were exposed to a range of concentrations of CNDAC for 24 hours and incubated for 10 to 11 days after washing out of drugs to allow colonies to grow. (B) p53+/+ HCT116 cells were transfected with siRNAs described in “Depletion of ATM by small interfering RNA,” and ATM protein levels were monitored by immunoblotting for 3 days. Cells were replated for clonogenic assay 1 day after transfection. Cells were exposed to CNDAC for 24 hours and incubated for 5 to 7 days after washing. (A-B) Representative data of 3 independent experiments. Points are mean ± SD of triplicate plates. (C) OCI-AML3 cells were treated with KU55933 (1μM or 5μM) 1 hour before addition of CNDAC at a variety of concentrations. Cells were incubated for an additional 24 hours before being washed into drug-free medium. Subsequently, cells were counted and diluted into methylcellulose with KU55933 (left) or drug-free medium (right) to form colonies in 8 to 9 days. The survival curves in each panel are mean ± SD values from 1 of 3 independent experiments. *P < .05; **P < .01 versus CNDAC alone. (D) AML primary cells from archived samples were incubated with KU55933 (2.5μM) 1 hour before addition of CNDAC at the indicated concentrations. Drugs were continuously present in methylcellulose medium during formation of colonies in 5 to 6 days. The survival curves are mean values of 4 patient samples (13-16), each with triplicates.
ATM is essential for cell survival in response to CNDAC. (A) AT-C (○, ATM-deficient), AT-V (Δ, AT-C transfected with vector), and AT-AT (♦, AT-C repleted with full-length ATM cDNA) cells were exposed to a range of concentrations of CNDAC for 24 hours and incubated for 10 to 11 days after washing out of drugs to allow colonies to grow. (B) p53+/+ HCT116 cells were transfected with siRNAs described in “Depletion of ATM by small interfering RNA,” and ATM protein levels were monitored by immunoblotting for 3 days. Cells were replated for clonogenic assay 1 day after transfection. Cells were exposed to CNDAC for 24 hours and incubated for 5 to 7 days after washing. (A-B) Representative data of 3 independent experiments. Points are mean ± SD of triplicate plates. (C) OCI-AML3 cells were treated with KU55933 (1μM or 5μM) 1 hour before addition of CNDAC at a variety of concentrations. Cells were incubated for an additional 24 hours before being washed into drug-free medium. Subsequently, cells were counted and diluted into methylcellulose with KU55933 (left) or drug-free medium (right) to form colonies in 8 to 9 days. The survival curves in each panel are mean ± SD values from 1 of 3 independent experiments. *P < .05; **P < .01 versus CNDAC alone. (D) AML primary cells from archived samples were incubated with KU55933 (2.5μM) 1 hour before addition of CNDAC at the indicated concentrations. Drugs were continuously present in methylcellulose medium during formation of colonies in 5 to 6 days. The survival curves are mean values of 4 patient samples (13-16), each with triplicates.
The dependency on ATM for survival strongly suggests that the HR pathway may play an important role in repair of CNDAC-induced damage, particularly DSBs. Because mutations in HR genes, such as BRCA1, BRCA2, RAD51, RAD54, XRCC2, or XRCC3, predispose cells to high levels of genetic instability and sensitivity to IR and cross-linking agents,35 we evaluated some components in the HR pathway for response to CNDAC. Increased chromatin association of Rad51 was observed in HeLa and HCT116 cells exposed to CNDAC for 1 day and 3 days, respectively (Figure 6A), consistent with the activity of HR in the CNDAC response process. The requirement for the other components of the HR machinery was further investigated. Testing genetically paired Chinese hamster cell lines proficient and deficient in 2 Rad51-associated proteins, Xrcc336 and Brca237 by clonogenic assays, cells lacking Xrcc3 and complemented with hXRCC3 exhibited approximately 100-fold and 4-fold sensitization, respectively, to CNDAC compared with the parental cells (Figure 6B,D), whereas cells lacking Brca2 were approximately 20-fold more sensitive to CNDAC, relative to the wild-type cells (Figure 6C-D). The lack of Rad51D conferred 60-fold sensitivity to CNDAC relative to cells complemented with Rad51D (Figure 6D). In addition, sensitivity of these lines to mitomycin C was studied for comparison with CNDAC and for validation purposes (Figure 6D). Together, these data demonstrate that Rad51 and 2 of its interacting proteins, Xrcc3 and Brca2, participate in HR repair of damage caused by CNDAC and that lack of any of these components sensitizes cell viability.
CNDAC-induced DNA damage is repaired through the homologous recombination pathway. (A-C) Rad51 and its 2 interacting proteins, Xrcc3 and Brca2, are involved in DNA damage response to CNDAC. (A) Exponentially growing HeLa (top) and HCT116 cells (bottom) were incubated with 4μM and 3μM CNDAC for 24 hours and 72 hours, respectively. Cells were harvested at indicated times and subjected to subcellular fractionation. Fractions: cytoplasmic (S1), nuclear soluble (S2), and chromatin binding (P2). Rad51, Orc2, and RhoA or β-tubulin in the indicated fractions were detected by immunoblotting. (B) AA8 (■, wild-type), irs1SF (ο, Xrcc3-deficient), and 1SFwt8 (▴, hXRCC3-complemented) cells were washed into drug-free medium after a 24-hour exposure to CNDAC at a range of concentrations and allowed to form colonies in 6 to 7 days. (C) V79 (■, wild-type) and V-C8 (ο, Brca2-deficient) cells were treated as in panel B. All points are mean ± SD of triplicate plates. (D) IC50 values of CNDAC and mitomycin C from clonogenic assays using paired hamster lines. (E) CNDAC induces higher levels of SCEs after the second S phase. Continuously exposed to bromodeoxyuridine, AA8 cells were treated with 1μM CNDAC for either 15 hours (1 cell cycle) or for 30 hours (2 cycles). Colcemid was added in the final 1.5 hours. Quantitation of SCEs was performed in a minimum of 20 metaphases per sample.
CNDAC-induced DNA damage is repaired through the homologous recombination pathway. (A-C) Rad51 and its 2 interacting proteins, Xrcc3 and Brca2, are involved in DNA damage response to CNDAC. (A) Exponentially growing HeLa (top) and HCT116 cells (bottom) were incubated with 4μM and 3μM CNDAC for 24 hours and 72 hours, respectively. Cells were harvested at indicated times and subjected to subcellular fractionation. Fractions: cytoplasmic (S1), nuclear soluble (S2), and chromatin binding (P2). Rad51, Orc2, and RhoA or β-tubulin in the indicated fractions were detected by immunoblotting. (B) AA8 (■, wild-type), irs1SF (ο, Xrcc3-deficient), and 1SFwt8 (▴, hXRCC3-complemented) cells were washed into drug-free medium after a 24-hour exposure to CNDAC at a range of concentrations and allowed to form colonies in 6 to 7 days. (C) V79 (■, wild-type) and V-C8 (ο, Brca2-deficient) cells were treated as in panel B. All points are mean ± SD of triplicate plates. (D) IC50 values of CNDAC and mitomycin C from clonogenic assays using paired hamster lines. (E) CNDAC induces higher levels of SCEs after the second S phase. Continuously exposed to bromodeoxyuridine, AA8 cells were treated with 1μM CNDAC for either 15 hours (1 cell cycle) or for 30 hours (2 cycles). Colcemid was added in the final 1.5 hours. Quantitation of SCEs was performed in a minimum of 20 metaphases per sample.
CNDAC increases the incidence of SCE
Homologous recombination has been indicated as a key molecular mechanism for SCE formation in vertebrates.38 To further elucidate the role of HR, we used the cytogenetic analysis of SCE formation in metaphase cells, which have been exposed to CNDAC or to mitomycin C (supplemental Figure 2). AA8 cells treated with 1μM CNDAC for 1 population doubling time (Figure 6E) presented 8.4 (± 3.2) SCEs per cell, compared with a background level of 5.9 (± 2.4) SCEs per cell in control. Strikingly, cells incubated in the same nonarresting concentration of CNDAC for 2 population doubling times manifested 17.6 (± 6.8) SCEs per metaphase. This is similar to that of cells treated 1 cycle with 10 ng/mL mitomycin C, which induces a similarly high frequency of SCE (15.2 ± 5.3 SCEs per metaphase). Consistent with results from γ-H2AX staining, comet assays and PFGE in Figures 2 and 3, cells acquire significantly greater numbers of DSB that were repaired by HR, reflected by increased SCE incidence, after progressing through a second S phase. These data support the conclusion that HR is the major mechanism for repair of CNDAC-induced DSBs.
Discussion
Nucleoside analogs are a major class of therapeutics that are effective in the treatment of hematologic malignancies. Although the mechanism of action of most nucleosides involves their incorporation into DNA,39,40 the formation of single-strand nicks by CNDAC and the cellular responses to this damage are unique. A summary of the cellular responses to this action of CNDAC is presented in Figure 7. At relatively high concentrations of CNDAC, cells activate the G2 checkpoint, which is initiated by ATR and DNA-PK, but not ATM, through the downstream Chk1-Cdc25C-Cdk1/cyclin B1 signaling pathway.41 However, the survival value of this checkpoint is probably small as CNDAC concentrations necessary to cause this response (> 1μM) greatly reduce clonogenic survival (Figure 1C). Cells appear to have the capacity to repair the analog-terminated lesion using the TC-NER pathway,20 which is a potential mechanism of resistance. Although DSBs may be formed from nicked DNA by endonucleolytic actions or cleavage by torsional forces, results of the current investigations using clinically relevant CNDAC concentrations1 indicate the likelihood that many of the thousands of nicks, which remain unrepaired at the time of a subsequent round of DNA replication, are converted to one-ended DSBs. Such lesions are characteristic of collapsed replication forks and are repaired by HR,8,9 whereas NHEJ requires both ends of a DSB as a substrate.12 This is consistent with the observed dependency of cell survival on ATM, but not DNA-PK, which indicates that HR is the major pathway for repair of CNDAC-induced DSBs.
A model for DNA repair proteins in response to CNDAC-induced DNA damage.
This conclusion is supported by the biochemical evidence of activation of H2AX phosphorylation and the physical indicators of DSB formation demonstrated by both PFGE and comet assays under neutral conditions at the time of progression to a second S-phase. This is consistent with the formation of SCEs after cells enter the second S-phase, the presence of which is diagnostic of the involvement of HR repair. Thus, we conclude that conversion of SSB to single-ended DSBs at the replication fork is probably the major mechanism of cell death. The great sensitization of clonogenic survival in cells lacking components of the HR repair pathway, but not in cells incapable of NHEJ, supports the conclusion that it is HR that is critical for viability, and probably for resistance to CNDAC.
CNDAC concentrations and durations of exposure used in the current experiments have little effect on DNA replication3 and do not activate checkpoint responses.21 However, either greater concentrations or longer exposures of to CNDAC treatment cause complex DNA damage. Studies in model systems showed that CNDAC incorporation blocks primer extension,7 and some transient accumulation of CNDAC-treated cells in the S-phase was evident.21 Thus, more extensive CNDAC incorporation may in part cause replication fork stalling, giving rise to structures that activate phosphorylation of H2AX, and subsequently DSBs, which is consistent with our observation that some DSBs were formed during the initial cell cycle (Figure 2).21
This mechanism of action of CNDAC and the cellular responses to the resulting DNA damage have implications for therapy. The fact that HR genes are deficient in some hematologic malignancies as well as solid tumors raises the possibility of creating synthetically lethal treatment strategies. Thus, we hypothesize that tumors lacking HR function would be selectively sensitized to CNDAC-based therapy. Several such opportunities exist. For instance, it is known that several genes that respond to DNA breaks (ATM, H2AX, Mre11, and Chk1) are located on long arm of chromosome 11, a region that is frequently deleted in some hematologic malignancies. This is a common occurrence in chronic lymphocytic leukemia, in which the incidence has been reported to be between 10% and 20% before treatment.42 Importantly, the residual ATM allele was found to be mutated in 26 of 72 (36%) leukemias with this deletion.43 Such tumor-associated loss of ATM function could confer a selective advantage to CNDAC-based therapy. In addition, because BRCA2 is the same protein as FANCD1,44 the evaluation of CNDAC in Fanconi anemia models is also warranted. Further, reduced expression of BRCA1, which acts with BRCA2 in homologous recombination, was recently reported to be frequent in AML with cytogenetic abnormalities, and in therapy-related AML, and was associated with promoter hypermethylation.45 A deficiency of the HR proteins BRCA1 and BRCA2, particularly in breast and ovarian carcinomas, could indicate a more susceptible cohort of patients. A synthetically lethal condition may also be created by a pharmacologic approach of administering the drug in combination with an inhibitor of ATM or another component of the HR pathway.46 Finally, treating indolent cells with combinations of alkylating agents or platinum derivatives allows nucleotide analog incorporation into repair patches.22,24 In such combinations, CNDAC incorporation could generate an SSB that would be converted to a DSB on a single replication as latent cells reenter the cell cycle. This approach to increase CNDAC-induced damage could be applied in both the genetic and pharmacologic contexts described in the previous paragraph.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Asha Multani and Walter Hittelman for their assistance in cytogenetic analysis, David Harris for clonogenic assay of primary samples, Billie Nowak for HPLC analysis of CNDAC metabolites, and Sherry Pierce for management of patient data.
This work was supported in part by the National Cancer Institute, Department of Health and Human Services (grants CA28596, CA81534, and CA100632, and Cancer Center Support Grant P50 CA16672).
National Institutes of Health
Authorship
Contribution: X.L. and Y.W. designed and performed research, analyzed data, and wrote the paper; S.B. performed research and analyzed data; A.M. contributed vital chemicals and wrote the paper; H.K. identified patients and reviewed the paper; Z.E. identified patients, conducted experiments, and reviewed the paper; and W.P. designed the research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: William Plunkett, Department of Experimental Therapeutics, University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: wplunket@mdanderson.org.
References
Author notes
X.L. and Y.W. contributed equally to this study.

![Figure 1. Identification of CNDAC and its metabolites in cells and DNA. (A) ML-1 cells were incubated with 0.3μM [3H]CNDAC for 18 hours and washed into fresh RPMI medium, and DNA was extracted and enzymatically hydrolyzed. Components of DNA hydrolysates were separated by reversed-phase HPLC, and radioactivity was quantitated. (Top panel) An authentic standard of CNDAC, CNddC, and CNDC; the ordinate unit represents ultraviolet absorption at 270 nm (AU). (Bottom panel) An HPLC analysis of DNA hydrolysis products degraded with DNase I, phosphodiesterase, and alkaline phosphatase; the ordinate represents radioactivity (CPM). (B) Number of CNddC molecules per cell, calculated as described in “Quantitation of CNDAC and its metabolites in cells and DNA.” (C) Cytotoxicity of CNDAC in ML-1 cells (left) and purified primary samples from AML patients (right) determined by clonogenic assay. Exponentially growing ML-1 cells were exposed to a range of concentrations of CNDAC for 18 hours, washed into fresh medium, and allowed to form colonies. The survival curve is mean ± SD values from triplicates. AML primary samples (patients 11-17) were incubated with CNDAC at indicated concentrations for 5 to 6 days without washing out of drug. The curves are mean values from duplicates or triplicates. (D) HPLC analysis of intracellular accumulation of CNDAC-TP in AML primary samples exposed to CNDAC at indicated concentrations for 2 hours (left, patients 1, 2, 4, 6, 7, 8, 9, and 10) or 5μM CNDAC for indicated times (middle, patients 3-10), and elimination of CNDAC-TP from AML primary cells over 10 hours after a 4-hour incubation with 10μM CNDAC (right, patients 6, 7, and 10).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/10/10.1182_blood-2009-05-220376/4/m_zh89991055780001.jpeg?Expires=1767704527&Signature=PwTYOZH3rVQJUY-WQvSyJZBziBUwACod1xSApfPXRWbetWj1NE4UDfoZIWJmNXxmsqhsAEzmZBEhTsCBbMB4hmrOz2a0ir4ek2G8HAuJQLSXjlDi1CcVyfx1GmjVkLX07Vy~3O3M~-XYFCoS9E0xJ4Pz-TKDbonwqEzoNEpXAGwCT-eQ~ZQ2szftcpTq4bV0d-2eDSuxtHvouUkVdZhuekG7~zqWIyCzVlAmmsEv5bzwWXBCyCJOCl0dKuVNZ0ovNhMpfnBJ2TU1xs-XLlvmmvUSc36SeA-aSZJ~rnwdmxfIMG9UX~al3mwbKNE5ljOY9i-sVaU8CDvwx9SMcUh98A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)