Abstract
Oxaliplatin and fludarabine have different but potentially complementary mechanisms of action. Previous studies have shown that DNA repair is a major target for fludarabine. We postulate that potentiation of oxaliplatin toxicity by fludarabine may be due to the inhibition by fludarabine of the activity of the DNA excision repair pathways activated by oxaliplatin adducts. To test this, we investigated the cytotoxic interactions between the 2 drugs in normal and chronic lymphocytic leukemia (CLL) lymphocytes. In each population, the combination resulted in greater than additive killing. Analysis of oxaliplatin damage revealed that fludarabine enhanced accumulation of interstrand crosslinks (ICLs) in specific regions of the genome in both populations, but to a lesser extent in normal lymphocytes. The action of fludarabine on the removal of oxaliplatin ICLs was explored to investigate the mechanism by which oxaliplatin toxicity was increased by fludarabine. Lymphocytes from patients with CLL have a greater capacity for ICL unhooking compared with normal lymphocytes. In the presence of fludarabine the extent of repair was significantly reduced in both populations, more so in CLL. Our findings support a role of fludarabine-mediated DNA repair inhibition as a mechanism critical for the cytotoxic synergy of the 2 drugs.
Introduction
Chronic lymphocytic leukemia (CLL) is the most frequent type of leukemia in Western countries and it accounts for approximately 25% of all leukemias.1 Although at the present there is no curative treatment, combinations of cytotoxic agents and of immunotherapies that generate high complete remission rates hold promise for altering the natural history of this disease.2-4 Alkylating agents and nucleoside analogues are 2 classes of anticancer agents important in the treatment of chronic lymphocytic leukemia. Indolent B-cell malignancies such as CLL exhibit a high DNA damage repair capacity.5 This biologic property provides a rationale for therapeutic strategies that combine DNA-damaging cytotoxic agents, such as oxaliplatin, with an inhibitor of DNA repair such as fludarabine.
Fludarabine (9-beta-D-arabinofuranosyl-2-fluoroadenine 5′-phosphate) is the most effective purine nucleoside analogue for the treatment of indolent lymphoproliferative disorders, including CLL, low-grade lymphoma, and prolymphocytic leukemia.6,7 On infusion into the bloodstream, fludarabine is dephosphorylated to the respective nucleoside (F-ara-A) which is a substrate for transporters.8 On entering the cell, fludarabine is anabolized to its triphosphate form, its major intracellular metabolite.9 The triphosphate can then become incorporated into DNA or RNA, actions which block further synthesis.10 Its inhibition of ribonucleotide reductase, an action that depletes the deoxynucleotide pools required for DNA repair and replication, may favor its increased incorporation into newly synthesized DNA.11 Further, its inhibitory actions against both DNA ligase12 and DNA primase13 are likely to antagonize the consequences of its incorporation. The finding that it is resistant to excision14 is likely to contribute to its effective inhibition of DNA synthesis. Finally, in vitro studies have shown that the loss of cell viability correlates directly with the level of incorporation of the analog into cellular DNA.
Oxaliplatin (DACH oxalate-platinum(II); Eloxatin) is a third-generation platinum compound with a different spectrum of activity and low cross-resistance with cisplatin.15 It is currently indicated for use with fluorouracil and leucovorin for the treatment of advanced cancer of the colon or rectum.16 Oxaliplatin has shown in vitro and in vivo supra-additive effects in combination with several other antitumor agents, including 5-fluorouracil, topoisomerase I inhibitors, thymidylate synthase inhibitors, paclitaxel, cisplatin, and carboplatin.15,17 The mechanism of action of oxaliplatin is mediated by the formation of DNA adducts. The main types of DNA lesions induced by oxaliplatin are intrastrand crosslinks covalently binding the platinum compound to guanine residues.18 The other type of lesions include DNA interstrand crosslinks (ICLs) and DNA protein crosslinks.19 Oxaliplatin-DNA adducts are suggested to exert their cytotoxicity by directly inhibiting DNA and RNA synthesis and inducing apoptosis.
The nonoverlapping side effect profiles of oxaliplatin and fludarabine and their different but potentially complementary mechanisms of action provide a basis for investigation of the activity of the drugs in combination. The rationale for combining oxaliplatin with fludarabine is based on preclinical data showing synergistic cytotoxicity between cisplatin in combination with the nucleoside analogs ara-C,20 gemcitabine,21,22 clofarabine,23 and fludarabine.24-26
It has been suggested previously that enhanced crosslink repair is a primary mechanism of resistance to nitrogen mustards in B-CLL.27 As such, we hypothesize that fludarabine may modulate oxaliplatin sensitivity by suppressing ICL removal, thereby resulting in synergistic cytotoxicity in CLL. Modulation of DNA repair represents one strategy to overcome cellular drug resistance to platinum compounds and achieve therapeutic gain. Such approaches focus on inhibiting the repair processes, thereby either subjecting the cells to the full consequence of the damage to DNA or by creating a new type of damage that the cell is incapable of repairing. In CLL, DNA repair has been suggested as the mechanism underlying drug resistance. It has been reported that CLL cells resistant to alkylating agents exhibit an increased nucleotide excision repair (NER) activity measured using the comet assay.28,29 We have hypothesized that this increased capacity for repair could favorably facilitate incorporation of fludarabine into the DNA repair patch.23 To that end, we have shown the synergistic cytotoxic interaction between oxaliplatin and fludarabine in normal and chronic lymphocytic leukemia lymphocytes and investigated the mechanistic basis for this finding.
Materials and methods
Drugs and solvents
To facilitate investigations, fludarabine (Berlex Biosciences, Richmond, CA) which is impermeable to cells, was dephosphorylated with alkaline phosphatase to generate the cell-permeable nucleoside that was used in these in vitro studies. It was dissolved in phosphate-buffered saline to a concentration of 2 mM and stored at –20°C. Oxaliplatin, provided by Sanofi-Synthelabo Research (Great Valley, PA), was dissolved in phosphate-buffered saline to a concentration of 1 or 5 mM. Drug solutions were prepared fresh for every experiment. All other reagents were of analytic grade and commercially available, and solutions were prepared using H2O from a MilliQ academic purification system (Millipore, Bedford, MA).
Preparation of normal and leukemic lymphocytes
A total of 39 samples were collected for this study, 15 from healthy persons and 24 from patients with CLL who had been selected for early-stage disease (Rai stage 0-1, 14; stage 2, 4; stage 4, 4). Only 2 patients had received prior fludarabine treatment. The median age was 62 years (range, 33-90 years). The median absolute lymphocyte count was 40.755 ×109/L (40 755/μL) (range, 3.828-226.080 ×109/L [3828-226 080/μL]), whereas the median proportion of lymphocytes in the white blood cell count was 0.86 (86%) (range, 0.06-0.96 [6%-96%]) with the median proportion of CD19/CD5 coexpressing lymphocytes being 0.899 (89.9%) (range, 0.08-0.98 [8%-98%]). Because of the labor-intensive nature of the procedures used in these investigations, we generally used separate cohorts of samples for the various experiments. We assumed that samples were derived from reasonably similar persons because the disease of most patients had not progressed to a state that required treatment, and volunteers were generally younger than age 25 years. Whole blood was drawn into heparinized tubes, diluted 1:5 with cold PBS, and layered over Fico/Lite LymphoH (specific gravity, 1.077; Atlanta Biologicals, Norcross, GA). The mixtures were then centrifuged at 430g for 20 minutes. Mononuclear cells were removed from the interphase, washed twice with cold PBS, and resuspended in 10 mL RPMI 1640 medium containing 10% fetal bovine serum. The cells were then counted, and their median volume was determined by using a Coulter Counter (Beckman Coulter, Fullerton, CA). Cells were diluted to a concentration of 2 × 106 cells/mL and incubated in plastic plates in a 5% CO2 humidified atmosphere at 37°C for 4 hours to remove the adherent monocytes and to acclimatize the cells to culture conditions. The lymphocytes were then carefully collected, counted, and resuspended at a concentration of 2 × 106 cells/mL. All donors provided written informed consent approved by the MD Anderson Cancer Center review board, and in accordance with the Declaration of Helsinki.
Quantitation of apoptotic-cell death
Apoptotic-cell death in normal or CLL lymphocytes was evaluated by flow cytometry analysis using Annexin V and propidium iodide (PI) double staining. Lymphocytes were treated with drugs alone (2.5, 5, 10 μM oxaliplatin; 2.5, 5, 10 μM fludarabine) or in combination simultaneously for 16, 24, or 36 hours. After the treatment, apoptotic-cell death was detected by FITC-conjugated Annexin V (BD Biosciences, San Jose, CA) staining. Following treatment, CLL cells were washed twice with PBS, resuspended in 200 μL binding buffer (10 mM HEPES/NaOH, pH 7.4, 140 mM NaCl, 2.5 mM CaCl2), and incubated with 10 μL Annexin V-FITC for 15 minutes at room temperature. After centrifugation, the pellets were resuspended with 500 μL binding buffer with 10 μL of 50 mg/mL PI. Samples were analyzed with a fluorescence-activated cell sorter flow cytometer (Becton Dickinson, Franklin Lakes, NJ). Data acquisition and analysis were performed by the CellQuest program (Becton Dickinson). Cells positive for Annexin V were considered apoptotic.
Detection of oxaliplatin DNA damage-DNA probes
Three different probes, all genomic inserts labeled by random priming, were used to detect oxaliplatin-induced ICLs in various regions of the genome. The 1.8-kb (kilobase) EcoRI fragment containing exons I and II of the DHFR gene was isolated from pBH31R1.8 plasmid probe (Dr V.A. Bohr, National Institute on Aging, NIH, Baltimore, MD) and labeled using [α-32P]dATP. The probe was used to detect a 22-kb fragment of the DHFR gene localized to the 5′ end of the gene. The plasmid pBR322 (Dr V.A. Bohr, National Institute of Aging, NIH) was digested with EcoRI and BamHI to generate the 0.8-kb δ-globin insert. The human δ-globin probe was used to detect the entire 18-kb gene. The plasmid pCRII-HI (Dr C.A. Filburn, National Institute of Aging, NIH) was digested with EcoRI to generate a 2574-bp (base pair) fragment of the human mitochondrial genome (nucleotides 652-3226). The mitochondrial fragment was used to detect the 16.5-kb human linearized BamHI mitochondrial DNA (mtDNA) genome.
Drug treatment of cells and DNA extraction
Cells were seeded in 15-cm dishes at a density of 5 × 106 cells/dish and incubated in the absence or presence of fludarabine for 2 hours before exposure to increasing concentrations of oxaliplatin for 7 hours. For the repair experiments, cells were incubated with oxaliplatin for 5 hours in the absence or presence of fludarabine, to induce damage, followed by treatment with 0.1 M thiourea for 1 hour to block the conversion of monoadduct to an ICL during the posttreatment incubation. The cells were then incubated for up to 8 hours in drug-free medium or in fludarabine-treated medium. Cells were subsequently washed twice with PBS, trypsinized, and pelleted by centrifugation for 5 minutes at 200 g. Pellets were stored at –80°C until required. The cell pellets were then resuspended in 200 μL PBS, and total genomic DNA was extracted using the Qiagen (Valencia, CA) QIAamp DNA blood isolation kit using the protocol provided, and the DNA concentration of the samples was determined. All experiments were performed at least 3 times.
Detection of gene-specific oxaliplatin interstrand crosslinks
To study the effect of fludarabine on the accumulation of oxaliplatin-induced ICLs in specific DNA genes, DNA (6 μg) from treated cells was restricted with HindIII (to release the 22-kb DHFR fragment and the 18-kb δ-globin gene), whereas 4 μg DNA was digested with BamHI (to linearize the mitochondrial genome) for 2 hours at 37°C. The DNA was extracted once with phenol, once with chloroform and precipitated with ethanol. The pellet was resuspended in 15 μL TE buffer (10 mM Tris-HCl, 0.1 mM EDTA, pH 7.5) and denatured by the addition of an equal volume of 100 mM NaOH and incubated for 20 minutes at 37°C. Loading buffer (4 μLof 10 × buffer: 10 mM EDTA, 26% Ficoll, 0.25% bromocresol green) was added to each sample, and the DNA was then electrophoresed through a 0.5% agarose gel in TAE buffer at 27 V overnight. The DNA was transferred to nylon (Hybond N+; Amersham, Piscataway, NJ), fixed to the membrane by baking for 2 hours at 80°C, and probed for the gene fragment of interest. Band visualization and quantitation were performed by PhosphorImager (Molecular Dynamics, Sunnyvale, CA) and Image Quant software (Version 4.2a; Molecular Dynamics). Assuming that crosslink formation is random along the DNA fragments, the Poisson distribution30 was used to calculate the average number of interstrand crosslinks present in DHFR, δ-globin, and mitochondrial DNA fragments (normalized to crosslink/10 kb) using the following formula,30 where N is the natural log of the fraction of DNA in the single-strand form:
Statistical analysis
Statistical significance was determined by using Student t test calculated with GraphPad Prism 4 software (GraphPad Software, San Diego, CA). A P value less than .05 was considered to be statistically significant.
Results
Toxicity of oxaliplatin and fludarabine to CLL lymphocytes alone and in combination
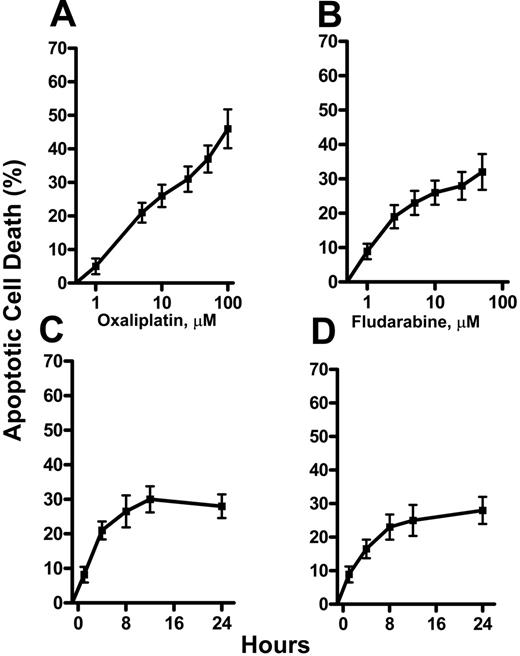
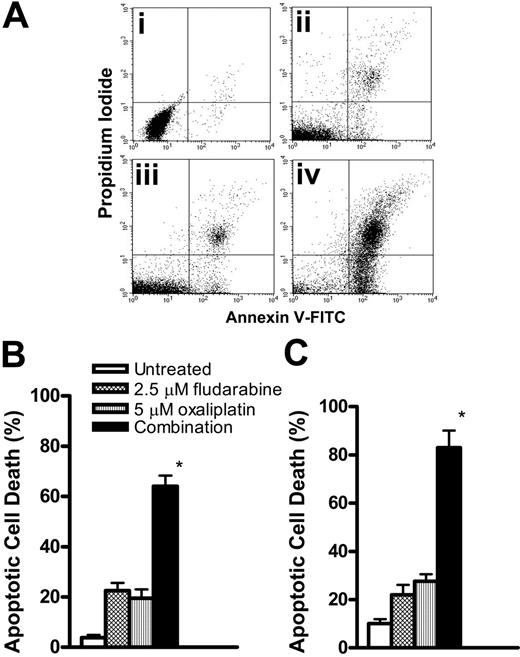
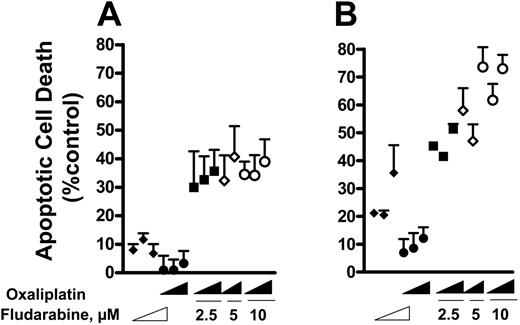
The induction of apoptosis by oxaliplatin and fludarabine as single agents was evaluated by flow cytometric analysis of Annexin V–stained CLL lymphocytes. Cell death was dependent on the concentration of oxaliplatin (Figure 1A) or fludarabine (Figure 1B). Induction of apoptosis was also time dependent, reaching maximum activity at approximately 12 hours for 5 μM oxaliplatin (Figure 1C) and 2.5 μM fludarabine (Figure 1D). Drug concentrations that induced 15% to 25% apoptosis were chosen for combination experiments. We then evaluated the ability of fludarabine and oxaliplatin as single agents or in combination for 24 hours to induce cell death. This is illustrated by the increase in number of cells staining positively for Annexin V (Figure 2A). The actions of oxaliplatin and fludarabine were then evaluated in lymphocytes from healthy donors (Figure 2B) and from patients with CLL (Figure 2C). Alone, each drug induced apoptosis, but treatment with the combination for 24 hours resulted in greater than additive killing in each population (P < .05). The interaction between fludarabine and oxaliplatin was further investigated by evaluating several concentrations of each drug at 16 hours and 36 hours (Figure 3). Although cell killing by each drug alone was dependent on time of incubation, this was more evident for fludarabine. No marked differences of the single drugs were seen in the concentration ranges investigated. Again, the combination generated greater than additive killing, which was more pronounced at 36 hours. This range of concentrations is relevant to the clinical use of fludarabine31 and oxaliplatin.32
Induction of apoptosis by oxaliplatin and fludarabine in CLL lymphocytes. CLL lymphocytes were treated with concentrations of oxaliplatin (A) (1, 5, 10, 25, 50, 100 μM) or fludarabine (B) (1, 2.5, 5, 10, 25, 50 μM) for 24 hours or with 5 μM oxaliplatin (C) or 2.5 μM fludarabine (D) for 24 hours. Annexin V–positive cells were considered apoptotic, presented as the percentage of apoptotic cells, and expressed as the mean ± SE. The results are the means of 3 experiments.
