Abstract
Natural killer (NK) cells contribute to control of HIV/SIV infection. We defined macaque NK-cell subsets based on expression of CD56 and CD16 and found their distribution to be highly disparate. CD16+ NK cells predominated in peripheral blood, whereas most mucosal NK cells were CD56+, and lymph nodes contained both CD56+ and CD16−CD56− (double-negative [DN]) subsets. Functional profiles were also distinct among subsets—CD16+ NK cells expressed high levels of cytolytic molecules, and CD56+ NK cells were predominantly cytokine-secreting cells, whereas DN NK possessed both functions. In macaques chronically infected with SIV, circulating CD16+ and DN NK cells were expanded in number and, although markers of cytoxicity increased, cytokine secretion decreased. Notably, CD56+ NK cells in SIV-infected animals up-regulated perforin, granzyme B, and CD107a. In contrast, the lymph node–homing molecules CD62 ligand (CD62L) and C-C chemokine receptor type 7 (CCR7), which are expressed primarily on CD56+ and DN NK cells, were significantly down-regulated on NK cells from infected animals. These data demonstrate that SIV infection drives a shift in NK-cell function characterized by decreased cytokine production, expanded cytotoxicity, and trafficking away from secondary lymphoid organs, suggesting that the NK-cell repertoire is not only heterogeneous but also plastic.
Introduction
Since their discovery in the 1970s, natural killer (NK) cells have been considered the major effector cells of the innate immune system because of their ability to kill virus-infected or neoplastic cells. Although NK cell–mediated killing does not require prior antigen sensitization, cell-to-cell contact between NK and target T cells occurs through a complex array of inhibitory and activating receptors. In humans, NK cells express both killer-cell immunoglobulin-like receptors (KIRs), which interact with major histocompatibility complex (MHC) class I molecules and can be either inhibitory or activating, and receptors belonging to the C-type lectin family such as natural killer group 2A (NKG2A), an inhibitory receptor that recognizes HLA-E and NKG2D, which recognizes the stress-induced ligands MHC class I chain-related gene A and B (MICA/MICB) and members of the ULBP family.1 Human NK cells also express various natural cytotoxicity receptors including NKp46, NKp30, and NKp44, for which the ligands remain incompletely characterized.2,3 However, increasing evidence suggests that the complexity of NK-cell function has been underappreciated and that in addition to cytolysis of aberrant T cells, NK cells also produce a wide array of cytokines, mediate tolerance to self-antigens, and regulate dendritic cell functions.4 Most recently, murine studies have suggested that NK cells may even display characteristics of adaptive immune responses.5
In humans, 2 primary phenotypically defined subsets of NK cells have been described, cytolytic CD56dimCD16+ and cytokine-secreting CD56brightCD16− subsets, of which the CD56dimCD16+ subset predominates in blood. However, efforts to identify comparable subsets of NK cells in nonhuman primate models used for HIV/AIDS research, such as rhesus macaques (Macaca mulatta), have been complicated by incomplete or erroneous phenotypic definitions. Initial reports variably defined rhesus macaque NK cells as either CD3−CD16+ or CD3−CD8αα+, and controversy has existed as to whether CD56, which is a primary marker of human NK cells, is expressed on rhesus NK cells at all.6-8 The definition of macaque NK cells as CD3−CD16+ cells in unprocessed whole blood has also been further complicated by “CD16 masking” by SIV/immunoglobulin G immune complexes, a phenomenon not observed in fractionated peripheral blood mononuclear cells (PBMCs).9 We have previously identified macaque NK cells as CD3−CD8αα+ CD20−/dimNKG2A+ cells, and determined that whereas the major NK-cell subpopulation is indeed CD16+CD56−/dim, 2 minor populations, CD16−/dimCD56hi and CD16−CD56− NK cells also exist in peripheral blood of rhesus macaques.10 This phenotypic definition of nonhuman primate NK cells has been verified in rhesus macaques by another group and also shown to apply to sooty mangabeys.11
The study of NK-cell biology has been further complicated by the fact that very little data exist on the nature of human NK cells outside peripheral blood, and in nonhuman primate models, virtually nothing is known. Fehniger et al12 first described NK cells in human lymph nodes as primarily CD56+CD16− cells that could regulate development of adaptive immune responses. NK cells of a similar phenotype and lacking cytolytic functions have been described in tonsils, the gastrointestinal and female reproductive tracts, and at the fetal-maternal interface where they regulate trophoblast invasion and help protect against infection.13-16 These data, although limited, are highly indicative of potential regulatory roles for primate NK cells and suggest they may be functionally compartmentalized.
In both humans and mice, NK cells are critical for defense against several viral infections including influenza, cytomegalovirus, varicella zoster virus, and herpes simplex virus, and an ever-expanding body of evidence suggests that NK cells are also important for control of HIV infection.17-21 Lysis of HIV-infected cells by NK cells is well documented, and can occur through a variety of different mechanisms including antibody-dependent cell-mediated cytotoxicity, indicating cooperative innate and adaptive immune responses in controlling infection.19-22 Subsets of NK cells also secrete relatively large amounts of the β-chemokines CCL3, CCL4, and CCL5, which are able to inhibit C-C chemokine receptor type 5 (CCR5)–dependent entry of HIV in vitro.23 Interestingly, acute HIV disease is associated with expansion of the CD56dimCD16+ NK-cell subset in humans, suggesting a virus-driven increase in cytolytic activity and long-term nonprogressors have increased NK-cell cytoxicity compared with viremic subjects.24,25 Furthermore, in large HIV-infected cohorts, subjects expressing killer cell immunoglobulin-like receptor 3DS1 (KIR3DS1) or KIR3DL1 and the cognate HLA-B Bw4 ligands had significantly slower rates of disease progression.26,27 Several studies also suggest that NK cells help mediate control of SIV infection in nonhuman primates. Acute infection with SIVmac251 results in activation and increased lytic capacity of NK cells that appear to be associated with control of SIV infection in both rhesus macaques and sooty mangabeys, and NK-cell expansion and infiltration have been associated with preventing development of neuro-AIDS in a pig-tailed macaque model of SIV infection.11,28-30 Interestingly, however, a recent study of monkeys acutely infected with SIV did not show any clear effect of depletion of the CD16+ subset of NK cells on either viral load or on loss of central memory CD4+ T cells.31
Efforts to obtain a clearer understanding of the specific role(s) NK cells may play during HIV infection are significantly hindered by a lack of data on NK cells in the genital and intestinal mucosae, where HIV transmission and replication primarily take place. Obtaining these tissues from humans is challenging and, therefore, necessitates rigorous functional and phenotypic characterizations of NK cells in multiple tissues from both virus-naive and SIV-infected macaques. In this study, we sought to comprehensively characterize the biology of macaque NK-cell subpopulations and their relative distribution in tissues outside peripheral blood and to ascertain the effects of chronic SIV infection on their phenotype and function.
Methods
Animals and infections
Indian rhesus macaques of either sex were used in this study. All animals were free of simian retrovirus type D, SIV, simian T-lymphotrophic virus type 1, and herpes B virus. All macaques were housed at the New England Primate Research Center and maintained in accordance with the guidelines of the Committee on Animals of the Harvard Medical School and the Guide for the Care and Use of Laboratory Animals. Institutional Animal Care and Use Committee approval for the animal studies was obtained from Harvard Medical School and the New England Primate Research Center. A total of 65 macaques were analyzed: 45 SIV-naive macaques and 20 macaques infected chronically with SIV. Of the 20, 5 were infected intravenously with SIVmac239 and 15 were infected with SIVmac251, either intravaginally (n = 5) or intrarectally (n = 10). The mean duration of infection was 293 days (range, 162-707 days).
Cell processing
Rhesus macaque PBMCs were isolated from ethylenediaminetetraacetic acid–treated venous blood by density gradient centrifugation over Lymphocyte Separation Medium (MP Biomedicals), and contaminating red blood cells were lysed using a hypotonic ammonium chloride solution. Tissue mononuclear cells were isolated from lymph nodes and rectal and vaginal biopsies as described previously,32 and after isolation all cells were washed and resuspended in phosphate-buffered saline supplemented with 2% fetal calf serum (Sigma-Aldrich) for subsequent assays.
Antibodies and flow cytometric analyses
Cell-surface staining was carried out using standard protocols for our laboratory as described previously.33 Except where noted, all antibodies were obtained from BD Biosciences and included fluorochrome-conjugated monoclonal antibodies to the following molecules: CCR5 (allophycocyanin [APC] conjugate, clone 3A9), CCR6 (phycoerythrin [PE] conjugate, clone 11A9), CCR7 (Alexa 700 conjugate, clone 150503, in-house conjugation), CD3 (APC, APC–cyanin 7 [Cy7], Pacific Blue and peridinin-chlorophyll-protein complex [PerCP]–Cy5.5 conjugates, clone SP34.2), CD8α (Qdot 605 conjugate, clone T8/7Pt-3F9 [National Institutes of Health Nonhuman Primate Reagent Resource Program] and APC-Cy7 conjugate, clone SK1), CD8β (PE conjugate, clone 2ST8-5H7; Immunotech) CD14 (PE-Cy7 conjugate, clone M5E2), CD16 (Alexa 700, PE, and fluorescein isothiocyanate [FITC] conjugates, clone 3G8), CD20 (PerCp-Cy5.5 conjugate, clone L27), CD27 (APC-H7 conjugate, clone M-T271), CD56 (PE-Cy7 conjugate, clone NCAM16.2), CD62 ligand (CD62L; FITC conjugate, clone SK11), CD69 (PE–Texas Red conjugate, clone TP1.55.3; Beckman-Coulter), CXC chemokine receptor 3 (CXCR3; PE-Cy5 conjugate, clone 1C6/CXCR3), CXCR4 (APC conjugate, clone 12G5), NKG2A (Pacific Blue conjugate, clone Z199, in-house conjugation), NKp30 (PE conjugate, clone Z25; Immunotech), and NKp46 (PE conjugate, clone BAB281; Immunotech). Intracellular staining for granzyme B (PE conjugate, clone GB12; Invitrogen), perforin (FITC conjugate, clone Pf-344; MabTech), and Ki67 (FITC conjugate, clone B56) was performed on mononuclear cells using Caltag Fix & Perm (Invitrogen) according to the manufacturer's suggested protocol. All acquisitions were made on an LSR II (BD Biosciences) and analyzed using FlowJo software (TreeStar Inc).
Whole-blood lymphocyte count assay
A whole-blood, no-wash, bead-normalized assay was used to enumerate total NK cells as described,34 and absolute counts of different NK subsets were determined by combining polychromatic flow cytometry–generated frequency data with these results.
NK-stimulation assay
We analyzed multiple functions of NK cells ex vivo after stimulation with the MICA/MICB-negative, MHC-devoid human cell line, 721.221. Prior to functional analyses, we enriched for NK cells by first depleting T cells with Pan–Immunoglobulin G Dynabeads (Dynal Biotech) coated with cross-reactive monoclonal anti-CD3 antibodies (clone 6G12; kindly provided by Johnson Wong, Massachusetts General Hospital). Next, 1 × 106 NK-enriched cells were resuspended in RPMI 1640 (Sigma-Aldrich) containing 10% FBS and stimulated at an effector-target ratio of 2.5:1 with 721.221 cells; phorbol myristate acetate (50 ng/mL) and ionomycin (1 μg/mL) or medium alone served as positive and negative controls, respectively. Anti–CD107a-PerCp-Cy5 (clone H4A3) was added directly to each of the tubes at a concentration of 20 μL/mL, and Golgiplug (brefeldin A) and Golgistop (monensin) were added at final concentrations of 6 μg/mL; then all samples were cultured for 12 hours at 37°C in 5% CO2. After culture, samples were surface stained using markers to delineate NK cells (CD3, CD8, NKG2A, CD16, CD56). Cells were then permeabilized using Caltag Fix & Perm and intracellular cytokine staining was performed for macrophage inflammatory protein 1β (MIP-1β; FITC conjugate, clone 24006; R&D Systems), interferon-γ (IFN-γ; APC conjugate, clone B27; Invitrogen), and tumor necrosis factor-α (TNF-α; Alexa 700 conjugate, Mab11; Pharmingen).
Plasma virus load quantification
RNA copy equivalents were determined in ethylenediaminetetraacetic acid–treated plasma using a standard quantitative real-time reverse-transcription polymerase chain reaction assay based on amplification of conserved sequences in gag.35 The limit of detection for this assay was 30 viral RNA copy equivalents per milliliter of plasma.
Statistical analyses
All statistical and graphic analyses were done using GraphPad Prism software (GraphPad Software Inc). Nonparametric Friedman and Mann-Whitney tests were used where indicated, and P values less than .05 were assumed to be significant in all analyses.
Results
Identification of multiple NK subsets in peripheral blood of rhesus macaques
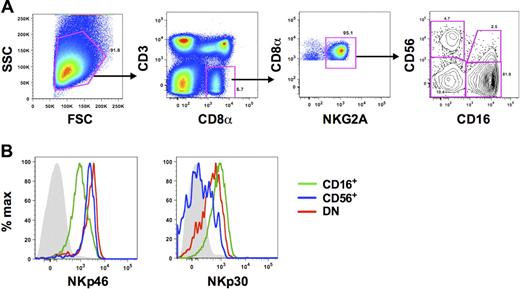
A major challenge of identifying NK cells in primates is the lack of a single lineage-specific maker. In humans, NK cells have generally been defined as being CD56+,36 and previous analyses of NK cells in rhesus macaques have used simplified definitions of CD3−CD8αα+ or CD3−CD16+. However, CD56, CD8α, and CD16 can also be expressed on monocytes,8 B cells,10 and dendritic cells,37 respectively. Therefore, using polychromatic flow cytometry, we were able to expand on previous work in our laboratory10 and refine a more detailed gating strategy that excludes these contaminating cell populations. NK cells were identified as medium-to-large lymphocytes that were CD3− and positive for CD8α and NKG2A (Figure 1). CD3−CD8αα+ NKG2A+ cells also lacked CD8β and CD14 expression (data not shown). In the CD3−CD8αα+NKG2A+ cell population, we identified 4 subsets based on expression of the NK-cell markers, CD56 and CD16. In peripheral blood the CD56−CD16+ subset (hereafter referred to as CD16+ NK cells) was by far the dominant subset among total blood NK cells (median frequency, 85.9%; n = 45), with low but variable frequencies of CD56+CD16− and CD56+CD16+ NK cells (hereafter referred to as CD56+ and double positive [DP], respectively). The DP NK subset was the least distinctly defined in peripheral blood and therefore was excluded from most phenotypic analyses. Interestingly, as has been shown previously by our laboratory, we also found low frequencies (median, 5.9%; n = 45) of NK cells expressing neither CD56 nor CD16 (double-negative [DN]). Using polychromatic flow cytometry, we then sought to confirm that each of these subsets was indeed composed of NK cells by examining coexpression of the natural cytotoxicity receptors NKp46 and NKp30. Although all subsets expressed these molecules on the cell surface, further confirming their identity as NK cells phenotypically, expression was highly disparate among the subsets (Figure 1B; supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The activating receptor NKp46 was expressed on all NK cells, but the mean fluorescence intensity, as an indicator of cell-surface density, was approximately 4-fold greater on CD56+ and DN NK cells than on CD16+ NK cells. In contrast, NKp30 was expressed at the highest levels on CD16+, but dull on CD56+ and DN NK cells.
Phenotypic characterization of NK-cell subsets in peripheral blood of rhesus macaques. (A) Representative flow cytometric plots defining NK cells in PBMCs. Macaque NK cells were identified using a broad side- versus forward-scatter gate and phenotypically defined as CD3−CD8α+NKG2A+. Subsets were further delineated by expression of CD56 and/or CD16. (B) Representative histogram overlays depict expression of NKp46 and NKp30 on each of the 3 primary NK subsets. Gray closed histograms are from isotype-matched controls. CD56+ indicates CD56+CD16−; CD16+, CD56−CD16+; and DN, CD56−CD16−. Examples are representative of 15 to 27 naive animals.
Phenotypic characterization of NK-cell subsets in peripheral blood of rhesus macaques. (A) Representative flow cytometric plots defining NK cells in PBMCs. Macaque NK cells were identified using a broad side- versus forward-scatter gate and phenotypically defined as CD3−CD8α+NKG2A+. Subsets were further delineated by expression of CD56 and/or CD16. (B) Representative histogram overlays depict expression of NKp46 and NKp30 on each of the 3 primary NK subsets. Gray closed histograms are from isotype-matched controls. CD56+ indicates CD56+CD16−; CD16+, CD56−CD16+; and DN, CD56−CD16−. Examples are representative of 15 to 27 naive animals.
Differential heterogeneity of NK subsets in lymphoid and mucosal tissues
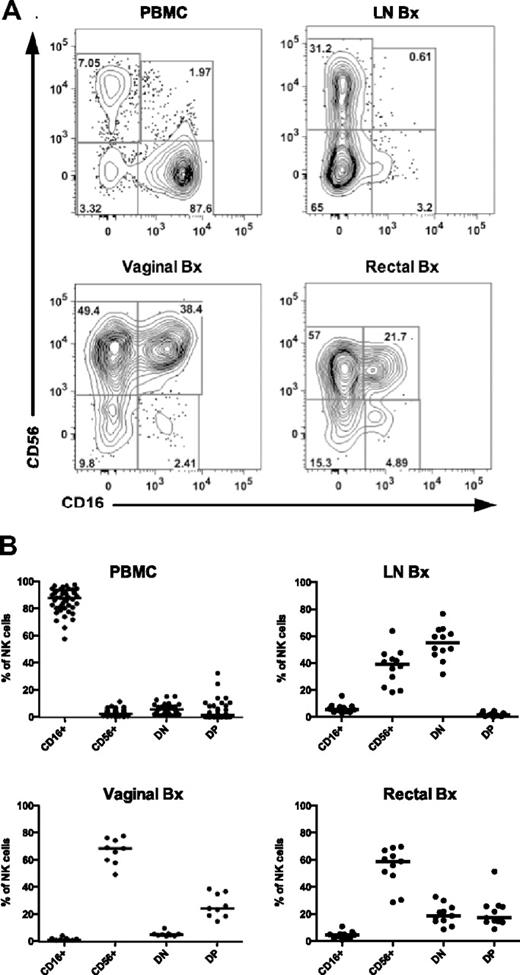
As noted previously, data on the phenotype and function of NK cells in tissues remain quite limited. Given the relative ease of access to tissues in rhesus macaques, we sought to phenotypically define NK-cell subsets in biopsies of peripheral lymph node, vagina, and rectum from normal rhesus macaques using the markers shown in Figure 1 while also adding a CD45 panleukocyte pregate (data not shown) to exclude epithelial cells and debris, which often contaminate mucosal cell suspensions (data not shown). Interestingly, the median frequency of total NK cells among lymphocytes in peripheral blood (median, 3.3%; range, 0.90%-11.2%; n = 45) was notably greater than that found in either lymph nodes (median, 0.38%; range, 0.06%-0.87%; n = 12) or in rectal biopsies (median, 1.2%; range, 0.48%-2.4%; n = 10), but was lower than what we observed in vaginal biopsies (median, 8.5%; range, 4.2%-16.8%; n = 8). Although most NK cells in peripheral blood were CD16+, the CD56+ subset was clearly enriched in lymph nodes, constituting 20% to 40% of the total population, and the CD16+ subset was virtually absent, similar to what has been described for humans12 (Figure 2). We found that in addition to the CD56+ subset, DN NK cells were also found at high frequencies in lymph nodes—nearly 80% of total lymph node NK cells in some animals. Most surprisingly, in vaginal and rectal mucosae, NK cells were generally more than 70% CD56+, and the DP subset, which was poorly defined in peripheral blood, was easily resolved in the mucosae and present at high frequencies.
Distribution of macaque NK-cell subsets in blood and tissues. (A) Representative flow cytometric plots showing distribution of NK subsets based on CD56 and CD16 expression in PBMCs and lymph node (LN), vaginal, and rectal biopsies (Bx) of uninfected rhesus macaques. (B) Frequencies of each of the NK-cell subsets are shown as a fraction of the total NK-cell population as defined in Figure 1. Horizontal bars represent medians of 9-15 animals.
Distribution of macaque NK-cell subsets in blood and tissues. (A) Representative flow cytometric plots showing distribution of NK subsets based on CD56 and CD16 expression in PBMCs and lymph node (LN), vaginal, and rectal biopsies (Bx) of uninfected rhesus macaques. (B) Frequencies of each of the NK-cell subsets are shown as a fraction of the total NK-cell population as defined in Figure 1. Horizontal bars represent medians of 9-15 animals.
Increased numbers of circulating CD56− NK subsets in macaques chronically infected with SIV
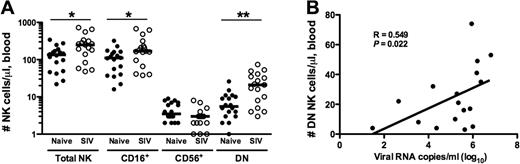
Standard methods for calculating absolute numbers of lymphocytes in peripheral blood are often inaccurate, and evaluations of cell frequencies can be misinterpreted when other cell types drastically change in number, such as CD4+ T cells during HIV and SIV infections. To address this problem, we have previously used a flow cytometric bead-normalized assay to calculate absolute numbers of NK cells in peripheral blood,34 and in this study applied the assay to enumerate NK subsets in both naive and SIV-infected macaques. In a cohort of 18 SIV-naive macaques, the median number of total NK cells was 135 cells/μL of blood (range, 22-341 cells/μL), similar to other reported numbers for rhesus macaques but somewhat lower compared with humans,38,39 and, as expected, the bulk of NK cells in peripheral blood were of the CD16+ subset (Figure 3A). Because both HIV and SIV infections have been reported to alter the numbers of NK cells in the circulation, we next examined absolute counts in chronically SIV-infected macaques. In 17 macaques infected with either SIVmac239 or SIVmac251, the median number of NK cells was 249 cells/μL of blood (range, 48-724 cells/μL)—significantly (P = .018, Mann-Whitney U test) higher than observed in naive animals. Furthermore, this increase was subset specific, with greater numbers in both the CD16+ and DN subsets, but slightly lower numbers of CD56+ NK cells in infected animals. Interestingly, although no relationship was found between plasma virus loads and circulating bulk, CD16+, or CD56+ NK cells, the numbers of circulating DN NK cells did significantly correlate with virus loads (Figure 3B).
Enumeration of absolute numbers of circulating NK-cell subsets in naive and SIV-infected macaques. Absolute numbers of total circulating NK cells were quantified in whole blood as previously described.34 NK subset counts were calculated as fractions of the total absolute counts using frequencies determined by polychromatic flow cytometry (Figure 1). (A) Counts from naive animals are compared with those from chronically infected animals, and only significant P values are shown. Horizontal bars indicate medians. Mann-Whitney test; *P < .05; **P < .01. (B) Absolute counts of peripheral blood DN NK cells correlated with plasma viral loads; Spearman correlation. P < .05 is considered significant.
Enumeration of absolute numbers of circulating NK-cell subsets in naive and SIV-infected macaques. Absolute numbers of total circulating NK cells were quantified in whole blood as previously described.34 NK subset counts were calculated as fractions of the total absolute counts using frequencies determined by polychromatic flow cytometry (Figure 1). (A) Counts from naive animals are compared with those from chronically infected animals, and only significant P values are shown. Horizontal bars indicate medians. Mann-Whitney test; *P < .05; **P < .01. (B) Absolute counts of peripheral blood DN NK cells correlated with plasma viral loads; Spearman correlation. P < .05 is considered significant.
Chronic SIV infection alters expression of tissue-homing molecules on NK subsets
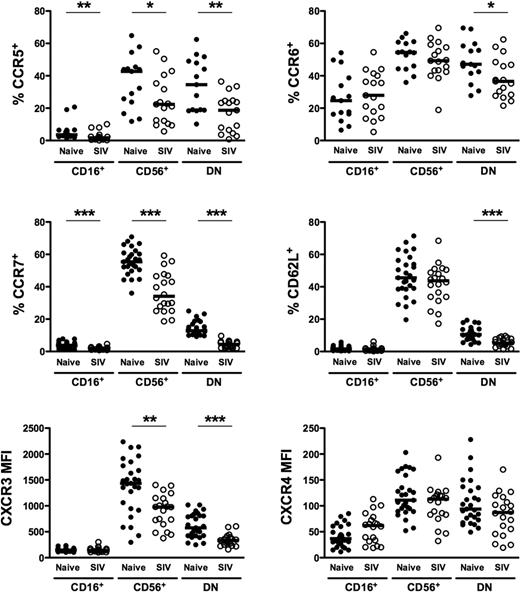
Given the high degree of heterogeneity and disparate distribution of macaque NK subsets in tissues, we next analyzed cell-surface molecules relevant to tissue trafficking on each of the subsets found in peripheral blood. As shown in Figure 4, the tissue-homing molecules CCR5 and CCR6 and lymph node–homing markers CCR7 and CD62L were expressed at the highest levels on CD56+ NK cells and were also found, albeit at variable levels, on DN NK cells. However, in stark contrast, among these molecules, only CCR6 was appreciably expressed on CD16+ NK cells, and CCR7 and CD62L were virtually absent, which could explain, in part, the lack of the CD16+ subset in lymph nodes (Figure 2). The highly disparate expression of tissue-homing markers on macaque NK cells is similar to what has been previously reported in both humans13 and rhesus macaques10 and suggests that each of the subsets have very different trafficking patterns. Similarly, the CXC chemokine receptor, CXCR3, was expressed on all NK subsets, but at disproportionately high levels on CD56+ compared with both CD16+ and DN cells, whereas expression of another chemokine receptor, CXCR4, was dull on all 3 subsets. Both CXCR3 and CXCR4 have been implicated in normal lymphocyte trafficking to spleen and bone marrow and recruitment to sites of inflammation.13,40-42
Comparison of homing markers and chemokine receptors on NK-cell subsets in naive and SIV-infected macaques. Percentages of positive cells above background (CCR5, CCR6, CCR7, CD62L) and mean fluorescence intensities (CXCR3, CXCR4) were compared between naive and SIV-infected macaques for peripheral blood CD56+, CD16+, and DN NK subsets as shown in Figure 1. Horizontal bars indicate medians. Only significant P values are shown for naive versus SIV-infected comparisons. Mann-Whitney test; *P < .05; **P < .01; ***P < .001.
Comparison of homing markers and chemokine receptors on NK-cell subsets in naive and SIV-infected macaques. Percentages of positive cells above background (CCR5, CCR6, CCR7, CD62L) and mean fluorescence intensities (CXCR3, CXCR4) were compared between naive and SIV-infected macaques for peripheral blood CD56+, CD16+, and DN NK subsets as shown in Figure 1. Horizontal bars indicate medians. Only significant P values are shown for naive versus SIV-infected comparisons. Mann-Whitney test; *P < .05; **P < .01; ***P < .001.
We next examined how chronic SIV infection affected expression of tissue-homing molecules on the different subsets of macaque NK cells. Strikingly, expression of CCR5, CCR7, CD62L, and CXCR3 on DN NK cells in chronically infected macaques was nearly half the levels observed in naive macaques and, although less dramatic, CCR6 expression was also significantly reduced (Figure 4). Similarly, CCR5, CCR7, and CXCR3 were all down-regulated on CD56+ NK cells in infected animals. It is interesting to note, however, that although the disparities between naive and infected macaques appeared to be global among NK cells for CCR5, CCR7, and CXCR3, the reduction in CD62L expression was subset specific to DN NK cells. These obvious alterations in the normal expression of trafficking markers on NK-cell subsets during chronic SIV infection indicate that the functional niches maintained by each subset in the tissues in which they reside could also be drastically altered.
Expression of activation and cytotoxic markers is increased on NK cells from SIV-infected animals
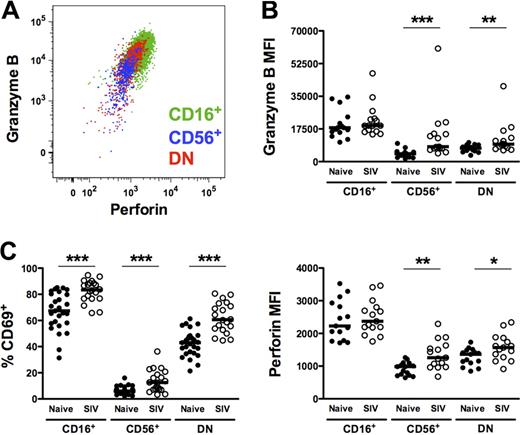
As shown in Figure 5A, all macaque NK subsets expressed intracellular perforin and granzyme B. However, not unexpectedly, and consistent with a more cytotoxic phenotype, intracellular levels of both cytotoxic enzymes were greatest in the CD16+ subset (Figure 5B), as was cell-surface expression of the activation marker CD69 (Figure 5C). However, whereas expression of perforin, granzyme B, and CD69 was generally low on CD56+ and DN NK cells, all 3 markers were significantly up-regulated in SIV-infected macaques, suggesting a generalized increase in NK-cell activation. Somewhat unexpected was the up-regulation of intracellular perforin and granzyme B in the CD56+ subset from infected animals. However, intracellular expression of both enzymes in CD56+ NK cells positively correlated with plasma virus load, indicating a virus-dependent change in phenotype and potential cytotoxic function (perforin, R = 0.75, P < .001; granzyme B, R = 0.69, P = .004).
Increased expression of cytotoxic and activation markers on macaque NK-cell subsets from SIV-infected animals. (A) Representative polychromatic flow cytometric overlay depicting coordinate but variable expression of intracellular granzyme B and perforin by all peripheral blood NK subsets. (B) Mean fluorescence intensities of granzyme B (top panel) and perforin (bottom panel) and (C) percentages of positive cells above background of cell-surface CD69 are shown for each of the NK subsets in naive and SIV-infected animals. Only significant P values are shown for naive versus SIV-infected comparisons. Mann-Whitney test; *P < .05; **P < .01; ***P < .001.
Increased expression of cytotoxic and activation markers on macaque NK-cell subsets from SIV-infected animals. (A) Representative polychromatic flow cytometric overlay depicting coordinate but variable expression of intracellular granzyme B and perforin by all peripheral blood NK subsets. (B) Mean fluorescence intensities of granzyme B (top panel) and perforin (bottom panel) and (C) percentages of positive cells above background of cell-surface CD69 are shown for each of the NK subsets in naive and SIV-infected animals. Only significant P values are shown for naive versus SIV-infected comparisons. Mann-Whitney test; *P < .05; **P < .01; ***P < .001.
NK cells from chronically SIV-infected macaques have decreased cytokine production but increased degranulation ex vivo
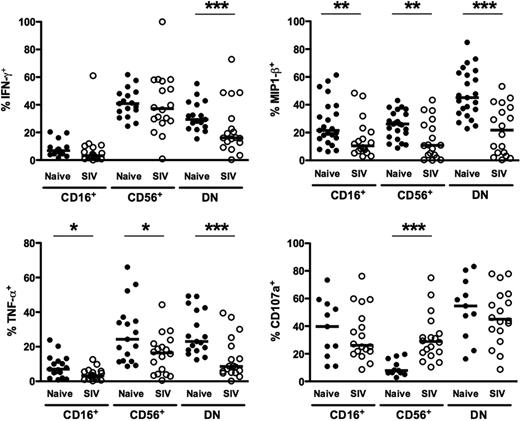
Because human NK-cell function varies significantly between CD56dimCD16+ and CD56brightCD16− subsets,36,43 we also wanted to examine NK-cell function in rhesus macaques, both in regard to cytokine production and, as an indicator of cytotoxicity, degranulation. Moreover, because chronic SIV infection dramatically altered the phenotype of macaque NK-cell subsets (Figures 4–5) and perturbation of NK-cell function has been reported for both HIV and SIV infections,25,44,45 we wanted to analyze NK-cell subset function comprehensively in both virus-naive and chronically SIV-infected animals. To do so, we stimulated enriched NK cells ex vivo with 721.221 cells and then measured intracellular expression of 3 cytokines produced by NK cells—IFN-γ, TNF-α, and CCL4 (MIP-1β), as well as expression of the degranulation marker, CD107a. In normal macaques, not unexpectedly, stimulated CD16+ NK cells produced little IFN-γ or TNF-α, whereas both CD56+ and DN NK cells expressed both cytokines at high frequencies (Figure 6). Interestingly, all 3 subsets produced CCL4, suggesting CD16+ NK cells could also mediate some chemokine-dependent regulatory functions. Furthermore, CD107a was up-regulated on both DN and CD16+ NK cells upon stimulation, with little expression on the CD56+ subset. These data are comparable with findings for human NK cells that have characterized CD56bright NK cells as primarily cytokine-producing cells and CD56dimCD16+ cells as more cytotoxic.36 Moreover, these data also highlight the uniqueness of the less clearly understood DN NK cells, which appear both to be potent cytokine-secreting cells and to possess strong cytotoxic potential.
Functionality of NK-cell subsets in naive and SIV-infected macaques. Percentages of peripheral blood NK-cell subsets positive for intracellular IFN-γ, TNF-α, MIP-1β, and CD107a in response to 721.221 cells. Horizontal bars indicate medians. Only significant P values are shown for naive versus SIV-infected comparisons. Mann-Whitney test; *P < .05; **P < .01; ***P < .001.
Functionality of NK-cell subsets in naive and SIV-infected macaques. Percentages of peripheral blood NK-cell subsets positive for intracellular IFN-γ, TNF-α, MIP-1β, and CD107a in response to 721.221 cells. Horizontal bars indicate medians. Only significant P values are shown for naive versus SIV-infected comparisons. Mann-Whitney test; *P < .05; **P < .01; ***P < .001.
Next, we compared the functions of NK subsets in naive macaques to those chronically infected with SIV. Strikingly, ex vivo production of both TNF-α and CCL4, after stimulation, was significantly impaired in all 3 subsets in infected compared with naive animals (Figure 6). Interestingly, although IFN-γ production was reduced by 100% in DN NK cells, CD56+ NK cells produced comparable levels in both naive and SIV-infected macaques. These data suggest an overall reduction in cytokine production by NK cells during chronic SIV infection, which is particularly dramatic in DN NK cells. Furthermore, although expression of the degranulation marker, CD107a, was comparable between naive and SIV-infected macaques on both CD16+ and DN NK cells, it was found at nearly 4-fold higher frequencies on CD56+ NK cells from SIV-infected compared with naive macaques. These data, coupled with the up-regulation of both intracellular granzyme B and perforin in CD56+ NK cells (Figure 5), are likely indicative of a SIV-induced shift in function, whereupon these normally cytokine-producing cells acquire cytolytic functions.
Discussion
Modeling HIV disease by experimentally infecting macaques with various SIV strains has provided valuable insights into both the pathogenesis and immune control of lentiviruses. However, a major deficit has been an incomplete understanding of the role of innate immunity, particularly NK cells, in disease control and how innate immune functions are subverted during virus-induced dysfunction. Here, we present 2 major bodies of data demonstrating that (1) NK cells in macaques are remarkably diverse, but can be divided into 4 primary subsets based on the classical NK markers CD56+ and CD16+ that are distinct in phenotype, function, and tissue distribution; and (2) chronic SIV infection significantly alters the phenotypes and functions ascribed to each of these NK subsets.
NK-cell heterogeneity in normal rhesus macaques
Here we have defined rhesus macaque NK cells as CD3−CD8αα+-NKG2A+, and in normal, SIV-naive rhesus macaques, total NK cells were found in frequencies and absolute numbers similar to previous reports for rhesus macaques,38 but slightly lower than what has been reported in healthy humans.39 Consistent with previous reports, we found the dominant NK subset in macaque peripheral blood was CD16+ accompanied by the minor CD56+ and DN subsets.10,38 Moreover, using polychromatic flow cytometry we confirmed prior data generated in our laboratory10 and validated unequivocally the identity of the minor subsets by demonstrating expression of NKp46 and NKp30 and, most convincingly, by showing each of these subsets respond to the MHC-devoid cell line 721.221.
Although all 3 peripheral blood NK-cell subsets responded to 721.221 cells, demonstrating generalized responsiveness to a lack of MHC, their functional profiles were distinct. CD56+ NK cells secreted copious amounts of cytokines with low levels of degranulation based on CD107a expression, whereas CD16+ cells expressed significantly higher levels of CD107a, but had limited cytokine secretion. These data suggest the 2 subsets are functionally analogous to the CD56brightCD16− and CD56dimCD16+ NK cells in humans. There were, however, some indications for functional overlap—CD56+ cells contained both perforin and granzyme B, highlighting that they do have potential for cell killing, and CD16+ NK cells secreted CCL4 at levels comparable with that of CD56+ NK cells. However, most interesting perhaps is the DN NK subset, which does not have an obvious human counterpart, but had the broadest spectrum of both cytokine-secreting and cytotoxic features. Collectively, we can infer that NK cells are highly plastic, and strict, simplistic definitions of “effector” versus “regulatory” functional roles for NK cells may be inappropriate. Furthermore, delineation of the phenotypic and functional heterogeneity among subsets may give some clues to their ontogeny. For example, we found expression of CD62L and CCR7 was highest on CD56+ and lowest on CD16+ NK-cell subsets, and some ex vivo human studies have suggested an inverse relationship between these 2 molecules and expression of CD16 to cytotoxicity.46-48 In these studies, interleukin-2 (IL-2) stimulation resulted in an up-regulation of CD16 and enhanced cytotoxicity accompanied by down-regulation of CCR7 and CD62L. In contrast, IFN-α, IL-12, IL-15, and IL-18 all induced up-regulation of CD62L and CCR7 with simultaneous CD16 down-regulation. These data are clearly in line with the phenotypic and functional differences we observed in macaque CD56+ and CD16+ NK cells and suggest the possibility that the multifunctional DN NK cells, which express intermediate levels of CD62L and CCR7 as well as the cytotoxic enzymes, perforin and granzyme B, are a precursor cell to the other NK subsets. Additional experiments will, however, be necessary to confirm this hypothesis.
In humans, lymph node NK cells have been characterized as primarily CD56bright and lacking CD16,12 and although data on NK cells in the reproductive and gastrointestinal mucosae are limited, most research suggests they have a similar phenotype.15,16 Here, we have shown that rhesus macaque NK cells are similarly compartmentalized, with CD16+ NK cells virtually absent from lymph nodes and most CD16+ NK cells in the mucosae coexpressing CD56. These data demonstrate the remarkable phenotypic heterogeneity of NK cells, but also underscore the fact that previous definitions of macaque NK cells as only CD16+, without including CD56+ cells, vastly underappreciate their systemic distribution and abundance, particularly because the majority of lymphocytes are found in the gastrointestinal tract. These data also complicate the interpretation of CD16+ NK-depletion studies in macaques,31,38 which would result in elimination of only small fractions of NK cells in tissues. Like human NK cells, the lymph node–homing molecules CCR7 and CD62L were expressed at the highest levels on CD56+ and DN NK cells, and the absence thereof likely explains the lack of CD16+ NK cells in lymph nodes.13,14 Interestingly, CD56+ cells also expressed the highest levels of CCR5, CCR6, and CXCR3 and CXCR4, all of which have been implicated in NK trafficking to a diverse array of tissues including spleen, skin, and gut-associated lymphoid tissue in humans and mice and may explain the enrichment of this subset in the mucosa.13,41,42,49 This expression pattern is mimicked on human CD56brightCD16− NK cells in blood. Moderate expression of CCR6 and CXCR3 was also found on CD16+ macaque NK cells in peripheral blood and could direct homing of these cells to mucosal sites.
Effect of SIV infection on NK-cell subsets
Although NK cells are known to suppress virus replication and kill infected cells, NK-cell dysfunction in HIV-infected subjects23-25,39,50 and, to a lesser degree, in SIV-infected macaques,28-30 is well documented. However, these studies have generally addressed only 1 or 2 functional parameters and, in the case of macaque studies, have limited their focus to only CD16+ NK cells. In particular, in vivo CD16 depletion in macaques showed minimal impact on SIV viremia and CD4+ T-cell loss.31 Because we have now shown that the bulk of macaque NK cells in the mucosa, where the virus primarily replicates, are not CD16+, NK-depletion experiments using anti-CD16 antibodies have significant caveats, and new reagents for in vivo NK-depletion experiments need to be developed. We also demonstrated that the number of DN and CD16+ NK cells in peripheral blood expanded during chronic SIV infection, with a modest but not significant reduction in CD56+ NK cells. Although this might reflect a net loss of CD56+ NK cells, it is equally possible that these cells have redistributed to tissues, such as the gut mucosa, where we have shown they are more prevalent and where virus replication primarily takes place. In general, chronic SIV infection resulted in a global down-regulation of homing molecules, which may indicate dysfunctional cellular trafficking and/or tissue redistribution. Down-regulation of CCR7 on CD56+ NK cells and of both CD62L and CCR7 on DN NK cells is highly suggestive of decreased homing of NK subsets to lymph nodes. The disparity in down-regulation of CCR7 but not CD62L on CD56+ NK cells indicates that the 2 subsets would not necessarily traffic to the same tissues during infection. Decreased expression of the cytokine receptors CCR5, CCR6, CCR7, and CXCR3 also suggests altered NK-cell trafficking patterns and implies NK cells are less responsive to the respective receptor ligands. Some of these chemokines are known to increase during HIV/SIV infections, and although their precise influence on NK-cell trafficking is unknown, decreased cell-surface expression of their receptors suggests NK cells may be more anergic to inflammatory mediators during chronic SIV infection.
Increased NK-cell activation and cytotoxicity have been reported during HIV infections,25,39 and a similar phenomenon is evident in our cohort of SIV-infected macaques—characterized by increased intracellular expression of perforin and granzyme B, increased degranulation, as evidenced by CD107a up-regulation, as well increased cell-surface expression of CD69. Similar to published reports in HIV-infected persons, we also observed an overall decrease in cytokine production by NK cells. However, most striking was that in the CD56+ subset of macaque NK cells, whose putative human counterpart has been characterized as primarily cytokine secreting,36 measures of cytoxicity were significantly increased, whereas measures of cytokine production decreased. Although it is possible that these alterations in function and phenotype are indicative of immune dysfunction, it is also likely that this is an inherent reshaping of the NK-cell repertoire in response to chronic viremic SIV infection.
Here we have shown that during chronic SIV infection NK-cell function is characterized by decreased cytokine production and increased cytotoxicity, likely driven by ongoing viral replication but not necessarily related to immune dysfunction. We also provide evidence for altered trafficking patterns directed away from the secondary lymphoid organs, perhaps to sites of SIV replication and/or transmission. Regardless, these data indicate not only that NK cells likely have a more complex role in lentivirus infections than previously recognized, but also that such infections provide indirect evidence of an inherent plasticity by triggering significant alterations in NK-cell functional and phenotypic repertoires.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Angela Carville and Elaine Roberts for dedicated animal care; Drs Dan Barouch, Ron Desrosiers, and David T. Evans for donating macaque blood samples; and Dr Jeff Lifson, Dr Michael Piatak Jr, and the Quantitative Molecular Diagnostics Core of the AIDS and Cancer Virus Program, SAIC Frederick Inc, NCI Frederick for plasma SIV RNA determinations.
This work was supported through National Institutes of Health grants AI062412, AI071306, and RR00168, and a Center for HIV/AIDS Vaccine Immunology/HIV Vaccine Trials Network Early Career Investigator award, grant number U19 AI 067854-04 (R.K.R.).
National Institutes of Health
Authorship
Contribution: R.K.R. and R.P.J. designed experiments; R.K.R., J.G., F.E.W., and Y.Y. processed samples and performed experiments; M.C. performed flow cytometric acquisitions and fluorochrome-conjugated “in-house” antibodies crucial to the study; and R.K.R. and R.P.J. analyzed the data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: R. Paul Johnson, Division of Immunology, New England Primate Research Center, Harvard Medical School, One Pine Hill Dr, Southborough, MA 01772-9102; e-mail: paul_johnson@hms.harvard.edu.