Abstract
NOTCH1 is activated by mutation in more than 50% of human T-cell acute lymphoblastic leukemias (T-ALLs) and inhibition of Notch signaling causes cell-cycle/growth arrest, providing rationale for NOTCH1 as a therapeutic target. The tumor suppressor phosphatase and tensin homolog (PTEN) is also mutated or lost in up to 20% of cases. It was recently observed among human T-ALL cell lines that PTEN loss correlated with resistance to Notch inhibition, raising concern that patients with PTEN-negative disease may fail Notch inhibitor therapy. As these studies were limited to established cell lines, we addressed this issue using a genetically defined mouse retroviral transduction/bone marrow transplantation model and observed primary murine leukemias to remain dependent on NOTCH1 signaling despite Pten loss, with or without additional deletion of p16Ink4a/p19Arf. We also examined 13 primary human T-ALL samples obtained at diagnosis and found no correlation between PTEN status and resistance to Notch inhibition. Furthermore, we noted in the mouse model that Pten loss accelerated disease onset and produced multiclonal tumors, suggesting NOTCH1 activation and Pten loss may collaborate in leukemia induction. Thus, in contrast to previous findings with established cell lines, these results indicate PTEN loss does not relieve primary T-ALL cells of their “addiction” to Notch signaling.
Introduction
The 4 mammalian Notch genes (NOTCH1-4) encode a family of highly conserved type I transmembrane receptors that are normally activated by ligands of the Delta/Serrate/Lag-2 family expressed on the surface of neighboring cells. Once activated by ligand, the Notch receptors undergo proteolytic cleavage first by a disintegrin and metalloprotease, then by a γ-secretase, which releases the intracellular domain (ICN) from the plasma membrane to translocate to the nucleus to stimulate transcription of downstream target genes in complex with the DNA-binding factor CBF1(RBPJ)/suppressor of hairless/Lag-1 and coactivators of the Mastermind family. Although regulated NOTCH1 signaling is important for normal T-cell development,1 it is frequently activated by mutation in the human cancer T-cell acute lymphoblastic leukemia (T-ALL).2 The potent oncogenicity of activated NOTCH1 has been demonstrated in murine bone marrow transduction/transplantation models and several transgenic mouse lines.3
Activating NOTCH1 mutations occur in more than 50% of primary human T-ALLs and cluster in the heterodimerization (HD) and C-terminal proline-, glutamic acid–, serine-, and threonine-rich (PEST) domains.4 HD mutations result in weakened association or complete dissociation of the receptor subunits, and thus lead to heightened/constitutive activation of the receptor.5 PEST domain mutations often generate premature stop codons that delete the PEST degron, and thus enhance signaling by reducing turnover/prolonging half-life of activated ICN.6 When present together, the HD and PEST mutations occur in cis, and stimulate signaling in a synergistic fashion.4 Interestingly, a similar overall frequency of Notch1 mutations (mostly PEST, but some HD) has been observed in various mouse models of T-ALL, underscoring the importance of Notch1 signaling in T-cell leukemogenesis. In addition, both human and murine T-ALL cells bearing NOTCH1 mutations are frequently sensitive to treatment with inhibitors of Notch signaling including γ-secretase inhibitors (GSIs) that induce G1 cell-cycle/growth arrest and in some cases apoptosis.4,7-10 Based on these findings, GSIs and other inhibitors of Notch signaling are being pursued for clinical use in patients with T-ALL.11
The phosphatidylinositol 3-kinase (PI3K)/phosphatase and tensin homolog (PTEN) pathway is one of the most commonly mutated signaling pathways in human cancer.12 Whereas PI3K activates downstream signaling through AKT by converting phosphatidylinositol 4,5-bisphosphate (PIP2) to phosphatidylinositol 3,4,5-triphosphate (PIP3), PTEN antagonizes PI3K activity by converting PIP3 back to PIP2. Pten-deficient mice13,14 are embryonically lethal, but heterozygotes display high incidence of spontaneous T-cell leukemia/lymphoma associated with loss of heterozygosity of the wild-type allele.13 Similarly, T-cell leukemia/lymphoma occurs in various conditional Pten-null models15,16 as well as in mice that received a transplant of Pten-deficient adult hematopoietic stem cells,17 further supporting an important contributing role for PI3K pathway activation in T-cell transformation. Finally, rapamycin can inhibit the serial transplantability of murine acute myeloid leukemia/T-ALL, suggesting a role for PI3K pathway activation in leukemia stem cell maintenance.17,18
PTEN protein loss is observed in 17% to 20% of human T-ALL cases, with chromosomal rearrangements involving the PTEN locus occurring in 15% and frameshift/truncating mutations in 5% to 8% of cases.19-21 More strikingly, more than 85% of cases exhibit hyperactivated PI3K/AKT signaling, which was correlated with reduced PTEN protein phosphatase activity either by casein kinase 2–mediated phosphorylation and/or reactive oxygen species–dependent oxidation.22 Interestingly, it was recently observed among human T-ALL cell lines that loss of PTEN was strongly correlated with GSI resistance, suggesting constitutive PI3K/AKT signaling could relieve dependence on Notch signaling.20 Because Notch inhibitors are currently being tested in patients with T-ALL and other cancers in which PTEN may be lost or mutated, we felt it was important to confirm whether PTEN loss indeed conferred resistance to NOTCH1 inhibition using primary T-ALL cells as opposed to established cell lines.
To this end, we generated primary murine leukemias on Pten-null versus wild-type backgrounds using a standard retroviral transduction/bone marrow transplantation approach. Given that most human T-ALLs lack CDKN2A, which encodes p16INK4A/p14ARF in human and p16Ink4a/p19Arf in mice,23-25 and that these tumor suppressors are known to play a major role in the G1 cell-cycle checkpoint, we also generated primary murine leukemias on a Pten, Ink4a/Arf double-null background. Both Pten single-null and Pten, Ink4a/Arf double-null leukemia cells exhibited reduced proliferation upon inhibition of Notch signaling by GSI treatment (GSI “sensitive”). Although we noted the degree of GSI sensitivity to vary between individual tumors, we were unable to detect any clear association between Pten loss and even partial GSI resistance. To ascertain the relevance of these findings to human disease, we examined a panel of 13 primary human T-ALL samples obtained at diagnosis for response to GSI treatment. We assayed patient cells directly and/or after expansion as xenografts in nonobese diabetic/severe combined immunodeficiency/Il2rg−/− (NSG) mice. Again, we observed variation among individual tumors, but could ascertain no significant correlation between PTEN loss and GSI resistance. Of note, we identified only 2 human T-ALL samples in our series (15%) that were GSI “resistant” (< 15% growth inhibition compared with control), suggesting primary resistance may not be encountered frequently in the clinic. These results demonstrate that constitutive PI3K/AKT signaling conferred by PTEN loss cannot replace or compensate for the pro-oncogenic effects of Notch signaling in established primary leukemias. Refractory/relapsed tumors that have undergone chemotherapy-induced mutation and/or selection may behave differently, but presumably will harbor many other genetic alterations besides PTEN loss.
Methods
Mice
Conditional Pten knockout (KO), conditional Ink4a/Arf KO, and Mx1-Cre mice have been described previously.26-28 The Ink4a/Arf allele contains loxP sites flanking exons 2 and 3, which are shared between p16Ink4a and p19Arf reading frames such that Cre-mediated excision results in deletion of both p16Ink4a and p19Arf. Polymerase chain reaction (PCR)–based genotyping and detection of Cre-mediated loxP excision were performed as described previously.26,28 All bone marrow donor animals were of (C57BL/6 × FVB) mixed background, whereas (C57BL/6 × FVB)F1 animals were used as recipients. NOD-scid/Il2rg−/− (NSG) mice were used as recipients for human T-ALL xenografts. Mice were housed in specific pathogen-free animal facilities according to institutional guidelines.
Retroviral transduction/bone marrow transplantation
High titer, replication-defective retroviral supernatants were produced by transient transfection of PlatE cells.29 Murine stem cell virus–based retroviral expression vectors included internal ribosomal entry site (IRES)–green fluorescent protein (GFP) cassettes for fluorescent tagging of transduced cells. The leukemogenic human NOTCH1 L1601P-ΔPEST mutant allele has been described previously.30 Retroviral transduction of 5-fluorouracil–treated bone marrow cells was performed as described previously.31 Lethally irradiated recipient mice were injected intravenously with 45 000 transduced GFP+ cells along with a minimum of 105 normal bone marrow cells to ensure hematopoietic reconstitution.
Clonality analysis
TCRβ.
T-cell receptor β (TCRβ) rearrangements were detected as described.32 Briefly, genomic DNA was isolated from freshly explanted murine tissues involved by leukemia and amplified by PCR with primers Dβ1.1ext and Jβ1.7ext, or Dβ2.1ext and Jβ2.7ext. Amplified products were then subjected to a second round of PCR using primers Dβ1.1int and Jβ1.7int, or Dβ2.1int and Jβ2.7int. Products were resolved by electrophoresis on 2% agarose gels and visualized by ethidium bromide staining.
Proviral integration.
Proviral integration sites were amplified by ligation-mediated PCR as described.33 Briefly, genomic DNA was digested with BamHI, ligated with BamHI linkers, and subjected to nested PCR using linker-primers followed by linker-primer/retrovirus-specific primers.
Human leukemia samples
Cryopreserved lymphoblast samples were provided by collaborating institutions in Canada (BC Children's Hospital), the United States (Karmanos Cancer Center), and France (Hopital Andre Trousseau). Primary samples were obtained at initial diagnosis with informed consent from patients or their legal guardians under approved institutional review board protocols and following guidelines established by the Declaration of Helsinki. Expansion of primary human T-ALL cells in irradiated NOD-scid/Il2rg−/− (NSG) mice has been described previously.7 NOTCH1, FBW7, and PTEN genomic loci were sequenced as described.4,20,34,35
In vitro culture of primary T-ALL cells
Primary human T-ALL cells were cultured either on MS5/MS5-DL1 stromal feeder cells as described,7 or without feeders in plastic dishes coated with immobilized DL1 ligand (Delta1ext-IgG, kind gift of Dr Irwin Bernstein, Fred Hutchinson Cancer Research Center)36 at a concentration of 0.5 μg/cm2. Primary mouse T-ALL cells were cultured without feeders or immobilized ligand in complete media with supplemental cytokines.
GSI treatment
In vitro.
Gamma-secretase inhibitor (GSI) compounds N-[N-(3,5-Difluorophenacetyl-L-alanyl)]-S-phenylglycine t-Butyl Ester (DAPT; Calbiochem) and compound E (Axxora) were dissolved in dimethyl sulfoxide (DMSO), prediluted in Hanks balanced salt solution, and added to culture media to achieve final concentrations of 10μM and 1μM, respectively. Control samples were treated with DMSO vehicle at equivalent final concentration (typically 0.01%-0.1% DMSO).
In vivo.
Freshly isolated splenocytes (86% GFP positive) from a primary leukemic mouse were transplanted into secondary recipients by tail vein injection (0.5 × 106 spleen cells = 0.43 × 105 tumor cells per mouse). At day 3 after transplantation, mice were treated by intraperitoneal injection with GSI (DAPT) versus DMSO vehicle control. All animals were killed for histologic analysis on day 3 after initiation of GSI treatment (day 6 after transplantation).
In vitro BrdU incorporation assay
Cultured cells were pulsed with 10μM bromodeoxyuridine (BrdU) for 2 hours in vitro prior to harvest. Staining was performed according to the manufacturer's instructions (allophycocyanin- or fluorescein isothiocyanate–BrdU kit; BD Biosciences). Human leukemia cells from xenografted mice were discriminated using anti–human CD45 antibody (eBioscience). Murine leukemia cells were discriminated by virtue of retroviral GFP expression.
RT-PCR
Total RNA was prepared using TRIzol reagent (Invitrogen). Reverse transcription was performed with SuperScript II RT (Invitrogen) from 1 μg of total RNA with a 1:1 mixture of oligo dT and random 15-mer primers. PCR was performed using Platinum Taq High Fidelity (Invitrogen) on a Dyad Disciple thermal cycler (Bio-Rad). Gene-specific primer sequences were as follows: human PTEN forward 5-GTT TAC CGG CAG CAT CAA AT, human PTEN reverse 5-CCC CCA CTT TAG TGC ACA GT; human p14ARF forward 5-TCG TGC TGA TGC TAC TGA GG, human p16INK4A forward 5-CAA CGC ACC GAA TAG TTA CG, human p16INK4A/p14ARF reverse 5-ACC AGC GTG TCC AGG AAG; human β-actin forward 5-CGC GAG AAG ATG ACC CAG AT, human β-actin reverse 5-GAT AGC ACA GCC TGG ATA GCA AC; mouse Pten forward 5-GTT TAC CGG CAG CAT CAA AT, mouse Pten reverse 5-TGG CAG GTA GAA GGC AAC TC; mouse β-actin forward 5-CTT CTA CAA TGA GCT GCG TGT G, and mouse β-actin reverse 5-TTG AAG GTC TCA AAC ATG ATC TGG. Human primers were confirmed not to amplify mouse sequences. Human PTEN primers were designed to prevent amplification of the PTEN pseudogene on chromosome 9p21.37
Flow cytometry
Intracellular PTEN staining was performed on formaldehyde-fixed, methanol-permeabilized cells38 using primary PTEN (Y184) antibody (1:100 dilution; Abcam) and secondary goat anti–rabbit AF647 antibody (1:200 dilution; Invitrogen). Intracellular human p16INK4A staining was performed according to the manufacturer's instructions (p16–fluorescein isothiocyanate kit; BD Biosciences). Routine phenotyping of mouse leukemias used the following antibodies: CD8a-allophycocyanin (53-6.7; Biolegend); CD4-phycoerythrin-Cy5.5 (L3T4; eBioscience); CD11b-biotin (M1/70; eBioscience); Gr1-biotin (RB6-8C5; eBioscience); and phycoerythrin-conjugated streptavidin (catalog no. 12-4317-87; eBioscience). Acquisition was performed on a FACSCalibur (BD Biosciences) and data were analyzed using FlowJo software (TreeStar).
Histopathology
Mouse tissues were fixed in 10% neutral buffered formalin, embedded in paraffin, and sectioned for hematoxylin and eosin (H&E) staining. Quantitative assessment of tumor burden was performed by automated image analysis of low-power photomicrographs using QuantityOne software (Bio-Rad). Three histologic fields were imaged per animal. Image thresholds were set by operators blinded to treatment groups to prevent observer bias. Statistical analysis was performed using GraphPad Prism 5 software.
Results
Generation of primary murine leukemias with activated NOTCH1 retrovirus
Murine bone marrow retroviral transduction/transplantation using activated forms of NOTCH1 is a well-established model for the study of T-ALL.3 Retroviral expression of the NOTCH1 intracellular domain (ICN), which mimics polypeptides derived from the classic but rare7,9 translocation, produces lethal T-ALL–like disease in mice with high penetrance and short latency. Similarly, HD/PEST mutations, which occur in more than 50% of human T-ALLs, also produce disease in mice albeit with prolonged latency and reduced penetrance compared with ICN, presumably due to weaker activation of signaling.30 For the present study, we elected to generate murine leukemias using NOTCH1 retroviruses with the frequently observed HD and PEST mutations to model human disease more closely. The L1601P-ΔPEST mutant contains a relatively common HD mutation, L1601P, that causes receptor subunit dissociation and subsequent ligand-independent activation,5 in cis with a frameshift mutation that creates a premature termination codon and truncation of the C-terminal negative regulatory PEST domain (ΔPEST). Importantly, the L1601P-ΔPEST allele is leukemogenic in mouse models on wild-type background,30 and is highly similar to HD/PEST mutations present in the PTEN-null, GSI-resistant human cell lines, MOLT 3 and PF382.20 Of note, activity of the L1601P-ΔPEST construct is blocked by γ-secretase inhibitors (GSIs) because, although subunit dissociation occurs, the transmembrane subunit cannot be cleaved, and thus the intracellular domain remains tethered to the plasma membrane and transcription of downstream target genes does not occur.
To begin to address the effects of Pten loss in the murine NOTCH1 leukemia model, we performed retroviral transduction/transplantation using marrow from conditional Pten KO donor mice. We selected a loxP-flanked conditional Pten allele28 in hopes of inducing Cre-mediated deletion in established leukemia cells; however, the interferon-inducible Mx1-Cre driver allele27 was sufficiently active after 5-fluorouracil (5-FU) treatment such that Pten was deleted in more than 50% of marrow progenitors before retroviral transduction (supplemental Figure 1A-B, available on the Blood website; see the Supplemental Materials link at the top of the online article). Consequently, wild-type and PTENf/f Mx1-Cre bone marrow (presumably containing a mixture of Ptenf/f, Ptenf/−, and Pten−/− genotypes) was transduced with L1601P-ΔPEST retrovirus, and 45 000 GFP+ cells were transplanted into sublethally irradiated recipients.
NOTCH1 leukemias generated on a Pten-null background exhibit increased penetrance and shortened latency
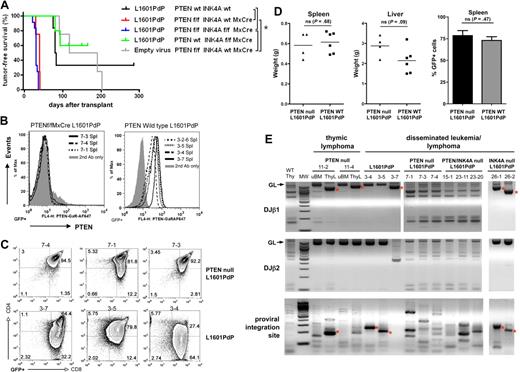
We observed mice that received a transplant of L1601P-ΔPEST–transduced, Ptenf/f, f/−, −/− marrow succumb to aggressive, disseminated T-ALL disease with 100% penetrance and median latency of 35 days (Figure 1A). All tumors were GFP+ and Pten protein–negative with complete deletion at the Pten locus (Figure 1B and supplemental Figure 1C). Although marrow progenitors with intact Pten were clearly present at the time of retroviral transduction, they are likely “outcompeted” by Pten-null cells during leukemogenic evolution. In contrast, mice that received a transplant of L1601P-ΔPEST–transduced, wild-type background marrow developed T-ALL with significantly reduced penetrance (66%, 4 of 6 at 285 days after transplantation) and prolonged latency (median, 81.5 days; P = .001, Figure 1A). Notably, tumors derived on Pten-null and wild-type backgrounds had similar immunophenotype, disease extent/organ distribution, and histology (Figure 1C-D, and supplemental Figure 1D). Consistent with prior studies, we also observed mice that received a transplant of empty vector–transduced, conditional Pten KO marrow (likely also containing Ptenf/f, Ptenf/−, and Pten−/− subsets) developed spontaneous Pten-null disease with 100% penetrance (Figure 1A) including isolated thymic lymphoma (n = 2) and disseminated biphenotypic acute T-myeloid leukemia/lymphoma (n = 2; supplemental Figure 2). Of note, these occurred with significantly longer latency (median, 153.5 days) than disease induced by transduction with L1601P-ΔPEST on the same background (median, 35 days; P = .007, Figure 1A).
Loss of Pten accelerates NOTCH1-induced leukemia. (A) Kaplan-Meier T-cell leukemia survival curves. Bone marrow from mice of indicated genotypes was transduced with mutated NOTCH1 (L1601P-ΔPEST) or empty retrovirus and transplanted into irradiated recipients. Median survivals are as follows: L1601P-ΔPEST retrovirus on Ptenwt Ink4a/Arfwt (81.5 days, n = 6), Ptenf/f Ink4a/Arfwt Mx1-Cre (35 days, n = 4), Ptenf/f Ink4a/Arf f/f Mx1-Cre (34 days, n = 12), and Ptenwt Ink4a/Arff/f Mx1-Cre (86.5 days, n = 2; 3 additional mice in cohort died of nonhematologic neoplasms) backgrounds; empty virus on Ptenf/f Ink4a/Arfwt Mx1-Cre background (153.5 days, n = 4). Indicated pairwise comparisons are significantly different (*log-rank test, P < .007). (B-C) Flow cytometric analysis of Pten protein expression (B) and CD4/CD8 phenotype (C) in GFP+ splenic cells from L1601P-ΔPEST leukemias on Ptenf/f Ink4a/Arfwt Mx1-Cre (7-x series) and Ptenwt Ink4a/Arfwt (3-x series) backgrounds. Analysis of bone marrow cells showed similar results (data not shown). (D) Tumor burden in spleen and liver as measured by whole organ weight and GFP+ fraction is not significantly different between Pten-null and wild-type background leukemias (Student t test). (E) Clonality assessment by TCRβ rearrangement and proviral integration site analysis. Dominant single bands indicative of monoclonality are highlighted by asterisk (*). Normal tissue samples including wild-type thymus (WT Thy) and uninvolved bone marrow (uBM) represent “polyclonal” controls for comparison. ThyL indicates thymic lymphoma; MW, molecular weight marker; and GL, germline. DNA was prepared from either bone marrow or spleen for cases of disseminated leukemia. Each numbered sample represents a different individual transplant recipient mouse. Please note, L1601P-ΔPEST is abbreviated as L1601PdP in the figure labels. Ink4a-Arf is abbreviated as INK4A in the figure labels.
Loss of Pten accelerates NOTCH1-induced leukemia. (A) Kaplan-Meier T-cell leukemia survival curves. Bone marrow from mice of indicated genotypes was transduced with mutated NOTCH1 (L1601P-ΔPEST) or empty retrovirus and transplanted into irradiated recipients. Median survivals are as follows: L1601P-ΔPEST retrovirus on Ptenwt Ink4a/Arfwt (81.5 days, n = 6), Ptenf/f Ink4a/Arfwt Mx1-Cre (35 days, n = 4), Ptenf/f Ink4a/Arf f/f Mx1-Cre (34 days, n = 12), and Ptenwt Ink4a/Arff/f Mx1-Cre (86.5 days, n = 2; 3 additional mice in cohort died of nonhematologic neoplasms) backgrounds; empty virus on Ptenf/f Ink4a/Arfwt Mx1-Cre background (153.5 days, n = 4). Indicated pairwise comparisons are significantly different (*log-rank test, P < .007). (B-C) Flow cytometric analysis of Pten protein expression (B) and CD4/CD8 phenotype (C) in GFP+ splenic cells from L1601P-ΔPEST leukemias on Ptenf/f Ink4a/Arfwt Mx1-Cre (7-x series) and Ptenwt Ink4a/Arfwt (3-x series) backgrounds. Analysis of bone marrow cells showed similar results (data not shown). (D) Tumor burden in spleen and liver as measured by whole organ weight and GFP+ fraction is not significantly different between Pten-null and wild-type background leukemias (Student t test). (E) Clonality assessment by TCRβ rearrangement and proviral integration site analysis. Dominant single bands indicative of monoclonality are highlighted by asterisk (*). Normal tissue samples including wild-type thymus (WT Thy) and uninvolved bone marrow (uBM) represent “polyclonal” controls for comparison. ThyL indicates thymic lymphoma; MW, molecular weight marker; and GL, germline. DNA was prepared from either bone marrow or spleen for cases of disseminated leukemia. Each numbered sample represents a different individual transplant recipient mouse. Please note, L1601P-ΔPEST is abbreviated as L1601PdP in the figure labels. Ink4a-Arf is abbreviated as INK4A in the figure labels.
NOTCH1 leukemias generated on a Pten-null background are multiclonal
We also performed TCRβ rearrangement and proviral integration site analysis on primary leukemias from individual mice to gain insight into the clonal diversity of tumors generated on the various genetic backgrounds. Using PCR amplification of genomic DNA to detect TCRβ rearrangements32 and proviral integration sites,33 we detected single dominant clones from spontaneous Pten-null thymic lymphomas (without L1601P-ΔPEST) and L1601P-ΔPEST acute T-cell leukemias induced on wild-type background. In contrast, we observed multiple clones from individual L1601P-ΔPEST leukemias on both Pten-null and Pten, Ink4a/Arf double-null backgrounds (Figure 1E). We also observed mice that received a transplant of L1601P-ΔPEST–transduced, Ink4a/Arff/f Mx1-Cre marrow (presumably containing a mixture of Ink4a/Arff/f, Ink4a/Arff/−, and Ink4a/Arf−/− genotypes) developed monoclonal tumors (2 of 5 mice died of T-cell leukemia; 3 of 5 died of nonhematologic malignancies; data not shown). Thus, we have observed L1601P-ΔPEST to induce multiclonal disease only on Pten-null backgrounds (with or without Ink4a/Arf) as opposed to monoclonal disease on Pten wild-type backgrounds (with or without Ink4a/Arf). In combination with our observation that L1601P-ΔPEST disease is accelerated on the Pten-null compared with Pten wild-type background (with or without Ink4a/Arf; Figure 1A), these findings suggest that Pten loss may cooperate with activated NOTCH1 in leukemia induction. One important caveat to this conclusion, however, is that Pten loss is known to promote cycling of hematopoietic progenitors,21 which could increase their retroviral transduction efficiency and contribute to disease acceleration and multiclonality. This issue is considered further in “Discussion.”
Primary murine NOTCH1 leukemia cells lacking Pten remain sensitive to GSI
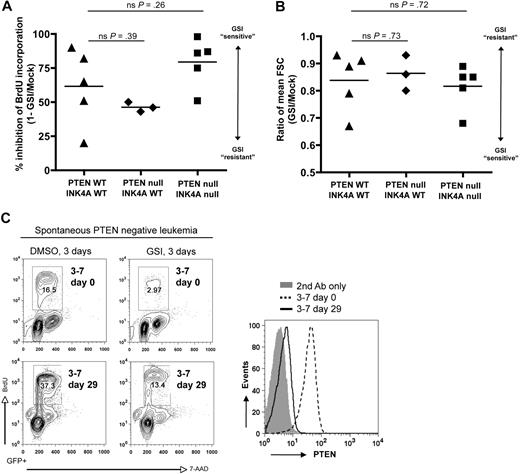
Consistent with the prior study linking PTEN loss with resistance to Notch inhibition,20 we observed striking correlation between PTEN loss and GSI resistance among human T-ALL cell lines (supplemental Table 1 and supplemental Figure 3). To determine whether this association is valid in primary T-cell leukemias derived on a defined genetic background, we treated freshly explanted L1601P-ΔPEST leukemia cells from both wild-type and Pten-null backgrounds with GSI in vitro for 3 days and measured proliferation, cell size, and apoptosis. Although we noted variation among individual tumors in the degree of response to GSI, both wild-type and Pten-null tumors were GSI sensitive, showing substantially decreased proliferation (Figure 2A and supplemental Figure 4) and reduced cell size (Figure 2B). Furthermore, quantitative comparison of GSI response failed to demonstrate any robust association between Pten loss and even partial GSI resistance (Figure 2A-B). We observed only minimal apoptosis (subG1 fraction) for both wild-type and Pten-null tumors within the 3-day GSI treatment period (supplemental Figure 4).
Murine NOTCH1 leukemias lacking Pten remain dependent on Notch signaling and are GSI sensitive. (A) Proliferation and (B) cell size analysis of primary mouse L1601P-ΔPEST leukemias on wild-type (WT) and Pten/Ink4a/Arf-null backgrounds treated with γ-secretase inhibitor (GSI) to block Notch signaling. Freshly explanted primary leukemia cells from different individual mice were cultured in vitro for 3 days with GSI (1μM compound E) or DMSO vehicle, pulsed with BrdU, and assayed by flow cytometry. Proliferation results are summarized from data plots presented in supplemental Figure 4. FSC indicates forward light scatter; ns, nonsignificant (Student t test). (C) Spontaneous loss of Pten protein expression occurred in one case (mouse nos. 3-7 as in Figure 1B) after 29 days of culture in vitro, yet it remained GSI sensitive. Pten protein expression was assessed by flow cytometry (right panel), and GSI response assayed by BrdU incorporation (left panel). Pten mRNA was detected by RT-PCR at both day 0 and day 29, and sequencing revealed no loss-of-function mutations (data not shown). Please note, only gated GFP+ events are depicted.
Murine NOTCH1 leukemias lacking Pten remain dependent on Notch signaling and are GSI sensitive. (A) Proliferation and (B) cell size analysis of primary mouse L1601P-ΔPEST leukemias on wild-type (WT) and Pten/Ink4a/Arf-null backgrounds treated with γ-secretase inhibitor (GSI) to block Notch signaling. Freshly explanted primary leukemia cells from different individual mice were cultured in vitro for 3 days with GSI (1μM compound E) or DMSO vehicle, pulsed with BrdU, and assayed by flow cytometry. Proliferation results are summarized from data plots presented in supplemental Figure 4. FSC indicates forward light scatter; ns, nonsignificant (Student t test). (C) Spontaneous loss of Pten protein expression occurred in one case (mouse nos. 3-7 as in Figure 1B) after 29 days of culture in vitro, yet it remained GSI sensitive. Pten protein expression was assessed by flow cytometry (right panel), and GSI response assayed by BrdU incorporation (left panel). Pten mRNA was detected by RT-PCR at both day 0 and day 29, and sequencing revealed no loss-of-function mutations (data not shown). Please note, only gated GFP+ events are depicted.
Interestingly, we also observed a few instances where L1601P-ΔPEST leukemia cells had lost Pten expression spontaneously after either extended culture in vitro (Figure 2C) or serial transplantation in vivo (supplemental Figure 5). In each of these cases, the leukemia cells remained responsive to GSI treatment despite Pten loss. Although the in vitro–cultured cells were ostensibly less GSI sensitive after Pten loss (2.8-fold response at day 29 versus 5.6-fold response at day 0), they also showed a 2.3-fold increase in proliferation without GSI treatment, suggesting rigorous selection for subclones that presumably have accumulated multiple genetic hits in addition to Pten loss. Thus, although Pten loss cannot be excluded as a potential contributing factor, these results clearly demonstrate that Pten loss is not sufficient to confer Notch independence in this genetically defined primary leukemia model.
Primary murine NOTCH1 leukemia cells lacking both Pten and Ink4a/Arf are also sensitive to GSI
Loss of cyclin-dependent kinase inhibitors p16INK4A and p14ARF is perhaps the most frequent genetic alteration in human T-ALL, either by chromosomal deletion or epigenetic silencing.23-25 Given that (1) all identified GSI-resistant human T-ALL cell lines are also null for p16INK4A/p14ARF,39 (2) these proteins play a crucial role in the control of cell cycle especially at the G1-S checkpoint, and (3) G1/G0 cell-cycle arrest is a characteristic response to GSI among sensitive cells,4,10 we considered the possibility that GSI resistance may require loss of both PTEN and INK4A/ARF. To address this issue, we generated primary L1601P-ΔPEST leukemias on a Pten, Ink4a/Arf double-null background and assessed GSI response. Again, leaky Mx1-Cre expression presumably during 5-FU treatment allowed deletion of Pten and Ink4a/Arf in some marrow progenitors that gave rise to all resulting leukemias (Figure 3A, supplemental Figure 6). Despite some tumor-to-tumor variability, leukemias were on the whole sensitive to GSI (Figure 3B) with no discernable degree of resistance conferred by coordinate loss of Pten and Ink4a/Arf (Figure 2A-B). Assessment of subG1 fraction revealed minimal apoptosis after the 3-day GSI treatment period (Figure 3B) similar to that noted for wild-type and Pten-null leukemias (supplemental Figure 4).
Murine NOTCH1 leukemias lacking both Pten and Ink4a/Arf remain dependent on Notch signaling and are GSI sensitive. (A) PCR analysis confirming homozygous deletion of both Ink4a/Arf (upper panel) and Pten (bottom panel) loci in L1601P-ΔPEST leukemias derived from Ptenf/f Ink4a/Arff/f Mx1-Cre bone marrow (n = 7). Analysis of 5-FU–treated bone marrow revealed a subpopulation of progenitors already deleted for Ink4a/Arf and/or Pten prior to retroviral transduction. (B) In vitro proliferation analysis of primary mouse L1601P-ΔPEST leukemias on Pten, Ink4a/Arf double-null background treated with GSI as in Figure 2A. Primary leukemia cells from 5 individual mice were assayed. Only gated GFP+ events are depicted. (C) In vivo assay for GSI sensitivity confirms in vitro results. Splenic tumor cells from a single Pten, Ink4a/Arf double-null L1601P-ΔPEST primary leukemia were serially transplanted by tail vein injection into secondary recipients. At day 3 after transplantation, mice were treated by intraperitoneal injection with GSI (100 mg/kg per day DAPT) for 1 day (n = 5) or 2 days (n = 3), or DMSO vehicle only (n = 4). Animals were then killed on day 3 after treatment initiation (day 6 after transplantation) and extent of tumor infiltration in liver was assessed by automated image analysis of H&E histology. Each data point represents a separately imaged histologic field; 3 fields were examined per mouse. Significantly fewer blasts were observed in GSI-treated mice (P < .001; 1-way analysis of variance with posttest linear trend analysis).
Murine NOTCH1 leukemias lacking both Pten and Ink4a/Arf remain dependent on Notch signaling and are GSI sensitive. (A) PCR analysis confirming homozygous deletion of both Ink4a/Arf (upper panel) and Pten (bottom panel) loci in L1601P-ΔPEST leukemias derived from Ptenf/f Ink4a/Arff/f Mx1-Cre bone marrow (n = 7). Analysis of 5-FU–treated bone marrow revealed a subpopulation of progenitors already deleted for Ink4a/Arf and/or Pten prior to retroviral transduction. (B) In vitro proliferation analysis of primary mouse L1601P-ΔPEST leukemias on Pten, Ink4a/Arf double-null background treated with GSI as in Figure 2A. Primary leukemia cells from 5 individual mice were assayed. Only gated GFP+ events are depicted. (C) In vivo assay for GSI sensitivity confirms in vitro results. Splenic tumor cells from a single Pten, Ink4a/Arf double-null L1601P-ΔPEST primary leukemia were serially transplanted by tail vein injection into secondary recipients. At day 3 after transplantation, mice were treated by intraperitoneal injection with GSI (100 mg/kg per day DAPT) for 1 day (n = 5) or 2 days (n = 3), or DMSO vehicle only (n = 4). Animals were then killed on day 3 after treatment initiation (day 6 after transplantation) and extent of tumor infiltration in liver was assessed by automated image analysis of H&E histology. Each data point represents a separately imaged histologic field; 3 fields were examined per mouse. Significantly fewer blasts were observed in GSI-treated mice (P < .001; 1-way analysis of variance with posttest linear trend analysis).
To validate these findings in vivo, tumor cells from a single Pten, Ink4a/Arf double-null L1601P-ΔPEST primary leukemia were transplanted into secondary recipients by tail vein injection and engrafted animals subsequently treated with GSI. We allowed a 3-day engraftment period prior to initiation of GSI therapy, as pilot experiments showed secondary recipients all died by 7 days after transplantation (data not shown). All animals were sacrificed on day 3 after initiation of GSI treatment (day 6 after transplantation), and liver parenchyma was assessed histologically for tumor burden. Tumor cells also engrafted bone marrow and spleen, but quantitation of liver involvement by automated image analysis was more robust. Consistent with in vitro findings, we observed significantly fewer lymphoblasts in livers of GSI-treated mice versus control (Figure 3C; P < .001). Thus, both in vitro and in vivo assays demonstrate that primary mouse NOTCH1 leukemia cells remain sensitive to GSI treatment despite loss of both Pten and Ink4a/Arf tumor suppressors.
Primary human T-ALL cells show no correlation between PTEN loss and Notch independence/GSI resistance
Although mouse models serve an important role in cancer research, it is of growing concern that there may be critical differences between mice and humans in terms of events required for cellular transformation and response to therapy. Our observation of GSI sensitivity among murine NOTCH1 leukemia cells regardless of Pten and Ink4a/Arf status differs from the situation noted in human cell lines20 (supplemental Table 1 and supplemental Figure 3). We felt it was critical therefore to investigate whether PTEN loss conferred Notch independence/GSI resistance using primary human T-ALL samples.
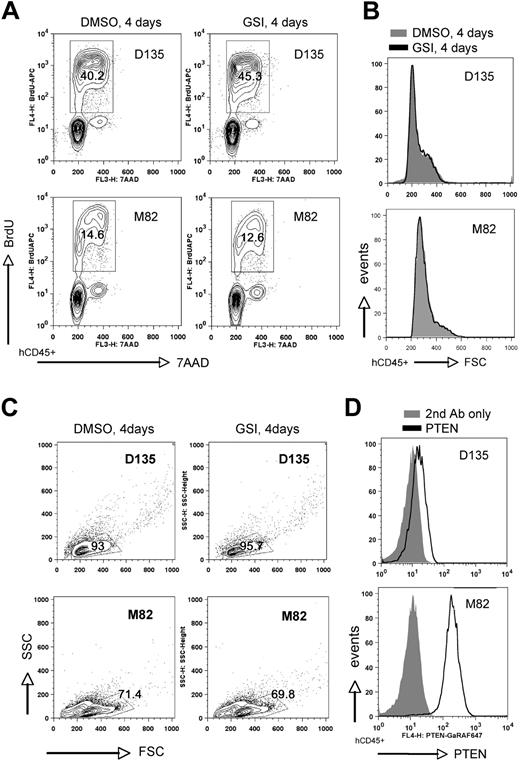
To this end, we assessed a total of 13 patient samples, including 7 cases where lymphoblasts came directly from the patient and 6 cases that were expanded first as xenografts in NOD-scid/Il2rg−/− (NSG) mice. All patient samples were obtained at initial diagnosis. We cultured human lymphoblasts in vitro on either MS5/MS5-DL1 feeders7 or immobilized DL1 ligand40 for 1 to 6 days prior to initiation of GSI treatment. Cultures were then exposed to GSI versus DMSO vehicle for a 4-day period and subsequently assayed for proliferation, cell size, and apoptosis (Figure 4 and supplemental Figure 7). The 13 cases included 7 PTEN-positive (6 of which were NOTCH1 mutated) and 6 PTEN-negative (4 of which were NOTCH1 mutated) (Table 1). We found 11 of these 13 cases to be GSI sensitive (6 PTEN positive, 5 PTEN negative) as illustrated by decreased proliferation and reduction in cell size (Figure 4 and supplemental Figure 7). We found only 2 cases to be GSI resistant, including 1 PTEN positive and 1 PTEN negative (Figure 5). As with the primary mouse leukemias, we observed only minimal apoptosis within the 4-day GSI treatment period. In these results, we observed no apparent bias to the particular method of culture used for in vitro assay or whether the cells had come directly from the patient versus passaged through NSG mice beforehand (Table 1). Of note, all of the cases were negative for p16INK4A and p14ARF mRNA by reverse-transcription (RT)–PCR and/or protein by flow cytometry (supplemental Figure 8). Thus, in keeping with the mouse results, we find no evidence for correlation between PTEN loss and GSI resistance in primary human T-ALL cells.
Primary human T-ALL cells are GSI sensitive regardless of PTEN status. In vitro proliferation and cell size analysis of primary human T-ALL samples maintained briefly (up to 6 days) on either MS5/MS5-DL1 murine stromal feeder cells (A,C) or immobilized Delta1 ligand (Delta1ext-IgG; B), and then treated with GSI (1μM compound E or 10μM DAPT) versus DMSO vehicle for 4 days. At the end of the treatment period, cultures were pulsed with BrdU and assayed by flow cytometry. Human T-ALL cells were discriminated from murine cells by costaining with hCD45. Each plotted data point in panels A and C represents a different primary human sample. Results are summarized from individual data plots presented in supplemental Figure 7. FSC indicates forward light scatter; ns, nonsignificant (Student t test).
Primary human T-ALL cells are GSI sensitive regardless of PTEN status. In vitro proliferation and cell size analysis of primary human T-ALL samples maintained briefly (up to 6 days) on either MS5/MS5-DL1 murine stromal feeder cells (A,C) or immobilized Delta1 ligand (Delta1ext-IgG; B), and then treated with GSI (1μM compound E or 10μM DAPT) versus DMSO vehicle for 4 days. At the end of the treatment period, cultures were pulsed with BrdU and assayed by flow cytometry. Human T-ALL cells were discriminated from murine cells by costaining with hCD45. Each plotted data point in panels A and C represents a different primary human sample. Results are summarized from individual data plots presented in supplemental Figure 7. FSC indicates forward light scatter; ns, nonsignificant (Student t test).
Summary of results for primary human T-ALL cells: no correlation between GSI response and PTEN status
| Sample ID . | NOTCH1 HD mutation . | NOTCH1 PEST mutation . | PTEN mutation . | PTEN protein . | FBW7 mutation . | GSI response . | GSI assay (fresh/NSG) . |
|---|---|---|---|---|---|---|---|
| K550 | L1597H | WT | WT | Positive | WT | Sensitive | NSG |
| KER | WT | WT | WT | Positive | R479Q | Sensitive | Both |
| M18 | WT | P2513L | WT | Positive | WT | Sensitive | NSG |
| M30 | WT | Q2520* | WT | Positive | WT | Sensitive | Both |
| M71 | L1586P | WT | WT | Positive | WT | Sensitive | Fresh |
| M78 | 1612 Del (HG) Ins (QIVVFKRDAHG) | WT | WT | Positive | WT | Sensitive | Fresh |
| D115 | WT | Q2459* | 245 YQFMFLVW* | Negative | WT | Sensitive | NSG |
| M22 | WT | WT | 229 KGTGRQVHVL*/233 PGKTSSCTLSSLSRY LCVVISK* | Negative | WT | Sensitive | NSG |
| M34 | WT | 2469 QGCHPRWSHP* | WT | Negative | WT | Sensitive | Both |
| M53 | WT | Q2417* | WT | Negative | WT | Sensitive | NSG |
| M69 | WT | WT | 233 (R)FSGKTSSCTLSS LSRYLCVVISK* | Negative | WT | Sensitive | Fresh |
| M82 | 1615 Del (QM) Ins (L) | 2515 RVP* | WT | Positive | WT | Resistant | Fresh |
| D135 | WT | 2506 DLLPP* | 233 EEKTSSCTLSSLSR YLCVVISK* | Negative | WT | Resistant | NSG |
| K388 | A1702P | 2402 QSSSSKACSRHHHH HSRTLA* | WT | Positive | WT | ND | |
| K390 | WT | WT | WT | Positive | WT | ND | |
| K419 | Y1717F | WT | WT | Positive | R465C | ND | |
| K424 | WT | WT | WT | Positive | WT | ND | |
| M62 | WT | 2444 RTCSQQTSSSSKACS RHHHHHSRTLA* | WT | Positive | WT | ND | |
| F1313 | L1586P | WT | WT | Positive | WT | ND | |
| U16017 | L1601P | WT | WT | ND | WT | ND |
| Sample ID . | NOTCH1 HD mutation . | NOTCH1 PEST mutation . | PTEN mutation . | PTEN protein . | FBW7 mutation . | GSI response . | GSI assay (fresh/NSG) . |
|---|---|---|---|---|---|---|---|
| K550 | L1597H | WT | WT | Positive | WT | Sensitive | NSG |
| KER | WT | WT | WT | Positive | R479Q | Sensitive | Both |
| M18 | WT | P2513L | WT | Positive | WT | Sensitive | NSG |
| M30 | WT | Q2520* | WT | Positive | WT | Sensitive | Both |
| M71 | L1586P | WT | WT | Positive | WT | Sensitive | Fresh |
| M78 | 1612 Del (HG) Ins (QIVVFKRDAHG) | WT | WT | Positive | WT | Sensitive | Fresh |
| D115 | WT | Q2459* | 245 YQFMFLVW* | Negative | WT | Sensitive | NSG |
| M22 | WT | WT | 229 KGTGRQVHVL*/233 PGKTSSCTLSSLSRY LCVVISK* | Negative | WT | Sensitive | NSG |
| M34 | WT | 2469 QGCHPRWSHP* | WT | Negative | WT | Sensitive | Both |
| M53 | WT | Q2417* | WT | Negative | WT | Sensitive | NSG |
| M69 | WT | WT | 233 (R)FSGKTSSCTLSS LSRYLCVVISK* | Negative | WT | Sensitive | Fresh |
| M82 | 1615 Del (QM) Ins (L) | 2515 RVP* | WT | Positive | WT | Resistant | Fresh |
| D135 | WT | 2506 DLLPP* | 233 EEKTSSCTLSSLSR YLCVVISK* | Negative | WT | Resistant | NSG |
| K388 | A1702P | 2402 QSSSSKACSRHHHH HSRTLA* | WT | Positive | WT | ND | |
| K390 | WT | WT | WT | Positive | WT | ND | |
| K419 | Y1717F | WT | WT | Positive | R465C | ND | |
| K424 | WT | WT | WT | Positive | WT | ND | |
| M62 | WT | 2444 RTCSQQTSSSSKACS RHHHHHSRTLA* | WT | Positive | WT | ND | |
| F1313 | L1586P | WT | WT | Positive | WT | ND | |
| U16017 | L1601P | WT | WT | ND | WT | ND |
NOTCH1, PTEN, and FBW7 status in 20 human T-ALL samples included in this study. GSI response was assessed in 13 of these using lymphoblasts directly from patients (fresh) and/or after expansion as xenografts in NSG mice (NSG). The 3 samples assessed in both situations yielded consistent results. PTEN protein was assessed by intracellular flow cytometry. Observed mutation frequencies among 18 unselected cases are similar to previous reports4,19,20,22 (D115 and D135 were specifically selected for NOTCH1 and PTEN mutation).
WT indicates wild type; and ND, not determined.
Premature stop codon.
Infrequent GSI-resistant primary human T-ALL samples include both PTEN-positive and -negative cases. In vitro analysis of 2 primary human T-ALL samples (D135 and M82) maintained briefly (2 days) on MS5-DL1 stromal feeders and then treated with GSI (1μM compound E) versus DMSO vehicle for 4 days. Cells were analyzed as in Figure 4 for proliferation (A), cell size (B), and viability by light scatter (C). PTEN protein expression was assessed by intracellular flow cytometry (D). FSC indicates forward light scatter; SSC, side scatter.
Infrequent GSI-resistant primary human T-ALL samples include both PTEN-positive and -negative cases. In vitro analysis of 2 primary human T-ALL samples (D135 and M82) maintained briefly (2 days) on MS5-DL1 stromal feeders and then treated with GSI (1μM compound E) versus DMSO vehicle for 4 days. Cells were analyzed as in Figure 4 for proliferation (A), cell size (B), and viability by light scatter (C). PTEN protein expression was assessed by intracellular flow cytometry (D). FSC indicates forward light scatter; SSC, side scatter.
One caveat to note regarding the in vitro culture system used here is that the majority of cases required some form of Delta1 ligand stimulation, whether expressed on the surface of stromal feeders or immobilized on plastic, to “nurture” the cells through the acute stress of transition to in vitro culture. As such, the addition of GSI to assess dependence on Notch signaling essentially withdraws an important aspect of the culture system itself. An alternative approach to assess Notch dependence could have been to plate the cells on MS5 versus MS5-DL1 feeders, or similarly, bare plastic versus immobilized DL1 ligand; however, neither of these methods adequately model withdrawal of NOTCH1 signaling because certain HD mutations presumably confer ligand-independent signaling.5 Moreover, addition of GSI to the cultures is arguably the most direct way to address the particular issue of GSI resistance.
Discussion
In the current study, we have examined the effect of combined NOTCH1 activation and Pten loss in T-cell leukemia induction and observed tumors to occur with shorter latency and exhibit multiclonality, whereas tumors provided with either NOTCH1 activation or Pten loss alone were observed to occur with longer latency and were typically monoclonal or oligoclonal. These observations suggest NOTCH1 activation and Pten loss function cooperatively in the leukemogenic transformation process; however, because our experimental model used retroviral transduction to deliver activated NOTCH1 into hematopoietic progenitors, we cannot exclude the possibility that enhanced retroviral transduction efficiency in cycling Pten-null progenitors21 may contribute to the accelerated disease/increased clonality we have observed. Of note, we did not observe a similar disease acceleration/increased clonality in leukemias derived on the Ink4a/Arf-null background, which is also likely to enhance cycling of hematopoietic progenitors.41,42 Because of this potential limitation, however, further studies will be required to address the issue of oncogenic collaboration in a more definitive manner.
We also addressed whether indeed PTEN loss confers resistance to Notch inhibition as has been suggested by cell line studies.20 Using primary human and murine T-cell leukemia samples, we observed no correlation to exist between loss of PTEN and GSI resistance. Of note, Cullion et al observed murine Tal1 leukemias lacking Pten also to remain sensitive to GSI treatment.43 These findings raise the question as to what complement of secondary genetic alterations can indeed relieve T-cell leukemias of their addiction to Notch signaling. It was proposed recently that FBW7 mutation may contribute to GSI resistance by reducing c-Myc protein turnover.35 Although there was only a single case among our assayed primary human T-ALL samples that carried an FBW7 mutation (KER, Table 1), we found this case to be sensitive to GSI (supplemental Figure 7). This particular sample is PTEN wild type, however, and because FBW7 mutations are notably present in several identified PTEN-null, GSI-resistant human cell lines, it remains an open question as to whether the combination of PTEN loss and FBW7 mutation may lead to GSI resistance. FBW7 mutation likely cannot be a requisite feature of GSI resistance, however, because FBW7 mutations are lacking from the 2 GSI-resistant primary human cases reported here and also from several GSI-resistant cell lines.34,35
Another opportunity to examine the issue of Notch independence in T-ALL is presented by mouse models in which Notch1 mutations do not occur. Although the majority do show frequent Notch1 mutations,3,9 T-cell leukemias arising in mice with activated β-catenin in thymocytes44 and deletion of Pten in hemangioblastic precursors15 are notable exceptions lacking Notch1 mutations. Leukemias in both models showed consistent up-regulation of c-Myc and activation of β-catenin, and have led to the intriguing hypothesis that the combination of Wnt/β-catenin, c-Myc, and PI3K activation can functionally replace the need for Notch1 signaling.15 In this regard, it is notable that NOTCH1 directly induces c-MYC expression45-47 and may also repress PTEN via HES1.20 Given the diversity of downstream Notch targets, it is perhaps surprising that we and others have observed c-MYC alone can rescue many murine T-cell leukemias and some, but not all, human T-ALL cell lines from GSI-induced growth arrest.46,47 It would seem these cases are likely to harbor other genetic hits that provide or compensate for multiple pathways including PI3K/AKT activation.
In setting up primary murine leukemias for in vitro assays, we noted 4 (44%) of 9 wild-type leukemias to undergo massive apoptosis within the first few days of culture, whereas 3 (100%) of 3 Pten-null leukemias were more than 90% viable. This suggests enhanced PI3K/Akt signaling may confer a survival advantage in the unique setting of initial transition to in vitro culture. Interestingly, we have noted leukemias induced by more potent NOTCH1 mutants (eg, ΔE or ICN alleles) also to fare better during initial setup in vitro, suggesting high amplitude Notch signaling can replace and/or compensate for critical signals received in vivo, possibly including but not necessarily limited to PI3K/Akt activation.
The high frequency of NOTCH1 mutation in human T-ALL has sparked considerable interest in Notch signaling inhibitors as targeted therapy. Although early clinical studies have identified goblet cell hyperplasia-associated secretory diarrhea as a dose-limiting toxicity, both intermittent dosing strategies43,48 and coadministration of glucocorticoids49 may help to ameliorate these gastrointestinal side effects yet allow a therapeutic window to be achieved. We observed GSI resistance to occur in only a small fraction of primary human samples (2/13 assayed; 15%), and we and others43 have encountered GSI resistance only infrequently in murine leukemias. With respect to the human samples, it is worth noting that all of the primary samples reported here were collected from patients at initial diagnosis. It will thus be of interest to determine whether GSI resistance is found at higher frequency in patient samples collected at relapse. Although the majority of human T-ALL cell lines have been derived from patient material obtained at relapse,39 including several GSI-resistant examples (eg, BE13, CCRF-CEM, Jurkat, MOLT3, MOLT16, PEER, PF382),20,34,35 it is not clear in most cases whether the critical set of mutations was present in the patients' original material or was acquired after culture in vitro. Chemotherapy-induced genomic instability and/or clonal selection could presumably provide a potent driving force for accumulation of additional genetic hits necessary for leukemia cells to break their addiction to Notch signaling. Because Notch inhibitors will find their greatest clinical utility in patients with refractory/relapsed disease, determining the incidence of GSI resistance and relevance of PTEN loss in this setting certainly warrants further study.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Julien Calvo and Bastien Gerby (Commissariat à l'energie atomique), Amina Kariminia (BC Children's Hospital), Ling Chen (M. D. Anderson Cancer Center), and Sonya Lam (BC Cancer Agency) for assistance with sample preparation and characterization, Dr Vincenzo Giambra for (BC Cancer Agency) for optimization of the ligation-mediated PCR method, Dr Jon Aster (Brigham and Women's Hospital) for the L1601P-ΔPEST construct, and Dr Irwin Bernstein (Fred Hutchinson Cancer Research Center) for Delta1ext-IgG ligand. Drs Paola Ballerini (Hopital Armand-Trousseau) and Thierry Leblanc (Hopital Saint-Louis) generously contributed human T-ALL samples to this study.
This work was supported by grants from the National Cancer Institute of Canada/Terry Fox Foundation (A.P.W.), US National Cancer Institute (K22 CA112538 [A.P.W.], CA76641 [L.H.M.]), St Baldrick's Foundation (L.H.M.), Leukemia & Lymphoma Society of Canada (A.P.W.), Cancer Research Society (A.P.W.), Association Laurette Fugain (F.P.), and Institut contre le Cancer (F.P.). H.M. is supported by a Human Frontier Science Program Fellowship. A.L.G. was supported by a training grant (T32 CA009531 from the US Public Health Service). M.J.Y. is supported by the Ladies Leukemia League, an American Cancer Society Institutional Research Grant, an Institutional Research Grant of University of Texas M. D. Anderson Cancer Center, and a Physician Scientist Award of University of Texas M. D. Anderson Cancer Center. A.P.W. is a Michael Smith Foundation for Health Research Scholar.
National Institutes of Health
Authorship
Contribution: H.M. and S.G. designed, performed, and analyzed experiments; F.A. and F.P. provided methodological expertise for expansion of human T-ALL samples; X.G. and Q.L. generated conditional knockout animals; F.A., A.L.G., L.H.M., K.R.S., and F.P. provided genetically characterized patient samples and discussed results; M.J.Y. and A.P.W. conceived and supervised the study; and H.M. and A.P.W. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mingjian James You, Department of Hematopathology-Unit 72, Division of Pathology and Laboratory Medicine, M. D. Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: mjamesyou@mdanderson.org; or Andrew P. Weng, BC Cancer Agency/Terry Fox Laboratory, 675 West 10th Ave, Vancouver, BC V5Z 1L3, Canada; e-mail: aweng@bccrc.ca.
References
Author notes
X.G., F.A., and S.G. contributed equally to this work.