Induction of apoptosis by oxaliplatin and fludarabine in CLL lymphocytes. CLL lymphocytes were treated with concentrations of oxaliplatin (A) (1, 5, 10, 25, 50, 100 μM) or fludarabine (B) (1, 2.5, 5, 10, 25, 50 μM) for 24 hours or with 5 μM oxaliplatin (C) or 2.5 μM fludarabine (D) for 24 hours. Annexin V–positive cells were considered apoptotic, presented as the percentage of apoptotic cells, and expressed as the mean ± SE. The results are the means of 3 experiments.
Cytotoxic interactions between oxaliplatin and fludarabine. (A) Apoptosis was compared in untreated CLL lymphocytes (i) or after incubation with 2.5 μM fludarabine (ii), 5 μM oxaliplatin (iii), or the combination (iv) for 24 hours. Apoptotic cell death was measured by Annexin V binding. Viable cells not undergoing apoptosis are Annexin V-FITC and PI negative; cells undergoing apoptosis are Annexin V-FITC positive and PI negative; cells already dead are Annexin V-FITC and PI positive. (B) Lymphocytes from healthy donors were incubated with 2.5 μM fludarabine, 5 μM oxaliplatin, or the combination for 24 hours. (C) Lymphocytes from patients with CLL were treated as in panel B. Values for Annexin V positivity are presented as the percentage of apoptotic cell death and expressed as the mean ± SE (n = 3). *P < .05 between the combination and the sum of fludarabine and oxaliplatin.
Cytotoxic interactions between oxaliplatin and fludarabine. (A) Apoptosis was compared in untreated CLL lymphocytes (i) or after incubation with 2.5 μM fludarabine (ii), 5 μM oxaliplatin (iii), or the combination (iv) for 24 hours. Apoptotic cell death was measured by Annexin V binding. Viable cells not undergoing apoptosis are Annexin V-FITC and PI negative; cells undergoing apoptosis are Annexin V-FITC positive and PI negative; cells already dead are Annexin V-FITC and PI positive. (B) Lymphocytes from healthy donors were incubated with 2.5 μM fludarabine, 5 μM oxaliplatin, or the combination for 24 hours. (C) Lymphocytes from patients with CLL were treated as in panel B. Values for Annexin V positivity are presented as the percentage of apoptotic cell death and expressed as the mean ± SE (n = 3). *P < .05 between the combination and the sum of fludarabine and oxaliplatin.
Fludarabine increases the accumulation of oxaliplatin-induced DNA interstrand crosslinks in both nuclear DNA and mtDNA
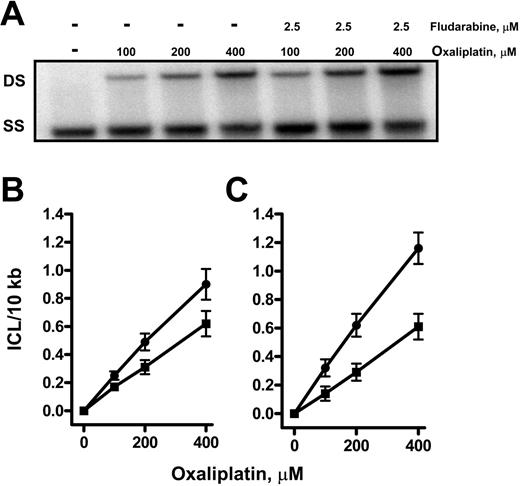
We then sought to investigate the actions of fludarabine on the accumulation of the important oxaliplatin lesion, the ICL. Normal or CLL lymphocytes were incubated in the absence or presence of fludarabine for 2 hours before exposure to several concentrations of oxaliplatin for 7 hours. Total genomic DNA was isolated and analyzed for the presence of the ICLs in a 22-kb fragment of the DHFR gene, the entire 18-kb δ-globin gene as well as the 16.5-kb mtDNA. DNA from control cells incubated in the absence of any drugs was completely denatured under the conditions used and migrated as single-strand DNA (Figure 4A). In contrast, there was an oxaliplatin concentration-dependent increase in double-strand crosslinked DNA in the DHFR gene from CLL lymphocytes. There was greater crosslinking in DNA from cells that were incubated with fludarabine prior to oxaliplatin addition. Quantitation of ICLs in 3 separate samples showed a linear increase as a function of oxaliplatin concentration within the range of 100 to 400 μM, reaching similar levels at 400 μM of 0.6 ICL/10 kb in normal (Figure 4B) and CLL lymphocytes (Figure 4C). Incubation of normal lymphocytes (Figure 4B) with oxaliplatin in the presence of fludarabine increased ICL levels by 40% (P < .05) at 400 μM. Similar results were obtained following analysis of ICLs in the transcriptionally inactive δ-globin gene and mtDNA. The amount of ICLs in the DHFR gene and the mitochondrial genome was similar, although fewer oxaliplatin lesions were detected in the δ-globin gene. In the CLL lymphocytes, incubation with 400 μM oxaliplatin in the presence of fludarabine nearly doubled ICL levels in the DHFR gene (Figure 4C). Again, similar findings were observed in both the δ-globin and the mtDNA in which the accumulation of ICLs was also increased by fludarabine (Table 1). These data support an association between the effect of fludarabine on oxaliplatin-induced ICL accumulation and the cytotoxic synergism observed in CLL lymphocytes.
Action of fludarabine on formation of oxaliplatin ICL in specific genes of CLL lymphocytes
. | . | ICL/10 kb* . | . | |
|---|---|---|---|---|
| Oxaliplatin, μM . | Gene size, kb . | -F-ara-A . | +F-ara-A . | |
| DHFR | 22 | |||
| 100 | 0.14 | 0.26 | ||
| 200 | 0.31 | 0.63 | ||
| 400 | 0.60 | 1.16† | ||
| δ-Globin | 18 | |||
| 100 | 0.08 | 0.19 | ||
| 200 | 0.19 | 0.42 | ||
| 400 | 0.43 | 0.87† | ||
| mtDNA | 16.5 | |||
| 100 | 0.12 | 0.22 | ||
| 200 | 0.27 | 0.56 | ||
| 400 | 0.51 | 1.05† | ||
. | . | ICL/10 kb* . | . | |
|---|---|---|---|---|
| Oxaliplatin, μM . | Gene size, kb . | -F-ara-A . | +F-ara-A . | |
| DHFR | 22 | |||
| 100 | 0.14 | 0.26 | ||
| 200 | 0.31 | 0.63 | ||
| 400 | 0.60 | 1.16† | ||
| δ-Globin | 18 | |||
| 100 | 0.08 | 0.19 | ||
| 200 | 0.19 | 0.42 | ||
| 400 | 0.43 | 0.87† | ||
| mtDNA | 16.5 | |||
| 100 | 0.12 | 0.22 | ||
| 200 | 0.27 | 0.56 | ||
| 400 | 0.51 | 1.05† | ||
Values represent ICL/10 kb. Data are from at least 3 blots each of 3 independent biologic experiments. Standard errors were in the range of 0 to 0.09 ICL per fragment.
P < .05 (significance evaluated at 400 mM oxaliplatin).
Influence of concentration and time of incubation on cell killing. CLL lymphocytes were treated with increasing concentrations of fludarabine (2.5, 5, 10 μM), oxaliplatin (2.5, 5, 10 μM), or the combinations (fludarabine 2.5 μM + oxaliplatin 2.5, 5, and 10 μM; fludarabine 5 μM + oxaliplatin 5 and 10 μM; fludarabine 10 μM + oxaliplatin 2.5, 5, and 10 μM) for 16 hours (A) or 36 hours (B). The range of concentrations is represented by a wedge. Annexin V–positive cells were considered apoptotic, presented as apoptotic cells (percentage of control). Each point is the mean ± SE of determinations on cell samples from 3 to 6 persons.
Influence of concentration and time of incubation on cell killing. CLL lymphocytes were treated with increasing concentrations of fludarabine (2.5, 5, 10 μM), oxaliplatin (2.5, 5, 10 μM), or the combinations (fludarabine 2.5 μM + oxaliplatin 2.5, 5, and 10 μM; fludarabine 5 μM + oxaliplatin 5 and 10 μM; fludarabine 10 μM + oxaliplatin 2.5, 5, and 10 μM) for 16 hours (A) or 36 hours (B). The range of concentrations is represented by a wedge. Annexin V–positive cells were considered apoptotic, presented as apoptotic cells (percentage of control). Each point is the mean ± SE of determinations on cell samples from 3 to 6 persons.
Fludarabine enhances the oxaliplatin-induced DNA interstrand crosslinks in the DHFR gene in normal or CLL lymphocytes. DNA from CLL lymphocytes incubated in absence or presence of fludarabine for 2 hours before exposure to increasing concentrations of oxaliplatin for 7 hours was isolated and analyzed for ICL formation. (A) Representation of HindIII-digested DNA denatured and electrophoresed through a 0.5% agarose gel, before being transferred to nylon and probed for a 22-kb DHFR gene fragment. DS indicates double-strand DNA; SS, single-strand DNA. The data shown were based on at least 3 independent experiments, and DNA samples from each biologic experiment were subjected to gel electrophoresis and Southern analysis at least twice. Crosslinks in the DHFR gene in normal lymphocytes (B) or CLL lymphocytes (C); this value was normalized to ICL/10 kb in the absence (▪) or presence (•) of 2.5 μM fludarabine, expressed as the mean ± SE (n = 3).
Fludarabine enhances the oxaliplatin-induced DNA interstrand crosslinks in the DHFR gene in normal or CLL lymphocytes. DNA from CLL lymphocytes incubated in absence or presence of fludarabine for 2 hours before exposure to increasing concentrations of oxaliplatin for 7 hours was isolated and analyzed for ICL formation. (A) Representation of HindIII-digested DNA denatured and electrophoresed through a 0.5% agarose gel, before being transferred to nylon and probed for a 22-kb DHFR gene fragment. DS indicates double-strand DNA; SS, single-strand DNA. The data shown were based on at least 3 independent experiments, and DNA samples from each biologic experiment were subjected to gel electrophoresis and Southern analysis at least twice. Crosslinks in the DHFR gene in normal lymphocytes (B) or CLL lymphocytes (C); this value was normalized to ICL/10 kb in the absence (▪) or presence (•) of 2.5 μM fludarabine, expressed as the mean ± SE (n = 3).
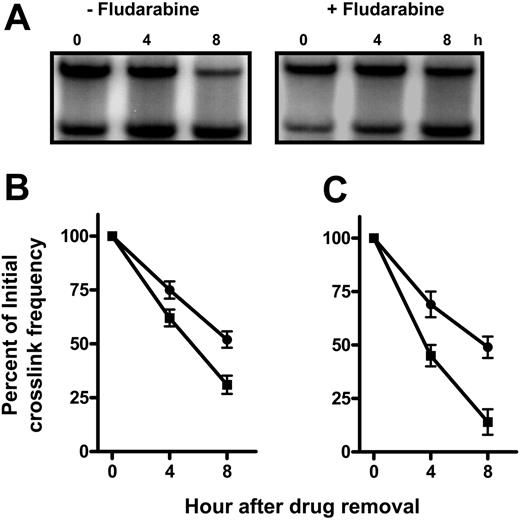
Fludarabine retards the removal of oxaliplatin-induced DNA interstrand crosslinks in specific regions of the genome
The action of fludarabine on the removal of oxaliplatin ICLs was examined to investigate the possibility that the apparent increase in ICL accumulation in the presence of fludarabine was due to suppression of oxaliplatin-induced DNA ICL removal. The recovery of the ability to denature crosslinked DNA, which measures the unhooking of the crosslink, was used to compare the repair proficiency in cells. That fludarabine affects the removal of ICLs can be seen in the analysis of the DHFR gene from normal lymphocytes after oxaliplatin treatment in the absence or presence of fludarabine (Figure 5A). Lymphocytes from healthy donors (Figure 5B) and from patients with CLL (Figure 5C) removed crosslinks with a half-life of about 4 hours. This reaction was retarded in cells that had been first treated with fludarabine.
Consistent with findings above (Figure 4), the amount of oxaliplatin-induced ICLs does not vary in specific genes between normal and CLL lymphocytes; however, the rate of ICL removal was different. Oxaliplatin ICLs were removed efficiently over 8 hours with 68% in normal lymphocytes (Figure 5B) and 85% in CLL cells (Figure 5C) crosslinks unhooked in the DHFR gene. Similar findings were observed in the δ-globin gene and the mtDNA (Figure 6; Table 2). This suggests that CLL lymphocytes have a greater capacity for DNA crosslink unhooking compared with normal lymphocytes.
Action of fludarabine on formation and removal of oxaliplatin ICL in specific genes of normal and CLL lymphocytes
. | Normal lymphocytes . | . | . | . | . | CLL lymphocytes . | . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | ICL/10 kb* . | . | Removal, %† . | . | Removal inhibition, %‡ . | ICL/10 kb* . | . | Removal, %† . | . | Removal inhibition, %‡ . | ||||||||
| Repair time, h . | Without fludarabine . | With fludarabine . | Without fludarabine . | With fludarabine . | . | Without fludarabine . | With fludarabine . | Without fludarabine . | With fludarabine . | . | ||||||||
| DHFR | ||||||||||||||||||
| 0 | 0.48 | 0.77 | — | — | — | 0.52 | 0.85 | — | — | — | ||||||||
| 4 | 0.29 | 0.53 | 40 | 31§ | 23 | 0.26 | 0.59 | 49 | 31§ | 37 | ||||||||
| 8 | 0.15 | 0.41 | 68 | 48§ | 29 | 0.08 | 0.42 | 85 | 51§ | 40 | ||||||||
| δ-Globin | ||||||||||||||||||
| 0 | 0.36 | 0.47 | — | — | — | 0.38 | 0.53 | — | — | — | ||||||||
| 4 | 0.26 | 0.36 | 29 | 23∥ | 21 | 0.19 | 0.33 | 51 | 38§ | 25 | ||||||||
| 8 | 0.20 | 0.30 | 45 | 37∥ | 15 | 0.13 | 0.25 | 65 | 53∥ | 19 | ||||||||
| mtDNA | ||||||||||||||||||
| 0 | 0.51 | 0.76 | 0 | 0 | — | 0.47 | 0.74 | — | — | — | ||||||||
| 4 | 0.34 | 0.56 | 34 | 26§ | 24 | 0.23 | 0.53 | 52 | 28§ | 46 | ||||||||
| 8 | 0.25 | 0.49 | 51 | 35§ | 25 | 0.12 | 0.37 | 75 | 50§ | 33 | ||||||||
. | Normal lymphocytes . | . | . | . | . | CLL lymphocytes . | . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | ICL/10 kb* . | . | Removal, %† . | . | Removal inhibition, %‡ . | ICL/10 kb* . | . | Removal, %† . | . | Removal inhibition, %‡ . | ||||||||
| Repair time, h . | Without fludarabine . | With fludarabine . | Without fludarabine . | With fludarabine . | . | Without fludarabine . | With fludarabine . | Without fludarabine . | With fludarabine . | . | ||||||||
| DHFR | ||||||||||||||||||
| 0 | 0.48 | 0.77 | — | — | — | 0.52 | 0.85 | — | — | — | ||||||||
| 4 | 0.29 | 0.53 | 40 | 31§ | 23 | 0.26 | 0.59 | 49 | 31§ | 37 | ||||||||
| 8 | 0.15 | 0.41 | 68 | 48§ | 29 | 0.08 | 0.42 | 85 | 51§ | 40 | ||||||||
| δ-Globin | ||||||||||||||||||
| 0 | 0.36 | 0.47 | — | — | — | 0.38 | 0.53 | — | — | — | ||||||||
| 4 | 0.26 | 0.36 | 29 | 23∥ | 21 | 0.19 | 0.33 | 51 | 38§ | 25 | ||||||||
| 8 | 0.20 | 0.30 | 45 | 37∥ | 15 | 0.13 | 0.25 | 65 | 53∥ | 19 | ||||||||
| mtDNA | ||||||||||||||||||
| 0 | 0.51 | 0.76 | 0 | 0 | — | 0.47 | 0.74 | — | — | — | ||||||||
| 4 | 0.34 | 0.56 | 34 | 26§ | 24 | 0.23 | 0.53 | 52 | 28§ | 46 | ||||||||
| 8 | 0.25 | 0.49 | 51 | 35§ | 25 | 0.12 | 0.37 | 75 | 50§ | 33 | ||||||||
—indicates not applicable.
Values represent ICL/10 kb. Data are from at least 3 blots each of 3 independent biologic experiments. Standard errors for each group were in the range of 0 to 0.08 ICL/fragment.
Values represent percentage of ICL removal.
Values represent percentage of inhibition of ICL removal at 4 and 8 hours, determined as 1 = 1 - (percentage of ICL removal + fludarabine) / (percentage of ICL removal - fludarabine).
P < .05
P = .07.
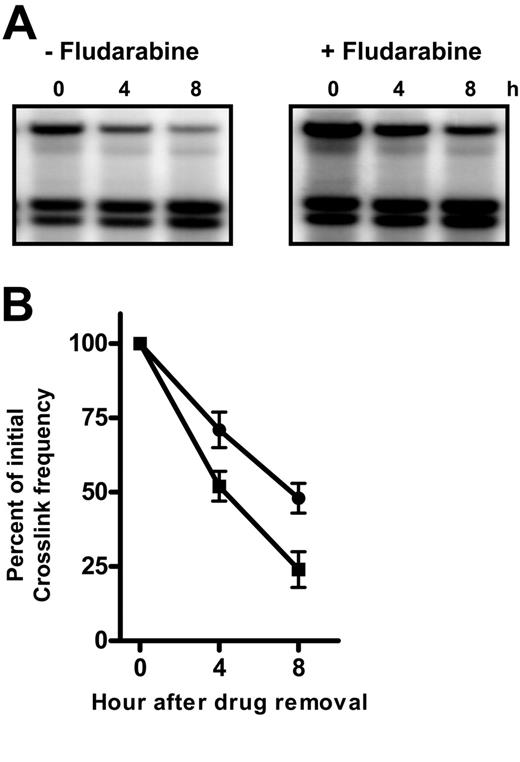
In cells treated with oxaliplatin alone and in combination with fludarabine, ICLs were removed from the mtDNA rapidly with few crosslinks remaining after 8 hours (Figure 6A). Quantitation revealed that CLL cells efficiently repaired oxaliplatin ICLs with 75% of damage removed within 8 hours. In the presence of fludarabine, however, the extent of repair was reduced to 50% (Figure 6B). In the presence of fludarabine, the extent of ICL removal in the DHFR gene was reduced to 51% in CLL (40% inhibition) and 48% in normal lymphocytes (29% inhibition) (Table 2). The actions of fludarabine on formation and removal of oxaliplatin ICLs in specific genes of normal or CLL lymphocytes are summarized in Table 2. Fludarabine action on the actively transcribed DHFR gene was similar to that observed in the mtDNA in normal or CLL lymphocytes; however, it exceeded that on the transcriptionally inactive δ-globin gene. These results suggest that suppression of the repair of oxaliplatin-induced ICLs by fludarabine may vary according to the state of condensation of chromatin.
Fludarabine retards the removal of oxaliplatin-induced DNA interstrand crosslinks in the DHFR gene in normal or CLL lymphocytes. Normal lymphocytes (A) or CLL lymphocytes (B) were incubated with 400 μM oxaliplatin in the absence or presence of 2.5 μM fludarabine for 5 hours, when cells were washed into drug-free medium. DNA from cells allowed to repair for up to 8 hours was analyzed for ICL remaining in the 22-kb DHFR gene fragment. (A) Representative ICL blot probed for the DHFR gene in peripheral-blood mononuclear cells (PBMCs). The plots represent a quantitation of oxaliplatin ICLs remaining in the DHFR gene of PBMCs (B) or CLL lymphocytes (C) as a function of repair time. Crosslinks remaining after repair in the absence (▪) or presence (•) of fludarabine were expressed as a percentage of initial ICL levels.
Fludarabine retards the removal of oxaliplatin-induced DNA interstrand crosslinks in the DHFR gene in normal or CLL lymphocytes. Normal lymphocytes (A) or CLL lymphocytes (B) were incubated with 400 μM oxaliplatin in the absence or presence of 2.5 μM fludarabine for 5 hours, when cells were washed into drug-free medium. DNA from cells allowed to repair for up to 8 hours was analyzed for ICL remaining in the 22-kb DHFR gene fragment. (A) Representative ICL blot probed for the DHFR gene in peripheral-blood mononuclear cells (PBMCs). The plots represent a quantitation of oxaliplatin ICLs remaining in the DHFR gene of PBMCs (B) or CLL lymphocytes (C) as a function of repair time. Crosslinks remaining after repair in the absence (▪) or presence (•) of fludarabine were expressed as a percentage of initial ICL levels.
Fludarabine impairs the removal of oxaliplatin-induced DNA interstrand crosslinks in the mitochondrial DNA (mtDNA). CLL lymphocytes were incubated with 400 μM oxaliplatin in the absence or presence of 2.5 μM fludarabine for 5 hours when cells were washed into drug-free medium. DNA from cells allowed to repair for up to 8 hours was analyzed for ICLs remaining. (A) Representative ICL blot probed for the 16.5-kb mtDNA. (B) Quantitation of oxaliplatin ICLs remaining in the mtDNA as a function of repair time. Crosslinks remaining after repair in the absence (▪) or presence (•) of fludarabine were expressed as a percentage of initial ICL levels.
Fludarabine impairs the removal of oxaliplatin-induced DNA interstrand crosslinks in the mitochondrial DNA (mtDNA). CLL lymphocytes were incubated with 400 μM oxaliplatin in the absence or presence of 2.5 μM fludarabine for 5 hours when cells were washed into drug-free medium. DNA from cells allowed to repair for up to 8 hours was analyzed for ICLs remaining. (A) Representative ICL blot probed for the 16.5-kb mtDNA. (B) Quantitation of oxaliplatin ICLs remaining in the mtDNA as a function of repair time. Crosslinks remaining after repair in the absence (▪) or presence (•) of fludarabine were expressed as a percentage of initial ICL levels.
Comparison of the actions of fludarabine with clofarabine and cytarabine on accumulation and removal of oxaliplatin ICLs in CLL lymphocytes
Finally, we extended our investigation to compare the action of cytarabine (ara-C) and clofarabine (Cl-F-araA) relative to fludarabine at equitoxic concentrations (2.5 μM and 10 μM, respectively) on ICL accumulation and removal kinetics. Under the same conditions used for fludarabine, oxaliplatin ICL accumulation and removal were assessed in the absence or presence of a nucleoside analogue in CLL lymphocytes. Clofarabine, but not cytarabine, significantly increased the level of ICL accumulation (Table 3). Furthermore, in the presence of clofarabine, the extent of ICL removal was retarded by nearly 65% in contrast to only 14% inhibition by cytarabine treatment (Table 3).
Action of various nucleoside analogs on formation and removal of oxaliplatin ICL in the DHFR gene of CLL lymphocytes
. | ICL/10, kb* . | Removal, %† . | Removal inhibition, %‡ . |
|---|---|---|---|
| Fludarabine | 40 | ||
| Without | 0.62 | 85 | |
| With | 1.16 | 51 | |
| Cytarabine | 14 | ||
| Without | 0.64 | 78 | |
| With | 0.79 | 67 | |
| Clofarabine | 63 | ||
| Without | 0.65 | 81 | |
| With | 1.41 | 30 |
. | ICL/10, kb* . | Removal, %† . | Removal inhibition, %‡ . |
|---|---|---|---|
| Fludarabine | 40 | ||
| Without | 0.62 | 85 | |
| With | 1.16 | 51 | |
| Cytarabine | 14 | ||
| Without | 0.64 | 78 | |
| With | 0.79 | 67 | |
| Clofarabine | 63 | ||
| Without | 0.65 | 81 | |
| With | 1.41 | 30 |
Values represent ICL/10 kb in the DHFR gene from CLL cells incubated with 400 μM oxaliplatin for 7 hours.
Values represent percentage of ICL removal at 8 hours.
Values represent percentage of inhibition of ICL removal at 8 hours. Data are from at least 3 blots each of 3 independent biologic experiments.
Discussion
Previous studies have shown that fludarabine is capable of inhibiting DNA repair induced by carboplatin33 or UV radiation34 in quiescent human lymphocytes that were stimulated by 4-hydroperoxycyclophosphamide in CLL lymphocytes,23 and cisplatin-generated damage in K562 cells.26 As an extension of those studies, the present investigations reveal a greater than additive cytotoxic effect of fludarabine in combination with oxaliplatin in both normal lymphocytes and CLL cells. These actions were associated with an inhibition of the rate of ICL removal in the presence of fludarabine, suggesting that this contributes to the increase in the cytotoxicity of the combination.
The action of fludarabine on the accumulation of oxaliplatin-induced DNA ICLs was investigated in specific DNA genes and measured directly using a denaturation and renaturation assay and Southern hybridization technique. This gene-specific assay has been previously used to explore the effects of fludarabine on the repair of cisplatin lesions24 and to investigate the actions of gemcitabine on cisplatin damage accumulation and repair.21 Although the requirement of quantitating ICLs in single gene DNA requires a relatively high level of ICLs throughout the entire genome, the results showed that fludarabine significantly increased oxaliplatin ICL accumulation in the nuclear DHFR gene and δ-globin gene as well as in the mtDNA in CLL lymphocytes (Table 1).
DNA repair proceeds at different rates throughout the genome depending on the function of the DNA. We selected the DHFR gene as an example of a transcriptionally active gene, in which it is known that repair processes proceed more rapidly than in transcriptionally inactive DNA, an example of which is the δ-globin gene.35 Mitochondrial DNA is compartmentalized away from the nuclear location of these genes. The formation of ICL in the DHFR gene and the mitochondrial genome was similar, consistent with the findings of others with cisplatin.21,36,37 Crosslinking of the δ-globin gene was less, possibly because that drug's accessibility to the DNA may have been limited. Fludarabine significantly inhibited the unhooking of interstrand crosslinks to similar extents in both the nuclear DNA and mtDNA. This action was greater in CLL cells than in normal lymphocytes, which may reflect the greater capacity for accumulating fludarabine triphosphate in CLL. Earlier studies showed that fludarabine also inhibited the removal of cisplatin ICLs in the total genome and in specific gene fragments of LoVo colon carcinoma cells using the denaturation and renaturation assay.24 Because these crosslinking assays detect an early event in the removal of crosslinks (the unhooking of an ICL and not necessarily complete excision of the ICL from both strands), the results indicate that the nucleoside analogue inhibits this step of ICL repair.
Repair of interstrand crosslinks in mammalian cells is not well understood but is suggested to involve the sequential actions of the nucleotide excision repair pathway and of homologous recombination.38 Thus, the inhibition of either of these pathways or their interaction with each other by fludarabine could explain the enhanced cytotoxicity with oxaliplatin. Previous investigations have shown that fludarabine triphosphate inhibits incision in the NER pathway, including activity of ERCC1.25 The activity of fludarabine may be related to blocking access of ATP to active binding sites on the repair enzymes. In this context, other excision proteins that use ATP such as the NER endonuclease ERCC1/XPF may be considered as a potential target for fludarabine. Indeed, it was striking that the purine nucleoside analog, clofarabine, inhibited ICL unhooking equally as did fludarabine and to a substantially greater extent than did the pyrimidine analogue, cytarabine (Table 3). Nevertheless, both DNA repair mechanisms require resynthesis of DNA to complete the repair process, so it would be expected that this would be sensitive to both pyrimidine and purine nucleoside analogues.
The use of platinum compounds and nucleoside analogs in cancer therapy is rich in rationale and practice. Indications of synergistic cell killing between cisplatin and cytarabine39 led to clinical trials in hematologic malignancies.40,41 With the demonstration of the activity of fludarabine in indolent diseases, this analog was used to augment combinations with cytarabine and cisplatin for the treatment of relapsed or transformed CLL and non-Hodgkin lymphoma.42-44 Although the latter protocols had activity, they were also associated with myelosuppression. This has been used to advantage in the design of conditioning regimens for marrow transplantation procedures.45-47 Recently, carboplatin was combined with fludarabine and evaluated in the therapy of high-risk acute leukemia.48 Results were judged promising, but marrow suppression was still a problem. Oxaliplatin has a different toxicity profile than either cisplatin or carboplatin with respect to myelosuppression. The development of a model that explains the synergistic cytotoxic interaction between fludarabine and oxaliplatin may aid in the design of more effective nucleoside analogues and novel platinum compounds with fewer side effects leading to better clinical outcomes. On the basis of this background, we have recently initiated a phase 1 study of oxaliplatin, fludarabine, cytarabine, and rituximab (OFAR) in patients with Richter transformation, prolymphocytic leukemia, or refractory or relapsed B-CLL.49
Authorship
Contribution: M.A.M. conceptualized the investigation, designed and conducted experiments, analyzed the data, and wrote the manuscript; D.S. designed and conducted experiments, analyzed the results, and wrote the manuscript; M.J.K. identified patients to participate and reviewed the manuscript; and W.P. conceptualized the investigation, designed the experiments, analyzed the results, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Prepublished online as Blood First Edition Paper, 9/5/2006; DOI 10.1182/blood-2006-05-023259.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank Min Du and Brenita Tyler for their assistance in obtaining blood samples and lymphocyte isolation, and Susan Lerner for providing information on patient characteristics and clinical laboratory evaluations.
This work was supported by grants from the National Cancer Institute, Department of Health and Human Services (CA81534 and Cancer Center support grant CA16672).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal