Abstract
The t(10;11) translocation results in a CALM-AF10 fusion gene in a subset of leukemia patients. Expression of a CALM-AF10 transgene results in leukemia, with prolonged latency and incomplete penetrance, suggesting that additional events are necessary for leukemic transformation. CALM-AF10 mice infected with the MOL4070LTR retrovirus developed acute leukemia, and ligation-mediated polymerase chain reaction was used to identify retroviral insertions at 19 common insertion sites, including Zeb2, Nf1, Mn1, Evi1, Ift57, Mpl, Plag1, Kras, Erg, Vav1, and Gata1. A total of 26% (11 of 42) of the mice had retroviral integrations near Zeb2, a transcriptional corepressor leading to overexpression of the Zeb2-transcript. A total of 91% (10 of 11) of mice with Zeb2 insertions developed B-lineage acute lymphoblastic leukemia, suggesting that Zeb2 activation promotes the transformation of CALM-AF10 hematopoietic precursors toward B-lineage leukemias. More than half of the mice with Zeb2 integrations also had Nf1 integrations, suggesting cooperativity among CALM-AF10, Zeb2, and Ras pathway mutations. We searched for Nras, Kras, and Ptpn11 point mutations in the CALM-AF10 leukemic mice. Three mutations were identified, all of which occurred in mice with Zeb2 integrations, consistent with the hypothesis that Zeb2 and Ras pathway activation promotes B-lineage leukemic transformation in concert with CALM-AF10.
Introduction
The rare but recurring chromosomal translocation t(10;11)(p12;q14) results in a CALM-AF10 fusion that is present in patients with both acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML).1-3 Using murine retroviral transduction and transplantation, Deshpande et al demonstrated that expression of a CALM-AF10 fusion led to AML with B-lymphoid characteristics.4 Subsequently, we demonstrated that mice that express a CALM-AF10 transgene in the hematopoietic compartment develop AML with lymphoid (both B-cell and T-cell) features.5 In general, these mice developed leukemia after a long latency period (median, 12 months) with incomplete penetrance (40%-50%). These findings suggest that additional genetic events are needed to complement the leukemogenic effect of the CALM-AF10 transgene, and are consistent with an emerging paradigm, which predicts that most, if not all, leukemic cells must undergo at least 2 collaborative oncogenic events to produce a malignant cell.6 As a consequence of these cumulative genetic and epigenetic events, the affected cell has a clonal advantage and progresses to a fully malignant cell.
Spectral karyotyping, comparative genomic hybridization, retroviral insertional mutagenesis, and targeted resequencing have been used to detect collaborative mutations associated with cancer.7,8 Retroviral insertional mutagenesis (RIM) is a whole-genome screening technique used to identify cancer-associated genes, including those involved in hematopoietic malignancies.9 Infection of murine hematopoietic cells with a retrovirus allows for integration of the viral DNA (provirus) into the host genome. Insertion of the provirus can lead to cellular transformation by several mechanisms, including activation of proto-oncogenes by viral long terminal repeats (LTRs), which can enhance transcription of nearby genes, or inactivation of tumor suppressor genes, through disruption of coding sequences.10 Dysregulation of proto-oncogenes and tumor suppressor genes results in a growth advantage and clonal expansion of transformed cells. Genes influenced by proviral integration can be identified through ligation-mediated polymerase chain reaction (LM-PCR) using viral sequence as PCR primers.
Because of the long latency period and incomplete penetrance observed in our initial CALM-AF10 study,5 we hypothesized that retroviral infection would accelerate the leukemia and allow us to identify genes that collaborate with the CALM-AF10 transgene. In addition, the 2 most differentially expressed genes in CALM-AF10 bone marrow (BM) are Meis1 and Hoxa95 (D.C., R.L.N., and P.D.A., unpublished data, June 2008). In this context, it is important to note that overexpression of HOXA cluster genes, particularly HOXA9 and MEIS1, is a common finding in both AML11 as well as B-lineage ALL associated with MLL fusions.12 Therefore, we speculated that the leukemias that develop in CALM-AF10 mice might identify genes that collaborate with other leukemias that overexpress MEIS1 and HOXA9, such as those associated with MLL fusions.
The MOL4070LTR is a recombinant retrovirus, derived from the Moloney murine leukemia virus and the 4070A virus, which has been shown to cause myeloid leukemia in approximately half of the infected mice, in contrast to Moloney murine leukemia virus, which is associated with a predominantly lymphoid phenotype.13 Using the MOL4070LTR virus, we show that retroviral insertional mutagenesis accelerates the onset of acute leukemia in CALM-AF10 transgenic mice and identifies genes that potentially collaborate with the CALM-AF10 fusion gene during leukemic transformation.
Methods
Retroviral infection
The MOL4070LTR retrovirus was propagated and virus harvested as described.13 Newborn CALM-AF10 transgenic and wild-type (WT) mice were inoculated intraperitoneally with 4 × 104 infectious particles. All animal experiments were conducted with the approval of the National Institutes of Health Intramural Animal Care and Use Committee.
Pathologic evaluation and immunophenotype
Mice were observed daily for signs of leukemia and killed when symptomatic. BM was harvested by flushing femora with Dulbecco medium containing 2% fetal calf serum (Invitrogen), hereafter referred to as HF2. Blood smears and cytospins were stained with May-Grunwald-Giemsa (Sigma-Aldrich). Tissues were fixed in 10% neutral buffered formalin (Sigma-Aldrich), paraffin embedded, sectioned, and stained with hematoxylin and eosin or incubated with the following monoclonal antibodies: antimyeloperoxidase (Dako North America), anti-CD3 (Dako North America), F4/80 (Caltag), and B220 (CD45R; BD Biosciences PharMingen). Single-cell suspensions from spleen and/or BM were incubated with fluorescein isothiocyanate-conjugated anti–mouse CD8, B220, and Gr1 or phycoerythrin-conjugated anti–mouse CD4 and CD11b (Mac1; BD Biosciences PharMingen) and analyzed by flow cytometry. Leukemias were classified according to the Bethesda proposals.14,15 Photomicrographs were taken on a BX51 Olympus microscope (Olympus America) with an Olympus DP12 camera system using 20× and 100× objectives and a 10× eyepiece objective. Digital images were imported into PowerPoint (Microsoft).
LM-PCR
Retroviral integration sites were cloned using LM-PCR.16 PCR products were purified (QIAGEN), ligated into pGEM-T Easy (Promega), and transformed into DH5α Escherichia coli cells (Invitrogen). Plasmid DNA was isolated and sequenced (Retrogen). Sequences were compared with National Center for Biotechnology Information mouse genome Build 36 (February 2006) using the UCSC genome browser (http://genome.ucsc.edu/).
Southern blot analysis
Genomic DNA from leukemic spleens was digested with EcoRI (for virus integrations or Igh gene rearrangements), XbaI (for Igh gene rearrangements), StuI (for Zeb2 gene rearrangements), BamHI or HindIII (for Nf1 gene rearrangements), size fractionated on agarose gels, denatured, neutralized, and transferred to nylon membranes (Schliecher & Schuell). The nylon membranes were hybridized (Ultrahyb; Ambion) to PCR-generated probes for either MOL4070LTR envelope, murine Zeb2, or murine Nf1. The murine Igh probe was a gift from Michael Kuehl (National Cancer Institute). Probes were labeled with 32P using Amersham Ready-To-Go DNA Labeling Beads (GE Healthcare). Primer sequences used to generate the murine Zeb2 and murine Nf1 probes are in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Western blot analysis
Spleens were lysed in RIPA (Radio-immunoprecipitation assay) buffer (1× Tris-buffered saline, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 0.004% sodium azide) supplemented with complete protease inhibitor cocktail (0.2mM phenylnethanesulphonyl fluoride and 0.1mM sodium orthovanidate; Santa Cruz Biotechnology) on ice for 30 minutes. Total protein (30 μg) was diluted in 2× sample buffer, boiled for 10 minutes, resolved on 6% and 10% Tris-glycine gels (Invitrogen), and transferred to nitrocellulose membranes (Invitrogen). Membranes were blocked with 1× Tris-buffered saline containing 0.1% Tween-20 (TBST) and 5% skim milk for 60 minutes at room temperature, and incubated with an anti-Nf1 murine monoclonal antibody (1:1000, McNFn27; Santa Cruz Biotechnology) or anti–α-tubulin rabbit polyclonal antibody (1:1000; Cell Signaling). Membranes were washed 3 times for 15 minutes in TBST at room temperature followed by a 60-minute incubation with horseradish peroxidase-conjugated anti–mouse or anti–rabbit antibodies (Pierce Chemical). Finally, the membranes were washed 3 times for 15 minutes each in TBST at room temperature and incubated with Enhanced Chemi-luminescence solution (Pierce Chemical). Band intensity was quantified with ImageJ software (National Institutes of Health).
Real-time quantitative PCR
Total RNA was isolated using TRIzol (Invitrogen); 1 μg was reverse transcribed with Superscript II reverse transcriptase and random hexamer primers (Invitrogen) in 20-μL reactions. Real-time reverse transcription PCR (RT-PCR) was performed on a 7500 Fast Real-Time TaqMan PCR system (Applied Biosystems) using aliquots of 1 μL from the first-strand cDNA as templates. TaqMan primer and probes sets were purchased from Applied Biosystems (supplemental Table 1). All reactions were in triplicate with 20-μL PCR reactions using Applied Biosystems default thermal cycling conditions. The −ΔCT mean and SE were calculated for each sample and normalized to the 18S rRNA value.
ZEB2 knockdown with siRNA
B-ALL1 (gift of David Chervinsky, Roswell Park Cancer Institute) and MV4-11 (ATCC) cell lines were cultured in Iscove modified Dulbecco medium (IMDM) media (Invitrogen) supplemented with 15% fetal bovine serum, 10 μg/mL penicillin/streptomycin, and 100mM l-glutamine (Invitrogen). A total of 3 × 105 MV4-11 or B-ALL1 cells were seeded in 100 μL of serum-free IMDM. A total of 1.5 μg of small interfering RNA (siRNA) specific to human ZEB2 or a nonspecific siRNA control (QIAGEN) weas mixed with 6 μL of HiPerFect transfection reagent (QIAGEN) in 100 μL of serum-free IMDM and incubated at room temperature for 10 minutes. The siRNA/lipid complexes were added to the cells and incubated at 37°C for 6 hours. Next, 600 μL of complete IMDM was added, and the cells were incubated at 37°C/5% CO2. At 24, 48, and 120 hours after transfection, cell numbers were determined by hemocytometer. Transfections were performed in triplicate, and the mean cell number and SD for each time point were determined. For quantitative RT-PCR analysis of ZEB2 expression, cells were harvested in Trizol (Invitrogen) at 24 hours after transfection and analyzed by quantitative RT-PCR as described in “Real-time quantitative PCR.”
Resequencing
Genomic DNA from leukemic spleens was amplified using primers listed in supplemental Table 1. PCR cycling conditions were: 94°C for 3 minutes, 40 cycles of 94°C for 30 seconds, 63°C (57°C for Kras exon 2 and 64.5°C for Nras exon 3) for 30 seconds, and 72°C for 60 seconds, followed by 72°C for 5 minutes. PCR products were purified using QIAGEN reagents and protocols and directly sequenced (Retrogen). Sequence chromatograms were manually evaluated for evidence of mutations.
Statistical analysis
Statistical significance for survival data was tested using the log-rank (Mantel-Cox) test; data for quantitative RT-PCR are expressed as mean plus or minus SD and were tested using an unpaired Student t test; data for relative expression by expression array are expressed as mean plus or minus SD, and groups are compared using an unpaired Student t test using GraphPad Prism, Version 5.00 for Windows (GraphPad Software). Data for genotype/phenotype and genotype/genotype correlations were tested using Fisher exact test.
Results
MOL4070LTR infection accelerates the onset of acute leukemia in CALM-AF10 transgenic mice
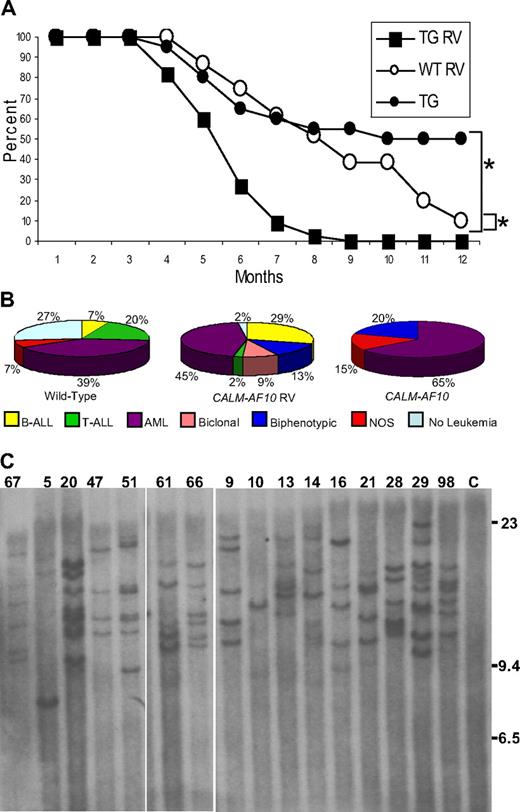
A total of 45 CALM-AF10 and 31 WT littermate control mice were infected at birth and monitored for signs of leukemia. Acute leukemia developed more rapidly in CALM-AF10–infected mice than in either the FVB WT mice infected with retrovirus (P < .001) or CALM-AF10 mice not infected with retrovirus (P < .001; Figure 1A). The mean latency of disease onset in CALM-AF10–infected mice was 5.5 months; by 9 months of age, all 45 transgenic mice either displayed symptoms of leukemia and were killed (41) or were found dead (4). Necropsy of leukemic mice typically revealed marked lymphadenopathy, hepatosplenomegaly, pale kidneys, and, in some cases, enlarged thymi.
MOL4070LTR infection accelerates leukemic transformation in CALM-AF10 mice. (A) Survival of MOL4070LTR-infected CALM-AF10 mice (■, N = 45), MOL4070LTR-infected WT (○, n = 31), and noninfected CALM-AF10 transgenic mice (●, N = 20). *P < .001, comparing infected CALM-AF10 with noninfected CALM-AF10, or infected CALM-AF10 with WT mice using a log-rank test. (B) Distribution of leukemia by immunophenotype in WT and CALM-AF10 mice infected with MOL4070LTR retrovirus. Note the predominance of B-cell and myeloid (AML) tumors, as well as the lack of pre-T LBL, in the CALM-AF10 mice compared with the WT control mice. (C) Retroviral integration analysis. Southern blot of EcoRI-digested DNA extracted from leukemic spleens hybridized to a viral env probe. Mouse identification numbers are indicated; C indicates germline control tissue; size standards are given in kilobases. The proviral genome is 8.7 kb and does not contain an EcoRI site.
MOL4070LTR infection accelerates leukemic transformation in CALM-AF10 mice. (A) Survival of MOL4070LTR-infected CALM-AF10 mice (■, N = 45), MOL4070LTR-infected WT (○, n = 31), and noninfected CALM-AF10 transgenic mice (●, N = 20). *P < .001, comparing infected CALM-AF10 with noninfected CALM-AF10, or infected CALM-AF10 with WT mice using a log-rank test. (B) Distribution of leukemia by immunophenotype in WT and CALM-AF10 mice infected with MOL4070LTR retrovirus. Note the predominance of B-cell and myeloid (AML) tumors, as well as the lack of pre-T LBL, in the CALM-AF10 mice compared with the WT control mice. (C) Retroviral integration analysis. Southern blot of EcoRI-digested DNA extracted from leukemic spleens hybridized to a viral env probe. Mouse identification numbers are indicated; C indicates germline control tissue; size standards are given in kilobases. The proviral genome is 8.7 kb and does not contain an EcoRI site.
CALM-AF10 mice develop B-lineage ALL and AML
CALM-AF10 mice infected with MOL4070LTR developed either acute lymphoid (B-cell or T-cell), myeloid, biphenotypic (myeloid and B-cell), or biclonal (myeloid and B-cell) leukemia, as determined by histologic and immunophenotypic analysis. Of the 4 mice that were found dead in their cages, 1 mouse did not have evidence of leukemia (mouse 52) and 2 mice (2 and 32) had leukemia by histology, but DNA was degraded and unsuitable for Southern blots or LM-PCR. Therefore, a total of 42 CALM-AF10 mice infected with MOL4070LTR were available for analysis. Leukemic blasts were typically evident in peripheral blood, spleen, BM, lymph nodes, liver, and kidney. The Bethesda proposals14,15 were used to classify the leukemias; in some cases, biclonal and biphenotypic diagnoses were applied. Biclonal leukemias represent 2 distinct clones with cell populations that express distinct surface antigens, which are not expressed on cells from the other clone. This is in contrast to biphenotypic leukemias, which are thought to arise in a multipotent progenitor cell capable of differentiating into both myeloid and lymphoid lineages. As shown in Figure 1, of the 42 mice, there was a smaller percentage of pre-T lymphoblastic leukemia/lymphoma (pre-T LBL; 2%), and a larger proportion of B-ALL (29%), biphenotypic (13%), and biclonal leukemias (9%) in CALM-AF10–infected mice compared with WT-infected mice. Leukemic spleens from these mice were also evaluated for clonal Igh gene rearrangements; 23 of 42 mice had clonal Igh rearrangements (supplemental Figure 1). Taken together, these findings indicate that CALM-AF10 mice infected with retrovirus developed a broader spectrum of leukemic phenotypes than did CALM-AF10 mice not infected with retrovirus, which developed predominantly myeloid leukemia.5
Clonality of leukemias in CALM-AF10 mice infected with retrovirus
Leukemic samples from infected CALM-AF10 mice were evaluated using Southern blots to determine clonality of the leukemias. As shown in Figure 1, between 1 and 9 integrations per tumor were detected (4.4 ± 0.5 integrations for the WT mice and 5.7 ± 0.5 integrations for the CALM-AF10 mice, P > .05), indicating that the tumors were clonal or oligoclonal, but not polyclonal. These findings suggest several possibilities. A single dominant band (such as mouse 10, Figure 1C) suggests that a clonal leukemia developed from a single integration event. A second pattern was seen in mouse 9 (Figure 1C), which had 5 unique bands, all of equal intensity, suggesting that the leukemia was composed of 1 major clone with 5 insertions. In contrast, mouse 13 developed a leukemia that contains 2 prominent bands and 3 less intense bands; this leukemia is probably oligoclonal, composed of a major clone (represented by the 2 intense bands) and a minor clone (represented by the 3 fainter bands).
LM-PCR identifies common retrovirus insertion sites
Using LM-PCR, a total of 262 unique insertions were identified in 40 mice (supplemental Table 2). Nineteen common insertion sites (CISs), defined as 2 or more insertion within a 100-kb window,17 were identified and are listed in Table 1. Nonrecurrent single integration sites that are of interest because of the function of nearby genes were also noted (supplemental Table 2).
Common integration sites in CALM-AF10 mice infected with the MOL4070LTR retrovirus
| Gene . | Incidence . | Occurrence in RTCGD* . | Mouse ID . |
|---|---|---|---|
| Nf1 | 12† | 30 | 13, 17, 23, 25, 26, 31, 36, 40, 42, 44, 96, 98 |
| Zeb2 | 11† | 14 | 5, 17, 21, 23, 25, 26, 27, 36, 51, 80, 98 |
| Mn1 | 6 | 12 | 25, 38, 39, 46, 67, 80 |
| Evi1 | 5 | 19 | 6, 14, 29, 31, 36 |
| Mllt3 | 3 | 2‡ | 14, 47, 69 |
| Plag1 | 2 | 8 | 21, 42 |
| Mpl | 2 | 0‡ | 10, 80 |
| Prkcz | 2 | 0‡ | 17, 31 |
| CX763467 | 2 | 0‡ | 25, 42 |
| Foxp1 | 2 | 1‡ | 35, 88 |
| Kras | 2 | 8 | 20, 27 |
| Rras2 | 2 | 35 | 39, 98 |
| Tle3 | 2 | 4‡ | 40, 69 |
| Phb | 2 | 1‡ | 69, 96 |
| Ift57/Cd47 | 2 | 2‡ | 36, 69 |
| Erg | 2 | 6 | 6, 69 |
| Vav1 | 2 | 0‡ | 15, 74 |
| Gata1 | 2 | 0‡ | 14, 47 |
| Cybb | 2 | 0‡ | 40, 8 |
| Gene . | Incidence . | Occurrence in RTCGD* . | Mouse ID . |
|---|---|---|---|
| Nf1 | 12† | 30 | 13, 17, 23, 25, 26, 31, 36, 40, 42, 44, 96, 98 |
| Zeb2 | 11† | 14 | 5, 17, 21, 23, 25, 26, 27, 36, 51, 80, 98 |
| Mn1 | 6 | 12 | 25, 38, 39, 46, 67, 80 |
| Evi1 | 5 | 19 | 6, 14, 29, 31, 36 |
| Mllt3 | 3 | 2‡ | 14, 47, 69 |
| Plag1 | 2 | 8 | 21, 42 |
| Mpl | 2 | 0‡ | 10, 80 |
| Prkcz | 2 | 0‡ | 17, 31 |
| CX763467 | 2 | 0‡ | 25, 42 |
| Foxp1 | 2 | 1‡ | 35, 88 |
| Kras | 2 | 8 | 20, 27 |
| Rras2 | 2 | 35 | 39, 98 |
| Tle3 | 2 | 4‡ | 40, 69 |
| Phb | 2 | 1‡ | 69, 96 |
| Ift57/Cd47 | 2 | 2‡ | 36, 69 |
| Erg | 2 | 6 | 6, 69 |
| Vav1 | 2 | 0‡ | 15, 74 |
| Gata1 | 2 | 0‡ | 14, 47 |
| Cybb | 2 | 0‡ | 40, 8 |
Data from retroviral insertion studies in hematologic malignancies include 2783 total insertions.
Includes integrations identified by Southern blot analysis.
Not previously identified as a common integration site.
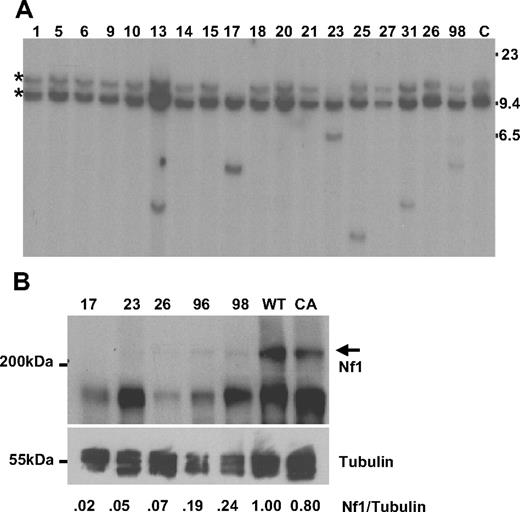
Retroviral integrations were identified within intron 36 of the tumor suppressor gene Nf1 in 4 mice by LM-PCR (17, 23, 36, and 98). The retroviral integrations determined by LM-PCR were confirmed by Southern blot analysis, and 8 additional integrations were identified (13, 25, 26, 31, 40, 42, 44, and 96, see representative examples in Figure 2A). Of note, there was loss of the germline allele in mice 17 and 23; this reduction to homozygosity, with loss of the WT Nf1 allele, is similar to the uniparental disomy seen in some NF1 patients with AML.18 Given the known role of Nf1 as a tumor suppressor gene, the observation that these insertions occurred far away from the Nf1 promoter, and the loss of theWT allele in some samples, we anticipated that Nf1 mRNA would be decreased in these mice. However, quantitative RT-PCR assays showed no consistent decrease of Nf1 mRNA expression in samples with Nf1 insertions; therefore, we evaluated the expression of Nf1 protein by Western blot in 5 mice with Nf1 integrations. As shown in Figure 2B, Nf1 protein expression was markedly decreased or absent compared with Nf1 protein expression in either WT or CALM-AF10 transgenic spleen.
Nf1 analysis. (A) Southern blot of BamHI-digested DNA from leukemic spleens hybridized to an Nf1 probe; 2 germline bands are seen as the probe contains a BamH1 site. Mouse identification numbers are indicated; C indicates germline control tissue; size standards are given in kilobases. *Germline bands. Note loss of the germline allele in mice 17 and 23. (B) Protein lysates from leukemic spleens with a known Nf1 CIS were compared with spleens from clinically healthy CALM-AF10 and WT mice. The Nf1 protein is indicated with  ; sizes are in kDa. The blot was reprobed with anti–α-tubulin as a loading control. Intensity of the Nf1 and α-tubulin signals were quantified with ImageJ software, and the ratio compared with that of the WT mouse.
; sizes are in kDa. The blot was reprobed with anti–α-tubulin as a loading control. Intensity of the Nf1 and α-tubulin signals were quantified with ImageJ software, and the ratio compared with that of the WT mouse.
Nf1 analysis. (A) Southern blot of BamHI-digested DNA from leukemic spleens hybridized to an Nf1 probe; 2 germline bands are seen as the probe contains a BamH1 site. Mouse identification numbers are indicated; C indicates germline control tissue; size standards are given in kilobases. *Germline bands. Note loss of the germline allele in mice 17 and 23. (B) Protein lysates from leukemic spleens with a known Nf1 CIS were compared with spleens from clinically healthy CALM-AF10 and WT mice. The Nf1 protein is indicated with  ; sizes are in kDa. The blot was reprobed with anti–α-tubulin as a loading control. Intensity of the Nf1 and α-tubulin signals were quantified with ImageJ software, and the ratio compared with that of the WT mouse.
; sizes are in kDa. The blot was reprobed with anti–α-tubulin as a loading control. Intensity of the Nf1 and α-tubulin signals were quantified with ImageJ software, and the ratio compared with that of the WT mouse.
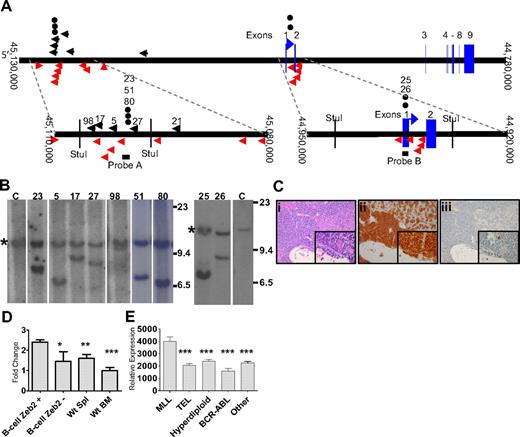
Zeb2 integrations were identified in 6 CALM-AF10 mice (5, 17, 21, 27, 36, and 98) by LM-PCR, as indicated in Figure 3A. This gene has been identified 14 times of 2783 retroviral integrations in mouse hematopoietic tumors reported in the Retroviral Tagging Cancer Genome Database (RTCGD; http://rtcgd.abcc.ncifcrf.gov/). Five of the 6 Zeb2 integrations we identified by LM-PCR occurred within a 12-kb cluster 5′ of Zeb2 (Figure 3A). To determine whether the Zeb2 integrations occurred in a major clone, as well as to identify additional Zeb2 integrations that might have been missed by LM-PCR, we used Southern blot analysis with a probe from the aforementioned 12-kb cluster (Figure 3A probe A). We were able to confirm the insertion in all 4 mice that had integrations within the range of this assay. In 3 of 4 mice (5, 17, and 27), the rearranged allele (Figure 3B) was of similar intensity to the germline allele, indicating that the leukemic clone, which contained the Zeb2 integration was the major, if not sole, leukemic clone in those mice. The Zeb2 rearranged band from mouse 98 was fainter than the germline band, suggesting that this leukemia represented a minor clone. Three additional leukemias (23, 51, and 80), which had not been identified by LM-PCR, were shown to have Zeb2 integrations by Southern blot.
Zeb2 integration analysis. (A) Mouse chromosome 2. Insertions identified by LM-PCR are indicated by black arrows. Insertion events reported in the RTCGD are indicated by red arrows. The vertical red arrow represents an insertion event for which the integration orientation was not reported in the RTCGD. (●) Insertion events identified by Southern blot. Probes A and B are indicated by black bars. Numbers above black arrows and circles represent mice with Zeb2 intregrations. (B) Southern blot of StuI-digested genomic DNA from infiltrated spleen hybridized to a Zeb2 probe. Mouse identifications are indicated; C indicates germline control tissue; size standards are given in kilobases. *Germline bands. Note the equal intensity of germline and rearranged bands indicating that the leukemias were predominately clonal, except for mouse 98. (C) Acute lymphoid leukemia (B-ALL) in CALM-AF10 mouse 17 infected with MOL4070LTR. Hematoxylin and eosin–stained liver (Ci inset), B220 stained liver (Cii inset), and myeloperoxidase-stained liver (Ciii inset). (D) mRNA levels determined by quantitative PCR of leukemic spleens with Zeb2 retrovirus insertions (N = 9). Zeb2 expression was normalized to the 18S ribosomal control and compared with WT spleen (N = 5 independent samples), WT bone marrow (N = 5 independent samples), or B-lineage ALL samples without Zeb2 integrations (N = 4, mice 8, 14, 38, and 39). *P < .03. **P < .01. ***P < .001. (E) Relative ZEB2 expression in pediatric B-lineage leukemias determined by expression array from supplemental data reported by Yeoh et al (http://www.stjuderesearch.org/data/ALL1).20 Asterisks denote comparison of each group to the MLL group; for all comparisons, P < .001.
Zeb2 integration analysis. (A) Mouse chromosome 2. Insertions identified by LM-PCR are indicated by black arrows. Insertion events reported in the RTCGD are indicated by red arrows. The vertical red arrow represents an insertion event for which the integration orientation was not reported in the RTCGD. (●) Insertion events identified by Southern blot. Probes A and B are indicated by black bars. Numbers above black arrows and circles represent mice with Zeb2 intregrations. (B) Southern blot of StuI-digested genomic DNA from infiltrated spleen hybridized to a Zeb2 probe. Mouse identifications are indicated; C indicates germline control tissue; size standards are given in kilobases. *Germline bands. Note the equal intensity of germline and rearranged bands indicating that the leukemias were predominately clonal, except for mouse 98. (C) Acute lymphoid leukemia (B-ALL) in CALM-AF10 mouse 17 infected with MOL4070LTR. Hematoxylin and eosin–stained liver (Ci inset), B220 stained liver (Cii inset), and myeloperoxidase-stained liver (Ciii inset). (D) mRNA levels determined by quantitative PCR of leukemic spleens with Zeb2 retrovirus insertions (N = 9). Zeb2 expression was normalized to the 18S ribosomal control and compared with WT spleen (N = 5 independent samples), WT bone marrow (N = 5 independent samples), or B-lineage ALL samples without Zeb2 integrations (N = 4, mice 8, 14, 38, and 39). *P < .03. **P < .01. ***P < .001. (E) Relative ZEB2 expression in pediatric B-lineage leukemias determined by expression array from supplemental data reported by Yeoh et al (http://www.stjuderesearch.org/data/ALL1).20 Asterisks denote comparison of each group to the MLL group; for all comparisons, P < .001.
Because there was a second, smaller cluster of viral integrations in the RTCGD surrounding the first exon of Zeb2 (Figure 3A), we speculated that there might be additional integrations in this region that were not identified by LM-PCR. A second Southern blot analysis was performed using a probe that assessed a region encompassing the first exon of Zeb2, and 2 additional mice (25 and 26) with Zeb2 integrations were identified. In sum, 11 of 42 (26%) mice had Zeb2 integrations, and in 8 of the 9 evaluable cases (integrations for mice 21 and 36 occurred outside of the regions assayed by the probes), the Zeb2 integration was present in the major leukemic clone. In contrast, we detected only 1 integration (in a very minor clone) in 15 WT mice infected with the MOL4070LTR (supplemental Figure 3), and Zeb2 integrations have been identified in only 14 of 2783, or less than 0.4%, of the integration sites deposited in the RTCGD (P < .001). Ten of 11 (89%) mice with a Zeb2 integration had B-cell ALL (a representative example is shown in Figure 3C) or biphenotypic leukemia with clonal Igh gene rearrangements. In contrast, only 13 of 33 (39%) leukemic mice without Zeb2 integrations had B-cell features (B-cell ALL, biphenotypic, or biclonal; P > .004).
Given that the leukemias we detected in this study were B-ALL, AML, biphenotypic, and biclonal leukemias, although BM alone may be a reasonable comparison for myeloid leukemias, it may not be an adequate comparison tissue for B-lineage leukemias. Therefore, in these and subsequent experiments, we present data for both WT BM and spleen as controls. Expression of Zeb2 in the leukemic samples was 2- to 3-fold higher than in WT spleen, WT BM, or B-lineage leukemias without Zeb2 integrations (Figure 3D), and there was no difference in Zeb2 expression between the upstream cluster (mean relative expression, 2.48; N = 7) and the downstream cluster (mean relative expression, 2.13; N = 2). Interestingly, ZEB2 was also up-regulated in MLL rearranged pediatric B-cell precursor ALLs (which, similar to CALM-AF10 fusions, also overexpress HOXA cluster genes) compared with other groups of pediatric B-cell precursor ALL (Figure 3E).19,20
Because the upstream cluster of integrations was approximately 140 kb 5′ of Zeb2, we considered the possibility that integrations at this cluster might affect another gene in addition to Zeb2. Based on National Center for Biotechnology Information mouse genome Build 37 (July 2007 Assembly), there was a spliced EST (BB633916) and a noncoding RNA (MMU56439) located near the upstream cluster (supplemental Figure 4A). We could not detect expression of the spliced EST (BB633916) using 4 sets of PCR primers (data not shown). Although the noncoding RNA (MMU56439) was expressed in all samples analyzed, its expression was not significantly higher in samples with Zeb2 integrations than without Zeb2 integrations (supplemental Figure 4B). These observations, together with the fact that Zeb2 mRNA was up-regulated in leukemias from both Zeb2 integration clusters, as well as the similarity in leukemic phenotype in mice from both Zeb2 integration clusters, indicates that Zeb2 is the target for these integrations.
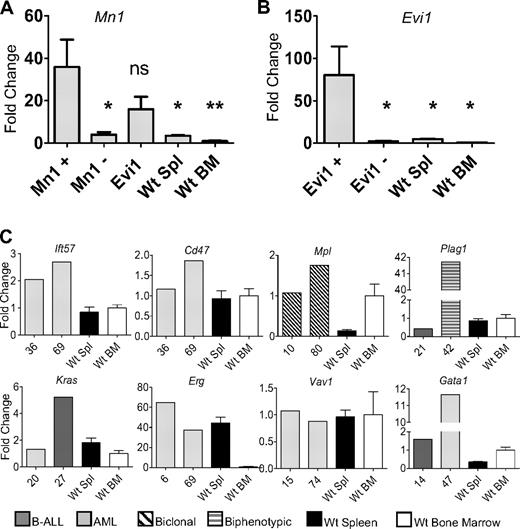
Mn1 insertions were identified in 6 mice (25, 38, 39, 46, 67, and 80). Three insertions were located in Mn1 intron 1, all in the forward direction. These 3 insertions occurred in the same intron as MN1-TEL fusions, which have been associated with AML.21 RT-PCR analysis revealed an in-frame Mn1-pol retroviral fusion transcript in all 3 mice with the intron 1 insertion (supplemental Figure 2). This fusion transcript encodes a protein that fuses the Mn1 N-terminus to the final 115-amino acid residues of the integrase peptide, encoded by the viral pol gene (accession no. AAC98548).16 To determine whether Mn1 mRNA levels were up-regulated by the insertions, we compared Mn1 levels in leukemic spleens from mice with Mn1 insertion to WT BM and spleen. There was a dramatic increase in Mn1 expression compared with WT spleen or BM (Figure 4A). In addition, we compared Mn1 levels in leukemic spleens from mice with Mn1 insertions with those without Mn1 insertions. As expected, mice without Mn1 insertions had lower levels of Mn1 expression, comparable with WT spleen or BM. Interestingly, mice without Mn1 insertions, but with Evi1 insertions, also had elevated levels of Mn1 expression (Figure 4A). These findings are consistent with reports that AML patients with EVI1 overexpression have increased expression of MN1.22
Increased expression of CIS genes. (A) Expression levels of Mn1 were evaluated in mice with Mn1 insertions (N = 4), mice without Mn1 or Evi1 insertions (N = 5), mice with Evi1 insertions but without Mn1 insertions (N = 5), WT spleen (N = 5), and WT BM (N = 5). *P < .03. **P < .01. ns indicates not significant. (B) Expression levels of Evi1 in mice with Evi1 insertions (N = 5), mice without Evi1 insertions (N = 4), WT spleen (N = 5), and WT BM (N = 5). (C) Evaluation of 8 additional CIS (Plag1, Mpl, Kras, Erg, Ift57, Cd47, Vav1, and Gata1) compared with either WT bone marrow (N = 5) or WT spleen (N = 5). Expression of each gene was determined by quantitative RT-PCR and normalized to the 18S ribosomal RNA.
Increased expression of CIS genes. (A) Expression levels of Mn1 were evaluated in mice with Mn1 insertions (N = 4), mice without Mn1 or Evi1 insertions (N = 5), mice with Evi1 insertions but without Mn1 insertions (N = 5), WT spleen (N = 5), and WT BM (N = 5). *P < .03. **P < .01. ns indicates not significant. (B) Expression levels of Evi1 in mice with Evi1 insertions (N = 5), mice without Evi1 insertions (N = 4), WT spleen (N = 5), and WT BM (N = 5). (C) Evaluation of 8 additional CIS (Plag1, Mpl, Kras, Erg, Ift57, Cd47, Vav1, and Gata1) compared with either WT bone marrow (N = 5) or WT spleen (N = 5). Expression of each gene was determined by quantitative RT-PCR and normalized to the 18S ribosomal RNA.
Five mice (6, 14, 29, 31, and 36) had insertions that were identified within intron 1 of Evi1; all of the insertions were determined to be in the opposite orientation of the gene. Four of the 5 mice had a marked increase in Evi1 mRNA compared with either WT spleen or BM (Figure 4B).
Two mice (36 and 69) had insertions that potentially affected the expression of Ift57 or Cd47. The integrations were 24 or 35 kb 3′ of Ift57 and 66 or 55 kb 5′ of Cd47. Because these insertions were located in close proximity to both Ift57 and Cd47, we considered the possibility that expression of either gene could be influenced by the insertion event. As shown in Figure 4, gene expression for Ift57 was elevated in both samples compared with WT spleen and BM, whereas Cd47 expression was similar to that of WT BM or spleen.
Insertion events 15-kb 5′ of Plag1 were identified in 2 mice (21 and 42), and there was a 40-fold increase in Plag1 expression for mouse 42 compared with WT BM or spleen (Figure 4). Two mice (10 and 80) were identified with insertion events 631 or 933 bp 5′ of the thrombopoietin receptor, Mpl, and were associated with modest increases in Mpl mRNA (Figure 4). Kras was identified as a CIS in 2 mice (20 and 27) and was up-regulated in 1 of the 2 mice (27, Figure 4). Retroviral insertions in the second intron of Erg were identified in 2 mice (6 and 69) and were associated with a 40- to 60-fold mRNA increase compared with WT BM (Figure 4). Two mice (14 and 47) had Gata1 insertions, either within the first intron (mouse 14) or 2.7 kb 5′ of the transcript initiation site (mouse 47); Gata1 expression was markedly elevated in mouse 47 (Figure 4).
Meis1 has commonly been identified by RIM, and Meis1 insertions were identified in 13 of 32 samples in a previous study using the MOL4070LTR virus.16 However, only a single Meis1 insertion was identified in this current study. Therefore, we screened the 42 leukemia samples for Meis1 insertion using Southern blot analysis, as previously described,16 but no additional Meis1 insertions were identified (data not shown).
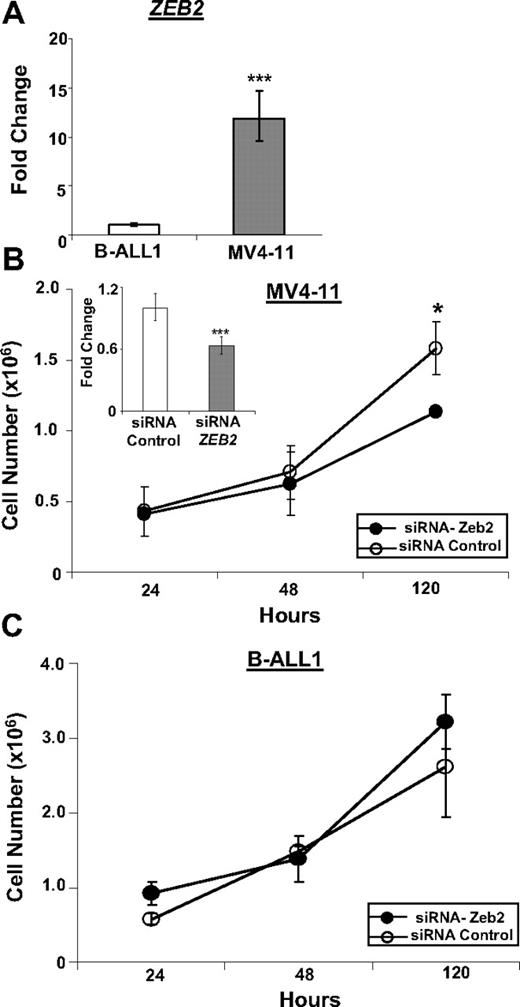
ZEB2 knockdown leads to decreased proliferation of the MV4-11 cell line
Because we know of no B-lineage cell lines that express a CALM-AF10 fusion, we studied MV4-11 cells,23 a human B-cell precursor ALL cell line that expresses an MLL-AF4 fusion, which, similar to the CALM-AF10 fusion, leads to up-regulation of HOXA9 and MEIS1, as a surrogate. To assess off-target effects of the ZEB2 siRNA in human B cells, MV4-11 cells were compared with B-ALL1 cells,24 a human B-cell precursor ALL cell line that does not express an MLL-AF4 fusion. Similar to the results shown in Figure 3E, ZEB2 was markedly overexpressed in MV4-11 cells compared with BALL-1 cells (Figure 5A). We used a ZEB2 siRNA to inhibit expression of ZEB2 in the MV4-11 cells, leading to decreased proliferation of MV4-11 cells, but not B-ALL1 cells, suggesting that ongoing ZEB2 expression was important for proliferation of the MV4-11 cells, but not the B-ALL1 cells (Figure 5B-C).
Inhibition of ZEB2 leads to decreased proliferation of MV4-11 cells. (A) Expression of ZEB2 mRNA in MV4-11 and B-ALL1 cell lines. (B) Inhibition of ZEB2 mRNA expression in MV4-11 cell line by siRNA leads to decreased proliferation. (Inset) ZEB2 quantitative RT-PCR assay at 24 hours. (C) Treatment of B-ALL1 cell line with ZEB2 siRNA does not lead to decreased proliferation. Quantitative RT-PCR values are plotted as minimum/maximum values from triplicate assays. *P < .03. ***P < .001. Error bars for 120-hour Zeb2 siRNA time point are very small and not visible.
Inhibition of ZEB2 leads to decreased proliferation of MV4-11 cells. (A) Expression of ZEB2 mRNA in MV4-11 and B-ALL1 cell lines. (B) Inhibition of ZEB2 mRNA expression in MV4-11 cell line by siRNA leads to decreased proliferation. (Inset) ZEB2 quantitative RT-PCR assay at 24 hours. (C) Treatment of B-ALL1 cell line with ZEB2 siRNA does not lead to decreased proliferation. Quantitative RT-PCR values are plotted as minimum/maximum values from triplicate assays. *P < .03. ***P < .001. Error bars for 120-hour Zeb2 siRNA time point are very small and not visible.
Ras pathway resequencing identifies spontaneous mutations
We hypothesized that the frequent Nf1 insertion resulted in Ras pathway activation. Given that point mutations of NRAS, KRAS, PTPN11, or FLT3 are common in AML25 and ALL26 patients, we evaluated CALM-AF10 retroviral-infected leukemic mice for spontaneous activating mutations in these genes. We sequenced Nras and Kras exons 2/3 in all 42 mice and found 2 mutations: a codon 12 GGT to GAT (Gly to Asp) mutation in mouse 25 and a codon 13 GGT to GAT (Gly to Asp) mutation in mouse 27 (Table 2). We also sequenced Ptpn11 exon 3 and Flt3 exon 20 in all 42 mice. We found 1 spontaneous point mutation in Ptpnll exon 3 in mouse 5 (supplemental Figure 5), a codon 72 GCC to GTC (Ala to Val) mutations, which has also been found in human ALL,26 AML,27 and juvenile myelomonocytic leukemia (JMML).28 No spontaneous point mutations were found in the tyrosine kinase domain of Flt3. Remarkably, all of the Ras pathway point mutations occurred in mice with Zeb2 insertions. In total, 8 of 11 mice with Zeb2 insertions also had Nf1 insertions or Ras pathway activating mutations, whereas only 6 of 31 mice without Zeb2 insertions had Nf1 insertions, and none of these mice had point mutations. This difference was highly significant (P < .003; Table 2).
Nf1 insertions and Ras pathway muations in CALM-AF10 mice with Zeb2 insertions
| ID . | Diagnosis . | IgH rearrangement . | Proviral insertion . | Gene mutation . |
|---|---|---|---|---|
| 5 | B-ALL | Yes | — | Ptpn11 |
| 17 | B-ALL | Yes | Nf1 | — |
| 21 | B-ALL | Yes | — | — |
| 23 | B-ALL | Yes | Nf1 | — |
| 25 | B-ALL | Yes | Nf1 | Kras |
| 26 | B-ALL | Yes | Nf1 | — |
| 27 | B-ALL | Yes | — | Kras |
| 36 | AML | No | Nf1 | — |
| 51 | Biphenotypic | Yes | — | — |
| 80 | Biphenotypic | Yes | — | — |
| 98 | B-ALL | Yes | Nf1 | — |
| ID . | Diagnosis . | IgH rearrangement . | Proviral insertion . | Gene mutation . |
|---|---|---|---|---|
| 5 | B-ALL | Yes | — | Ptpn11 |
| 17 | B-ALL | Yes | Nf1 | — |
| 21 | B-ALL | Yes | — | — |
| 23 | B-ALL | Yes | Nf1 | — |
| 25 | B-ALL | Yes | Nf1 | Kras |
| 26 | B-ALL | Yes | Nf1 | — |
| 27 | B-ALL | Yes | — | Kras |
| 36 | AML | No | Nf1 | — |
| 51 | Biphenotypic | Yes | — | — |
| 80 | Biphenotypic | Yes | — | — |
| 98 | B-ALL | Yes | Nf1 | — |
Eight of 11 mice with Zeb2 insertions also had Nf1 insertions and/or Ras pathway activating point mutations. Only 6 of 31 mice without Zeb2 insertions have Nf1 insertions, and none of these mice had Ras pathway activating point mutations (P < .003).
— indicates no Nf1 insertion or gene mutation was identified.
Discussion
CALM-AF10 transgenic mice develop leukemia after a long latency period with incomplete penetrance, suggesting the need for complementary events.5 In contrast, infection of mouse BM with a CALM-AF10 retrovirus led to AML in 100% of mice after a short latency.4 The AMLs in that study were clonal, as demonstrated by clonal Igh gene rearrangements, suggesting the possibility that a cooperative event had been introduced by the retrovirus used to carry the CALM-AF10 cDNA. Although the authors did not detect any CIS among the 18 retroviral insertions cloned in that study, it remains possible that collaborating insertions were not identified, or that the insertions identified, even though not known CIS, collaborated with the CALM-AF10 fusion to produce AML. We used retroviral insertional mutagenesis to identify candidate genes that could complement a CALM-AF10 fusion.
The most common retroviral integration in CALM-AF10 mice was located within intron 36 of Nf1. Neurofibromatosis type 1 is an autosomal disorder caused by inactivating mutations of NF1.29 Children with NF1 are at increased risk for developing malignant myeloid disease, including AML and JMML, which is often associated with monosomy 7, suggesting a collaboration between monosomy 7 and NF1.30,31 NF1 encodes a protein that serves as a GTPase-activating protein which accelerates Ras-GTP hydrolysis, converting Ras-GTP to its inactive form Ras-GDP.32 Several lines of evidence emphasize the role of Nf1 as a tumor suppressor gene. Nf1+/− heterozygous mice developed a myeloproliferative disease, in year 2 of life, which was associated with LOH at the Nf1 locus in myeloid cells,33 similar to findings in patients with JMML. Transplantation of Nf1−/− fetal liver cells leads to a myeloproliferative syndrome that resembles JMML.34 Finally, analysis of retroviral integration sites in BXH2 mice that developed leukemias identified common integrations within Nf1 intron 36, often with loss of Nf1 protein expression.35
In the current series, Nf1 was identified as a CIS in 12 of 42 mice. The leukemic cells from 2 mice (17 and 23) demonstrated loss of the germline Nf1 allele, consistent with the known role of Nf1 as a tumor suppressor gene. Similar to previous reports, although Nf1 mRNA expression was consistently detected in mice with Nf1 insertions, Nf1 protein expression was absent or markedly decreased. Given the frequent association of NF1 gene inactivation and monosomy 7, which itself is associated with marked overexpression of HOXA9,36 it is interesting to note that Hoxa9 expression is also markedly elevated in BM from CALM-AF10 mice,5 supporting the hypothesis that Nf1 inactivation collaborates with Hoxa9 gene activation.
Zeb2 (Sip1 or Zfhx1b) was identified as a CIS in 11 of 42 mice. However, Zeb2 has only rarely been identified in previous RIM studies and has not previously been implicated in leukemic transformation. Much of what is known about ZEB2 function stems from studies in patients with Mowat-Wilson syndrome,37 a developmental disorder of neural crest-derived tissue that results from germline mutations of ZEB2.38 Interestingly, several lines of evidence are consistent with the hypothesis that ZEB2 may be involved in malignant transformation. The ZEB2 protein interacts with several members of the SMAD protein family, including SMAD1, SMAD2, SMAD3, and SMAD539 ; SMAD proteins have been shown to play important roles in normal and malignant hematopoiesis.40 In addition, Zeb2 has been shown to act upstream of Wnt signaling,41 and recent evidence implicates Wnt signaling in hematopoietic stem cell self-renewal and progenitor development,42 including B lymphocyte proliferation.43 Furthermore, overexpression of ZEB2 in primary epithelial cells promotes epithelial-mesenchymal transition, a transdifferentiation process observed in breast cancer and renal cell carcinoma.44 Finally, a preliminary report indicated that ZEB2 was overexpressed in leukemia patients with MLL gene rearrangements.45
Zeb2 integrations were present in the dominant leukemic clone in most of the mice with this CIS. In addition, mouse 5 had a single retroviral integration in the dominant clone (Figure 1); this integration involved Zeb2 (Figure 3), strongly suggesting that the Zeb2 integrations were important for leukemic transformation, and were not merely “passengers.” Furthermore, we noted that most of the mice with Zeb2 insertions developed B-cell or biphenotypic leukemia, suggesting that Zeb2 collaborates with CALM-AF10 to promote B-cell leukemia. The only mouse (36) that had neither a B-cell phenotype nor a clonal Igh gene rearrangement did not overexpress Zeb2, and had an integration site outside of the region assayed by either of the Southern blot probes. Therefore, it is possible that the leukemic clone containing a Zeb2 integration from mouse 36 represented a minor clone, or that integration outside of the 2 clusters did not lead to overexpression of Zeb2. The mice with Zeb2 integrations had increased expression of Zeb2 at the mRNA level compared with WT spleen. Increased expression of ZEB2 is also present in pediatric ALLs with rearranged MLL compared with other pediatric ALLs. The high incidence of Zeb2 insertions in our CALM-AF10 model and the consistent overexpression of ZEB2 in MLL rearranged ALLs suggest that increased ZEB2 may collaborate with HOXA9/MEIS1 overexpression in ALL patients.
Mn1 was identified 6 times as a CIS. MN1 forms a fusion transcript with TEL in some AML patients,21 and MN1 overexpression is associated with a poor response to induction chemotherapy, increased relapse rate, and decreased overall survival.46 In addition, Mn1 was reported as a CIS in NUP98-HOXD13 transgenic mice, which overexpress Hoxa7, 9, and 10 in the BM, and a MN1-TEL fusion has been shown to collaborate with HOXA9 to induce AML in a murine BM transplantation model.47 Given that Hoxa9 is up-regulated in CALM-AF10 mouse BM, these findings collectively suggest a collaborative role between overexpression of Hoxa9 and Mn1 in leukemic transformation.
Activating point mutations in Ras pathway genes developed spontaneously in mice with 2 additional oncogenic events (CALM-AF10 transgene and retroviral insertion), reinforcing the importance of Ras pathway activation in leukemogenesis. In addition, there is a remarkably strong correlation between Ras pathway activation, via Nf1 inactivation or spontaneous point mutation (Kras or Ptpn11), and Zeb2 activation via proviral integration in our study.
Of note, we did not identify a CIS near Meis1, a transcription factor that collaborates with HOX genes during leukemic transformation.48,49 Meis1 has often been identified as a CIS through a retroviral mutagenesis screen in mice50 and was identified as a CIS in 14 of 32 NUP98HOXD13 transgenic mice that were infected with the MOL4070LTR.16 However, we previously demonstrated that Meis1 was markedly overexpressed in BM from CALM-AF10 mice.5 Therefore, there may be no selective advantage for Meis1 insertions in CALM-AF10 mice because CALM-AF10 mice without Meis1 insertions overexpress Meis1.
Although CALM-AF10 fusions are rare, these translocations share many important biologic features with MLL fusions, including a tendency to produce both myeloid and lymphoid malignancy. Using retroviral insertional mutagenesis and targeted resequencing, we have identified potential collaborative oncogenic pathways that complement CALM-AF10 fusions, and, by extension, similar translocations during leukemic transformation. The frequent correlation of Zeb2 integrations and Ras pathway activating mutations in CALM-AF10 mice suggests that Zeb2 and Ras pathway activation collaborate with CALM-AF10 (and potentially other leukemic fusions that activate HOXA cluster genes) to induce leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Helge Hartung, Sarah Beachy, Sheryl Gough, Yang Jo Chung, R. Mark Simpson, and Siba Samal for insightful discussions and Christine Perella for expert technical assistance with mouse retroviral infection and necropsies.
This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
National Institutes of Health
Authorship
Contribution: D.C. and D.P.H. designed and performed research, analyzed data, and wrote the initial drafts of the manuscript; R.L.N., R.M.P., and C.S. designed and performed research and analyzed data; L.W. designed research and supervised the retroviral infections; and P.D.A. designed research, analyzed data, and wrote the final draft of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Peter D. Aplan, NIH/NCI/CCR/Genetics Branch, 8901 Wisconsin Ave, Navy 8, Rm 5101, Bethesda, MD 20889-5105; e-mail: aplanp@mail.nih.gov.
References
Author notes
D.C. and D.P.H. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal