Abstract
High-level expression of the cytokine receptor-like factor 2 gene, CRLF2, in precursor B-cell acute lymphoblastic leukemia (pB-ALL) was shown to be caused by a translocation involving the IGH@ locus or a deletion juxtaposing CRLF2 with the P2RY8 promoter. To assess its possible prognostic value, CRLF2 expression was analyzed in 555 childhood pB-ALL patients treated according to the Acute Lymphoblastic Leukemia Berlin-Frankfurt-Münster 2000 (ALL-BFM 2000) protocol. Besides CRLF2 rearrangements, high-level CRLF2 expression was seen in cases with supernumerary copies of the CRLF2 locus. On the basis of the detection of CRLF2 rearrangements, a CRLF2 high-expression group (n = 49) was defined. This group had a 6-year relapse incidence of 31% plus or minus 8% compared with 11% plus or minus 1% in the CRLF2 low-expression group (P = .006). This difference was mainly attributable to an extremely high incidence of relapse (71% ± 19%) in non–high-risk patients with P2RY8-CRLF2 rearrangement. The assessment of CRLF2 aberrations may therefore serve as new stratification tool in Berlin-Frankfurt-Münster–based protocols by identifying additional high-risk patients who may benefit from an intensified and/or targeted treatment.
Introduction
Despite major improvements, for approximately 20% of children with acute lymphoblastic leukemia (ALL) therapy still fails and surviving patients often experience significant toxicities.1-3 Therefore, an improved assessment of a patient's risk of relapse is necessary to adapt treatment accordingly and enhance the chance of cure.
In the international Berlin-Frankfurt-Münster (BFM) study group trial ALL-BFM 2000, risk-adapted treatment stratification was mainly determined by the measurement of the in vivo treatment response.4-8 Response was assessed cytomorphologically (blast reduction in peripheral blood after 7 days of treatment [prednisone response, PR], blast clearance from bone marrow [BM] after induction therapy at week 5 [response on treatment day 33]), and molecularly by the measurement of minimal residual disease (MRD) at week 5 and after induction consolidation at week 12. Besides positivity for BCR-ABL or MLL-AF4 rearrangements, patients were stratified into the high-risk group (HR) by a poor PR, nonresponse by treatment day 33 (> 5% BM blasts), and a high MRD (> 10−3) load after induction consolidation at week 12. Whereas the relative number of relapses is greatest in the HR group, more than one half of relapses still occur in patients not classified as HR (ie, intermediate risk, standard risk).4 If identified early, these patients may benefit from an intensified HR treatment (ie, by application of a more intensive conventional chemotherapy, by addition of stem cell transplantation, or, ideally, by addition of a specific targeted treatment). However, current strategies fail to identify these patients and indicate the need for new prognostic markers.
Recently, we and other groups identified a novel subgroup of childhood precursor B-cell ALL (pB-ALL) characterized by high-level expression of the cytokine receptor-like factor 2 gene (CRLF2) caused by a translocation involving the immunoglobulin heavy chain locus (IGH@) locus on chromosome 14q32.3 and/or an interstitial deletion centromeric to CRLF2 juxtaposing CRLF2 with the P2RY8 promoter.9-11 The incidence of these abnormalities in pB-ALL was estimated at approximately 7%.9,10 We hypothesized that this subgroup of pB-ALL has distinct properties and that a high CRLF2 expression might be associated with treatment outcome. To test this hypothesis, we analyzed CRLF2 gene expression in an unselected population of 555 pB-ALL patients treated according to the ALL-BFM 2000 protocol.
Methods
Patients
In accordance with institutional review board regulations, clinical samples were obtained from children with ALL before treatment. The study was 4approved by the institutional review board of the Hannover Medical School and informed consent was obtained from patients and/or their legal guardians in accordance with the Declaration of Helsinki. Diagnostics, risk group assignment, and treatment were performed according to the ALL-BFM 2000 protocol.5,12 Between July 1999 and December 2004, 1933 patients with pB-ALL (aged ≤ 18 years) were enrolled into the ALL-BFM 2000 trial. In the present study, patients were included when spare diagnostic specimens containing more than 80% blasts were available from the German ALL-BFM biological specimen bank.
Real-time quantitative PCR
RNA isolation and real-time quantitative polymerase chain reaction (PCR) were performed as previously described.13 The succinate dehydrogenase complex subunit A (SDHA) gene was chosen for normalization. QuantiTect Primer Assays were used (CRLF2 [QT00210987], SDHA [QT00059486]; QIAGEN). Each sample was tested in duplicate. The expression ratio was calculated as 2n, where n was the CT value difference normalized by the CT difference of a calibrator sample.
Statistical analysis
Event-free survival (EFS) was calculated from date of diagnosis to last follow-up or to the first event (no complete remission [CR] as event on day 0, relapse, secondary malignancy, or death of any cause). Rates were calculated according to Kaplan-Meier and compared by log-rank test.14,15 Cumulative incidence of relapse functions were constructed by the method of Kalbfleish and Prentice and compared with the Gray test.16,17 Cox regression analysis was used for multivariate analysis.18 Proportional differences between patient groups were analyzed by χ2 or Fisher exact tests. Depending on the distribution of variables, correlation analyses were performed by computing contingency tables, Pearson, or Spearman correlation coefficients.
Genetic analysis
Fluorescence in situ hybridization (FISH) was performed on cells left from cytogenetic analysis according to routine methods. For detection of breakpoints in the IGH@ locus, the LSI IGH BAP probe was applied (Abbott/Vysis). Detection of breakpoints affecting the CRLF2 locus for a microdeletion upstream to CRLF2 was performed as previously described.9 In addition, reverse transcription PCR to detect the CRLF2-P2RY8 fusion and sequencing of CRLF2 to detect the CRLF2F232C mutation, JAK1 (exons 13 and 14), and JAK2 (exons 16, 20, 21) were performed as previously described.11,19
Results and discussion
CRLF2 gene expression was measured in diagnostic specimens of 555 patients (Figure 1A). Comparing characteristics of samples included in the present study and of those not analyzed, more patients older than 10 years of age (25.2% vs 20.8%, P = .03), with a greater white blood cell (WBC) count at diagnosis (>10 000/μL: 66.8% vs 39.9%, P < .001), and with an MLL-AF4 rearrangement (0.2% vs 0.9%, P = .05) were included. No significant differences were observed with respect to sex, presence of TEL-AML1 or BCR-ABL rearrangements, PR, MRD, and final risk stratification (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
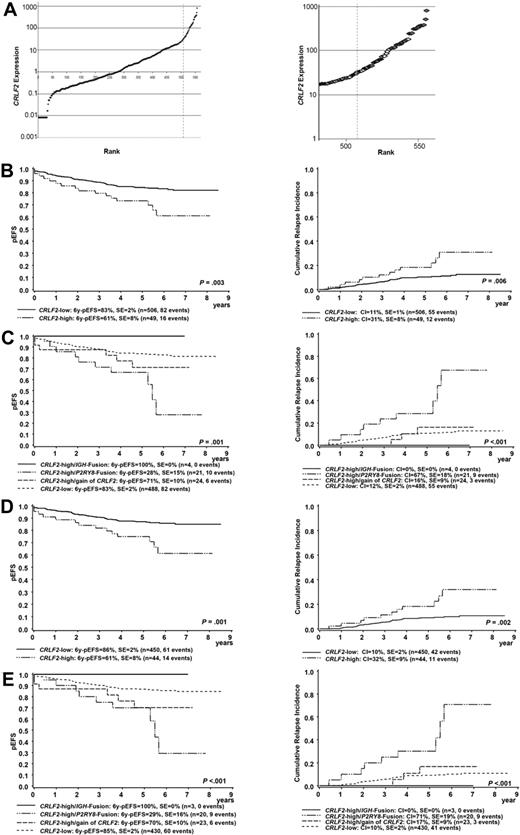
CRLF2 gene expression, underlying genomic alterations, and their association with treatment outcome. (A) Expression of CRLF2 in 555 patients with precursor B-cell ALL (left) is shown relative to the median expression of all samples. The dashed line indicates the cutoff between a CRLF2 high- and CRLF2 low-expression group. (Right) Zoom on the 70 cases with greatest CRLF2 expression analyzed for underlying genomic CRLF2 aberrations. Cases with P2RY8-CRLF2 or IGH-CRLF2 rearrangement (red), additional copies of the CRLF2 gene locus (yellow), negative for the P2RY8-CRLF2 rearrangement but without FISH analysis (blue), and without CRLF2 aberrations (green) are shown. (B) Kaplan-Meier estimate of EFS (left) and CRI (right) at 6 years according to CRLF2 expression in all patients analyzed. (C) Kaplan-Meier estimate of EFS (left) and CRI (right) at 6 years according to CRLF2 expression and detected underlying genomic CRLF2 aberrations in all patients analyzed. For EFS, the P value comparing the CRLF2 high-/P2RY8 fusion-positive and the CRLF2 low-expression group is shown. (D) Kaplan-Meier estimate of EFS (left) and CRI (right) at 6 years according to CRLF2 expression in non-HR patients only. (E) Kaplan-Meier estimate of EFS (left) and CRI (right) at 6 years according to CRLF2 expression and detected underlying genomic CRLF2 aberrations in non-HR patients only. For EFS, the P value comparing the CRLF2 high/P2RY8 fusion-positive and the CRLF2 low-expression group is shown.
CRLF2 gene expression, underlying genomic alterations, and their association with treatment outcome. (A) Expression of CRLF2 in 555 patients with precursor B-cell ALL (left) is shown relative to the median expression of all samples. The dashed line indicates the cutoff between a CRLF2 high- and CRLF2 low-expression group. (Right) Zoom on the 70 cases with greatest CRLF2 expression analyzed for underlying genomic CRLF2 aberrations. Cases with P2RY8-CRLF2 or IGH-CRLF2 rearrangement (red), additional copies of the CRLF2 gene locus (yellow), negative for the P2RY8-CRLF2 rearrangement but without FISH analysis (blue), and without CRLF2 aberrations (green) are shown. (B) Kaplan-Meier estimate of EFS (left) and CRI (right) at 6 years according to CRLF2 expression in all patients analyzed. (C) Kaplan-Meier estimate of EFS (left) and CRI (right) at 6 years according to CRLF2 expression and detected underlying genomic CRLF2 aberrations in all patients analyzed. For EFS, the P value comparing the CRLF2 high-/P2RY8 fusion-positive and the CRLF2 low-expression group is shown. (D) Kaplan-Meier estimate of EFS (left) and CRI (right) at 6 years according to CRLF2 expression in non-HR patients only. (E) Kaplan-Meier estimate of EFS (left) and CRI (right) at 6 years according to CRLF2 expression and detected underlying genomic CRLF2 aberrations in non-HR patients only. For EFS, the P value comparing the CRLF2 high/P2RY8 fusion-positive and the CRLF2 low-expression group is shown.
To define the best cutoff to distinguish a CRLF2 high- from a CRLF2 low-expression group, samples were screened for known CRLF2 involving genomic aberrations beginning with those having the greatest CRLF2 expression. The cutoff was set between positivity and negativity for P2RY8-CRLF2 and IGH@-CRLF2 rearrangements (Figure 1A). Screening for a P2RY8-CRLF2 rearrangement was performed in 70 samples; additional information on the IGH@-CRLF2 rearrangement by FISH was available in 32 of 49 (65%) samples negative for the P2RY8-CRLF2 rearrangement. P2RY8-CRLF2 rearrangements were detected in 21 and IGH@-CRLF2 rearrangements in 4 samples. Remarkably, 24 of the 28 samples showed supernumerary copies of the CRLF2 locus in the absence of a CRLF2-fusion, with 16 of them also having gains of the IGH@ locus. This finding could be explained at least in part by hyperdiploidy (as determined by a DNA index > 1.16 or by cytogenetics), which was observed in 9 of 12 cases with information on either DNA index (n = 10) or cytogenetics (n = 2) available. In none of the patients was a hereditary syndrome with constitutional gain of either chromosome X or chromosome Y described. In 17 of 70 samples a P2RY8-CRLF2 rearrangement could be excluded by reverse transcription PCR, but no cells were available for additional FISH analyses.
On the basis of the aforementioned results, 49 of 555 samples (9%) were included in the CRLF2 high-expression group (Figure 1A; supplemental Table 2): 21 cases characterized by the P2RY8-CRLF2 fusion, 4 cases by an IGH@-CRLF2 rearrangement, and 9 samples by additional CRLF2 copies. Two samples (both positive for BCR-ABL) did not show any CRLF2-involving abnormality, and in 13 samples an IGH@-CRLF2 rearrangement could not be excluded because no cells were available for additional FISH analyses. Interestingly, none of the 25 samples with the greatest CRLF2 expression was characterized by additional copies of the CRLF2 locus (supplemental Table 2). JAK2 mutations were observed in 5 P2RY8-CRLF2–positive cases and 1 case with an IGH@-CRLF2 rearrangement; the CRLF2F232C mutation was detected in 2 cases with a P2RY8-CRLF2 rearrangement. Neither CRLF2 nor JAK mutations were found in cases with a gain of the CRLF2 locus (supplemental Table 2).
When we compared the CRLF2 high- and low-expression groups, we observed no significant differences for sex, age and WBC at diagnosis, NCI risk groups, or the different measures of treatment response (Table 1). As expected, the number of Down syndrome–ALL (DS-ALL) patients was greater in the CRLF2 high- compared with the CRLF2 low-expression group (14.2% vs 1.4%, P < .001). There were no cases with TEL-AML1 or MLL-AF4 rearrangement in the CRLF2 high-expression group in contrast to 146 (29.7%) and 5 (1.0%) cases, respectively, in the CRLF2 low-expression group. Two BCR-ABL–positive cases showed a high CRLF2 expression but were not characterized by any of the known CRLF2 involving genomic aberrations. Within the CRLF2 high-expression group, patients with a P2RY8-CRLF2 rearrangement had a greater WBC count at diagnosis (> 50 000/μL, 48% vs 13%, P = .02) and a greater prevalence of NCI-HR status (62% vs 21%, P = .01) compared with patients with additional copies of the CRLF2 gene.
Patient characteristics and response to treatment according to CRLF2 expression in 555 patients with childhood precursor B-cell acute lymphoblastic leukemia
| . | CRLF2 low, n (%) . | CRLF2 high, n (%) . | P* . |
|---|---|---|---|
| Number of patients | 506 (100) | 49 (100) | |
| Down syndrome | < .001 | ||
| Yes | 7 (1.4) | 7 (14.2) | |
| No | 499 (98.6) | 42 (85.8) | |
| Sex | .65 | ||
| Male | 267 (52.8) | 24 (49.0) | |
| Female | 239 (47.2) | 25 (51.0) | |
| Age at diagnosis, y | .30 | ||
| 1 to less than 10 | 375 (74.1) | 40 (81.6) | |
| 10 or older | 131 (25.9) | 9 (18.4) | |
| Presenting WBC count, cells/μL | .23 | ||
| Less than 10 000 | 168 (33.2) | 16 (32.7) | |
| 10 000 to less than 50 000 | 223 (44.1) | 19 (38.8) | |
| 50 000 to less than 100 000 | 70 (13.8) | 5 (10.2) | |
| More than 100 000 | 45 (8.9) | 9 (18.4) | |
| BCR/ABL | .19 | ||
| Positive | 7 (1.4) | 2 (4.1) | |
| Negative | 499 (98.6) | 47 (95.9) | |
| MLL/AF4 | > .999 | ||
| Positive | 5 (1.0) | 0 (0.0) | |
| Negative | 501 (99.0) | 49 (100.0) | |
| TEL/AML1 | < .001 | ||
| Positive | 146 (28.9) | 0 (0.0) | |
| Negative | 345 (68.2) | 45 (90.5) | |
| Unknown | 15 (2.9) | 4 (9.5) | |
| NCI risk group | .76 | ||
| Standard | 295 (58.3) | 30 (61.2) | |
| High | 211 (41.7) | 19 (38.8) | |
| Prednisone response† | > .999 | ||
| Good | 466 (92.1) | 46 (93.9) | |
| Poor | 38 (7.5) | 3 (6.1) | |
| No result | 2 (0.4) | 0 (0.0) | |
| MRD‡ | .42 | ||
| Less than 10−3 | 462 (91.3) | 42 (85.7) | |
| More than 10−3 | 19 (3.8) | 3 (6.1) | |
| No result | 25 (4.9) | 4 (8.2) |
| . | CRLF2 low, n (%) . | CRLF2 high, n (%) . | P* . |
|---|---|---|---|
| Number of patients | 506 (100) | 49 (100) | |
| Down syndrome | < .001 | ||
| Yes | 7 (1.4) | 7 (14.2) | |
| No | 499 (98.6) | 42 (85.8) | |
| Sex | .65 | ||
| Male | 267 (52.8) | 24 (49.0) | |
| Female | 239 (47.2) | 25 (51.0) | |
| Age at diagnosis, y | .30 | ||
| 1 to less than 10 | 375 (74.1) | 40 (81.6) | |
| 10 or older | 131 (25.9) | 9 (18.4) | |
| Presenting WBC count, cells/μL | .23 | ||
| Less than 10 000 | 168 (33.2) | 16 (32.7) | |
| 10 000 to less than 50 000 | 223 (44.1) | 19 (38.8) | |
| 50 000 to less than 100 000 | 70 (13.8) | 5 (10.2) | |
| More than 100 000 | 45 (8.9) | 9 (18.4) | |
| BCR/ABL | .19 | ||
| Positive | 7 (1.4) | 2 (4.1) | |
| Negative | 499 (98.6) | 47 (95.9) | |
| MLL/AF4 | > .999 | ||
| Positive | 5 (1.0) | 0 (0.0) | |
| Negative | 501 (99.0) | 49 (100.0) | |
| TEL/AML1 | < .001 | ||
| Positive | 146 (28.9) | 0 (0.0) | |
| Negative | 345 (68.2) | 45 (90.5) | |
| Unknown | 15 (2.9) | 4 (9.5) | |
| NCI risk group | .76 | ||
| Standard | 295 (58.3) | 30 (61.2) | |
| High | 211 (41.7) | 19 (38.8) | |
| Prednisone response† | > .999 | ||
| Good | 466 (92.1) | 46 (93.9) | |
| Poor | 38 (7.5) | 3 (6.1) | |
| No result | 2 (0.4) | 0 (0.0) | |
| MRD‡ | .42 | ||
| Less than 10−3 | 462 (91.3) | 42 (85.7) | |
| More than 10−3 | 19 (3.8) | 3 (6.1) | |
| No result | 25 (4.9) | 4 (8.2) |
MRD indicates minimal residual disease; NCI, National Cancer Institute; and WBC, white blood cell.
Fisher exact test comparing the CRLF2 high and CRLF2 low groups.
Good: < 1000 leukemic blood blasts/μL on treatment day 8; poor: > 1000/μL.
After induction consolidation at week 12, MRD > 10−3 qualifies for the high-risk group.
Association of CRLF2 expression and treatment outcome
First, we analyzed the association of CRLF2 expression and treatment outcome in the entire set of patients. Patients with a high CRLF2 expression had a worse 6-year EFS probability compared with patients with a low CRLF2 expression (61% ± 8% vs 83% ± 2%, P = .003). This effect was mainly related to a greater cumulative relapse incidence (CRI; 31% ± 8% vs 11% ± 1%, P = .006; Figure 1B). No differences between CRLF2 high- and low-expression cases were seen with respect to time to relapse (< 30 months after initial diagnosis, 41.7% vs 47.3%; > 30 months, 58.3% vs 52.7%, P = .76) or site of relapse (isolated BM relapses: 75.0% vs 63.6%, central nervous system relapses, 25.0% vs 18.2%; combined relapses, 0% vs 18.2%; P = .28).
Next, we were interested whether there were differences detectable in clinical outcome between those CRLF2 high-expression cases with presence of a CRLF2 rearrangement and those with additional copies of the CRLF2 locus. Whereas all 4 patients with an IGH@-CRLF2 rearrangement remained in long-term CR, the 6-year EFS in patients with a P2RY8-CRLF2 rearrangement was 28% plus or minus 15% only, compared with 71% plus or minus 10% in cases with CRLF2 high-expression and additional copies of the CRLF2 locus and 83% plus or minus 2% in the CRLF2 low-expression group (P = .001). This association was again mainly attributable to a different CRI (67% ± 18% vs 16% ± 09% vs 12% ± 2%, P < .001; Figure 1C). Notably, all 13 CRLF2 high-expression patients without P2RY8-CRLF2 rearrangement and unavailable FISH data are in long-term CR. The exclusion of cases with MLL-AF4, BCR-ABL, or TEL-AML1 rearrangements and/or DS-ALL did not significantly change these results (supplemental Figures 1-3). Moreover, although the number of DS-ALL with high CRLF2 expression is limited (n = 7) and does not allow any statistical analysis, those with a P2RY8-CRLF2 rearrangement also appear to have a worse prognosis compared with those without it and low CRLF2 expression (3 of 6 relapses vs 0 of 7 relapses).
Because CRLF2 expression was associated with outcome but not with measures of treatment response, we next tested for potential effect modification by stratifying the analysis by risk groups (non-HR vs HR). We observed that the prognostic effect of a high CRLF2 expression was mainly attributable to the non-HR group. Non-HR patients with a high CRLF2 expression had an EFS probability of 61% plus or minus 9% compared with 86% plus or minus 2% for those in the low-expression group (P = .001). This effect was again mainly related to a greater CRI (32% ± 9% vs 10% ± 2%, P = .002; Figure 1D). By analyzing clinical outcome in non-HR CRLF2 high-expression cases with presence of CRLF2 rearrangements and those with supernumerary copies of the CRLF2 locus separately, we observed an extremely poor outcome in P2RY8-CRLF2–positive cases (EFS 29% ± 16%, CRI 71% ± 19%) compared with cases with more than 2 copies of the CRLF2 locus (EFS 70% ± 10%, CRI 17% ± 9%) or those with low CRLF2 expression (EFS 85% ± 2%, CRI 10% ± 2%, P < .001 for EFS and CRI, respectively; Figure 1E). Remarkably, all 8 P2RY8-CRLF2–positive relapses with information on MRD were negative for MRD after induction consolidation treatment at week 12. Only 5 of the 49 patients with a high CRLF2 expression were stratified as HR according the ALL-BFM 2000 protocol; 2 of them were positive for BCR-ABL, and only 1 patient (positive for BCR-ABL) experienced a relapse.
Altogether, patients with CRLF2 aberrations appear to be sensitive to in vivo treatment as measured by MRD. Therefore, most of them were not identified as being HR for relapse. When we examined outcome in NCI HR and NCI low-risk patients, we found that the CRLF2 high-expression status was associated with a poor outcome in both groups (supplemental Figures 1-3). We detected JAK2 mutations in 6 and CRLF2F232C mutations in 2 cases with CRLF2 rearrangement (supplemental Table 2): only 1 patient with JAK2R683S and 1 patient with CRLF2F232C mutation experienced a relapse. On the basis of the limited number of patients no statement can be given whether there are differences in outcome between patients with aberrant CRLF2 expression with or without JAK or CRLF2 mutations.
In a multivariate analysis considering initial WBC count, age at diagnosis, presence of TEL-AML1 rearrangement, presence of BCR-ABL or MLL-AF4 rearrangement, and MRD after induction consolidation in addition to either a high CRLF2 expression or presence of the P2RY8-CRLF2 rearrangement, the presence of a P2RY8-CRLF2 rearrangement but not a high CRLF2 expression irrespective of the underlying aberration provided independent prognostic information (risk ratio for relapse 3.11, 95% confidence interval 1.40-6.92, P = .005; supplemental Table 3).
In summary, high-level CRLF2 expression was associated with a poor EFS in childhood pB-ALL treated according to the ALL-BFM 2000 protocol. Similar observations were recently made by other groups.11,19,20 However, in contrast to published studies, we witnessed that this effect was mainly related to a greater CRI in non-HR patients with the presence of the P2RY8-CRLF2 rearrangement. Once confirmed independently, the assessment of CRLF2 status may, therefore, serve as a new stratification tool on BFM treatment regimens by identifying additional patients who are deemed HR for relapse. On the basis of the data presented here, however, it is not yet clear whether a high CRLF2 expression independent of the underlying aberrations per se or specifically the detection of the P2RY8-CRLF2 rearrangement is the decisive prognostic factor. However, with the detection of CRLF2 aberrations, there is for the first time a prognostic marker for a relative large group of patients (5%-10% of pB-ALL) with a HR of relapse who currently remain unrecognized by the stratification regimen because 90% of them are regularly stratified and treated as standard-risk or intermediate-risk patients. Whether these cases, which in the majority are sensitive to treatment (as measured by MRD), may benefit from an intensification of conventional therapy or whether they need the addition of hematopoietic stem cell transplantation has to be evaluated in future clinical trials. Moreover, high-level CRLF2 expression and/or aberrant CRLF2/JAK signaling may serve as therapeutic targets for this important subgroup of patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Birthe Fedders and Christian Bretscher from the ALL-BFM laboratory at the Department of Pediatrics and Magret Ratjen, Ursula Schnaidt, and Dorit Schuster from the Institute of Human Genetics in Kiel for their excellent technical assistance.
This work was supported by the Deutsche Krebshilfe, the Kinderkrebsinitiative Buchholz/Holm-Seppensen, and the Madeleine-Schickedanz-Kinderkrebsstiftung.
Authorship
Contribution: G.C., M.Z., and M. Stanulla designed the study, analyzed the data, and wrote the manuscript; R.R. and A.S. performed gene expression and MRD analysis and contributed to the writing of the manuscript; A.M. contributed to data analysis and interpretation and to the writing of the manuscript; S.G., J.H., and R.S. performed FISH analysis and contributed to the writing of the manuscript; I.V. performed sequencing and contributed to the writing of the manuscript; and S.I., T.A., M.J.S.D., R.S., and M. Shrappe made initial observations, were involved in the initiation of the study, and contributed to data interpretation and the writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gunnar Cario, MD, Department of Pediatrics, University Medical Center Schleswig-Holstein, Campus Kiel, Arnold-Heller-Str 3, 24105 Kiel, Germany; e-mail: g.cario@pediatrics.uni-kiel.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal