Abstract
A consistently increased mRNA expression of the adhesion receptor CD11b is a hallmark of the reported genomewide gene expression changes in precursor B-cell acute lymphoblastic leukemia (PBC-ALL) after 1 week of induction therapy. To investigate its clinical relevance, CD11b protein expression in leukemic blasts has been prospectively measured at diagnosis (159 patients) and during therapy (53 patients). The initially heterogeneous expression of CD11b inversely correlated with cytoreduction rates measured at clinically significant time points of induction therapy in the ALL–Berlin-Frankfurt-Münster 2000 protocol. CD11b positivity conferred a 5-fold increased risk of minimal residual disease (MRD) after induction therapy (day 33) and of high-risk group assignment after consolidation therapy (day 78). In the multivariate analysis CD11b expression was an independent prognostic factor compared with other clinically relevant parameters at diagnosis. During therapy, CD11b expression increased early in most ALL cases and remained consistently increased during induction/consolidation therapy. In more than 30% of MRD-positive cases, the CD11b expression on blast cells exceeded that of mature memory B cells and improved the discrimination of residual leukemic cells from regenerating bone marrow. Taken together, CD11b expression has considerable implications for prognosis, treatment response monitoring, and MRD detection in childhood PBC-ALL.
Introduction
Microarray-based gene expression profiling is a powerful tool for basic research, target discovery, and identification of novel molecular biomarkers.1-4 This method has been used to examine gene expression profiles of malignant cells, and recent studies have identified signatures characteristic of various hematologic malignancies.1,4 Gene expression profiling in childhood acute lymphoblastic leukemia (ALL) has been performed to describe cytogenetic subgroups and to identify differences in gene expression at diagnosis associated with differential treatment resistance.5-9 In the ALL–Berlin-Frankfurt-Münster (BFM) protocol, treatment resistance is quantified by the minimal residual disease (MRD) load at days 8, 33, and 78, which provides major criteria for patient risk stratification.10,11
In view of its crucial clinical significance, MRD assessment has become central to the clinical management of patients with ALL.10,12 Flow cytometry (FCM) is among the methods available for MRD monitoring and provides, in addition to blast number quantification, cell-biologic information on immunophenotypic features of MRD cells. ALL cells differ from normal lymphoid precursors by quantitative aberrations in their tissue-, differentiation-, and cytogenetic-associated antigenic expression profiles.13-15 However, the expression of major antigens used for FCM-MRD identification, CD10, CD34, CD45, CD20, CD19, TdT, is modulated during therapy.16,17 This phenomenon could be due to a selection of leukemic subclones that exhibit the different antigenic profiles already at diagnosis and sustain the first period of chemotherapy better than the bulk of leukemia cells.18 Alternatively, we considered that a drug-induced regulatory modulation of the cellular machinery of antigen expression is the predominant cause of the phenomenon, rather than subclone selection or apoptosis-related artifacts.19 Independent from the underlying mechanism, the investigation of cells persisting during therapy may provide valuable information on the molecular factors involved in differential therapy response and prognosis in ALL. We directly addressed this issue by comparing genome-wide gene expression analysis of leukemic blast cells at diagnosis and after 7 days of induction therapy with prednisolone.20 This allowed us to identify genes potentially involved in the cellular responses to treatment at functional levels of cell cycling, differentiation, and survival. These changes collectively indicated an expression shift in day 8 (d8) cells toward resting mature B cells. In addition to the well-known changes of B-cell differentiation markers, the list of the genes with altered expression in d8 blasts showed an up-regulation of the CD11b cell surface molecule, which has not been previously reported in context of B-lineage–specific development.20 CD11b belongs to the α subunits of the integrin receptor family and acts as a transmembrane molecule, critical in cellular adhesion and signal transduction.21,22 Integrins mediate multiple cellular functions, including adhesion, migration, complement binding, and cell survival. Given their frequent involvement in cancer development and therapy resistance, integrins are considered as attractive candidates for targeted treatment approaches in oncology. In immunophenotyping, CD11b is a leukocyte-specific receptor and is regarded as a marker for monocyte/macrophages, granulocytes, and natural killer cells.23-25 It is also expressed on a memory subset of normal B cells, but its expression and clinical significance have not been addressed in precursor B-cell (PBC)–ALL.26
In the present study we explored the significance of CD11b expression in PBC-ALL. Because CD11b expression has been initially described in cells persisting under therapy, we asked whether CD11b+ cases may be present already at diagnosis and clinically differ from CD11b− cases. The protein expression at diagnosis was quantified by FCM and analyzed in context of the laboratory and clinical data available so far. Furthermore, we addressed applicability of CD11b as a MRD-specific marker in PBC-ALL. To this end, a CD11b antibody has been included into a 9-color, single-tube panel (a nuclear stain Syto16 and fluorochrome-conjugated monoclonal antibodies to CD19, CD20, CD10, CD34, CD45, CD58, CD3), which has been used for MRD monitoring in PBC-ALL at clinically significant MRD time points of the ALL-BFM protocol (days 8, 15, 33, and 78). With the use of the same panel, CD11b expression in ALL was compared with that in normal mature B cells and B-cell precursors. Our data indicated inverse correlation of CD11b expression at diagnosis with cytoreduction rates and usefulness of this antigen in the MRD detection by FCM.
Methods
Patients
Patients with PBC-ALL were enrolled in the BFM multicenter trial ALL-BFM 2000 for the treatment of childhood ALL (age, 1-18 years). The data on CD11b expression at diagnosis were collected from September 2005 to August 2007. The monitoring of FCM-MRD with the use of the antibody panel with CD11b antibody was performed from November 2006 to February 2008. The investigations were approved as a part of the international Italian Association for Pediatric Hematology and Oncology and BFM Study Group (AIEOP–BFM) ALL trial by all participating review boards. The patients or legal guardians gave informed consent in accordance with the Helsinki protocol. The diagnosis of ALL was based on morphologic as well as cytochemical and immunophenotypic features. The criteria for the subclassification of PBC-ALL (pro-B, common, and pre-B-ALL) were adopted from the guidelines proposed by the European Group for the Immunological Characterization of Leukemias.27 Assignment to the risk groups (standard, intermediate, and high) after induction and consolidation therapy was performed according to the ALL-BFM 2000 protocol.10,11
CD11b mRNA expression at diagnosis and at day 8
The data on CD11b mRNA expression have been obtained from the genomewide gene expression study of patients with ALL at diagnosis and at day 8 of induction therapy reported by Rhein et al.20 In brief, blast cells were isolated by flow sorting (FACSVantage; Becton Dickinson) from intraindividual sequentially paired peripheral blood (PB) samples at time points day 0 and day 8 of 18 patients with PBC-ALL. Total RNA from isolated cells was extracted with the use of the RNeasy Micro Kit (QIAGEN). Each RNA sample was linearly amplified by 2 rounds of in vitro transcription and labeled with biotin with the use of the MessageAmp aRNA Kit (Ambion) and biotin-11–cytidine triphosphate and -16–uridine triphosphate (Perkin-Elmer). Biotin-labeled cRNA was hybridized to Affymetrix GeneChip HG U133A chips (Affymetrix).
CD11b protein expression at diagnosis
CD11b expression at diagnosis was investigated by FCM prospectively in 159 patients. Individual clinical features of the patients, including age, sex, immunophenotype, white blood cell (WBC) counts, DNA index, and molecular translocations are given in Table 1. The CD11b antibody (clone Bear1; Beckman Coulter), conjugated with phycoerythrin (PE)–cyanin 5, was used in combination with CD19, CD10, and CD34 antibodies, and viable cells were gated with the use of forward versus side scatter plots. Detailed data on the antibody panels, sample preparation, and staining have been described recently.28 The acquisition of cells was performed with the use of a FC500 cytometer (Beckman Coulter). CD11b expression values were quantified with the use of Dako FluoroSpheres (Beckman Coulter) with assigned values of molecules of equivalent soluble fluorochrome (MESFs). The final values were corrected for background staining with the use of negative nonleukemic subpopulations.20
FCM-MRD and CD11b protein expression
MRD quantification by FCM (FCM-MRD) has been performed within the international AIEOP-BFM ALL 2000 study at clinically significant MRD time points in bone marrow (BM; days 0, 15, 33, and 78) and PB (day 8) as described.28,29 At least 50 000 events at diagnosis and 300 000 events for MRD measurements were acquired. FCM measurements provided both relative MRD values (as percentage of nucleated cells) and absolute blast counts (BCs; blasts per μL). Absolute BCs were calculated with the cell count of the sample (assessed with a conventional hemocytometer) and the relative MRD estimate from the FCM analysis. According to the standard operating procedure of the Flow-MRD in the AIEOP-BFM ALL 2000 study, the minimum number of cells required for the sample MRD positivity has been set to 30 blasts; this corresponds to the sensitivity of 10−4 if 300 000 cells have been acquired. Blast reduction rates (BRRs) have been calculated as absolute BC changes in relation to the initial blast burden, for example, for day 8 (d8): BRRd8 = (BCd0 − BCd8)/BCd0 × 100. The FCM-MRD status in the day-33 BM was considered as categorical parameter with the dichotomous outcome values “MRD-positive” or “MRD-negative.”
In the present study, the method of the FCM-MRD approach has been modified by up-grading of the simultaneous parameter acquisition from 4 to 9 fluorescence colors (CyanADP flow analyser; Beckman Coulter). This enabled MRD analysis with the use of a single tube approach with the cyanin 5–conjugated CD11b antibody (Beckman Coulter) and a backbone combination of 7 monoclonal antibodies against CD10, CD45, CD34, CD3, CD19, CD58, and CD20 (conjugated with PE, PE–Texas red, PE–cyanin 7, cascade yellow, pacific blue, allophycocyanin, and allophycocyanin–cyanin 7, respectively; all from Beckman Coulter, with the exception of CD20, Becton Dickinson). In the ninth fluorescence channel a cell- permeable nucleic acid stain Syto 16 (Molecular Probes) has been used to distinguish nucleated cells from debris.
Statistics
Mean values of continuous variables are given as mean plus or minus SE. Comparison of mean values was performed by the Mann-Whitney U test. Associations of continuous variables were evaluated with bivariate nonparametric Spearman correlation statistics. Comparison of categorical variables was performed with the Fisher exact test or the χ2 test, including risk estimation and calculation of the odds ratio with a 95% confidence interval. Cutoff values for CD11b expression as predictive parameter were calculated with receiver operating characteristic (ROC) curve analysis. The predictive contribution of CD11b in the multiparametric setting was investigated with the binary logistic regression model. All calculations were performed with SPSS (Version 16.0; SPSS Inc).
Results
CD11b mRNA expression at diagnosis correlates with early response to therapy
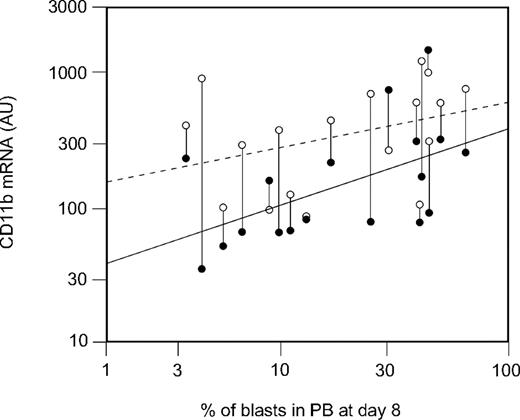
CD11b mRNA data at days 0 and 8 have been analyzed for correlation with the treatment response parameters percentage of blast cells (day 8%) and cytoreduction rate (BRR day 8) at day 8. CD11b expression at diagnosis showed significant correlations with both day 8% (Spearman coefficient = 0.61, P = .007; Figure 1) and BRR day 8 (Spearman coefficient = −0.53, P = .025; data not shown). As Figure 1 shows, CD11b mRNA levels have been consistently increased in most patients with ALL (15 of 18; 83%). In contrast to the expression levels in untreated ALL blasts at diagnosis, CD11b expression on the blasts persisting at day 8 did not significantly correlate with the day 8% (Spearman coefficient = 0.38, P = .12; Figure 1).
Correlation of CD11b mRNA at diagnosis with percentage of blasts at day 8. Blast cells from paired day 8 to day 0 samples were purified by flow sorting, and mRNA was isolated, amplified, and hybridized to oligonucleotide chips. CD11b mRNA expression at diagnosis in arbitrary units (AUs; ●) correlates significantly with blast percentage in PB at day 8 (Spearman coefficient = 0.61, P = .007). The CD11b expression at day 8 (○) is consistently increased and does not correlate with blast percentage at day 8 (Spearman coefficient = 0.38, P = .12).
Correlation of CD11b mRNA at diagnosis with percentage of blasts at day 8. Blast cells from paired day 8 to day 0 samples were purified by flow sorting, and mRNA was isolated, amplified, and hybridized to oligonucleotide chips. CD11b mRNA expression at diagnosis in arbitrary units (AUs; ●) correlates significantly with blast percentage in PB at day 8 (Spearman coefficient = 0.61, P = .007). The CD11b expression at day 8 (○) is consistently increased and does not correlate with blast percentage at day 8 (Spearman coefficient = 0.38, P = .12).
CD11b protein expression at diagnosis correlates with response to induction therapy
In a series of 61 patients, CD11b has been measured at diagnosis in both PB and BM samples. There was a slightly higher level of expression in PB than in BM (805 ± 166 vs 611 ± 116 MESFs; P = .028) with a strong correlation (Spearman coefficient = 0.85, P < .001) between PB and BM samples (data not shown).
To investigate the clinical relevance of CD11b expression, CD11b has been prospectively measured in diagnostic PB samples from a total of 159 patients with ALL. CD11b showed a broad range of expression intensity between 0 and 77 800 MESFs with mean expression value of 1840 MESFs. MESF values that corresponded to the 25th, 50th, and 75th percentiles were represented by 103, 360, and 1288 MESF units, respectively. Table 1 shows CD11b expression in context of initial laboratory, cytogenetic, and diagnostic parameters (age, sex, WBC count, immunophenotype, molecular translocations, DNA index) and the corresponding subgroups, which have been considered to be of clinical relevance.30 There were no significant differences for CD11b expression between patient groups subdivided by sex (female/male [f/m]), age (10 years), WBC count (50.0 × 109/L [50 000/μL]), DNA index (1.16), or immunophenotype (pro-B, common, and pre-B ALL). The CD11b expression differences were not significant also between BCR/ABL-positive and -negative cases. Interestingly, TEL/AML1-positive ALLs were almost totally CD11b− and significantly differed from the TEL/AML1-negative samples (260 ± 37 vs 2500 ± 720 MESFs; P = .002).
Initial laboratory and molecular parameters and CD11b expression
| . | No. of patients . | CD11b, MESF units, mean ± SE . | P . |
|---|---|---|---|
| Sex | |||
| Male | 88 | 2390 ± 910 | .24 |
| Female | 71 | 1170 ± 250 | |
| Age | |||
| 1-9 y | 123 | 1930 ± 650 | .76 |
| 10-18 y | 36 | 1540 ± 480 | |
| WBC count at diagnosis | |||
| Less than 50.0 ×109/L (50 000/μL) | 132 | 2010 ± 620 | .50 |
| More than 50.0 ×109/L (50 000/μL) | 24 | 1020 ± 370 | |
| Immunophenotype | |||
| Pro | 1 | 1752 | |
| Common | 137 | 2000 ± 600 | .49* |
| Pre | 16 | 770 ± 340 | |
| Other† | 5 | ||
| BCR/ABL | |||
| Positive | 7 | 3420 ± 1100 | .52 |
| Negative | 152 | 1780 ± 540 | |
| MLL/AF4 | |||
| Positive | 1 | 1960 | |
| Negative | 138 | 2010 ± 590 | |
| TEL/AML1 | |||
| Positive | 43 | 260 ± 37 | .002 |
| Negative | 113 | 2500 ± 720 | |
| DNA index‡ | |||
| Less than 1.16 | 118 | 1700 ± 680 | .50 |
| Greater than 1.16 | 33 | 2600 ± 490 |
| . | No. of patients . | CD11b, MESF units, mean ± SE . | P . |
|---|---|---|---|
| Sex | |||
| Male | 88 | 2390 ± 910 | .24 |
| Female | 71 | 1170 ± 250 | |
| Age | |||
| 1-9 y | 123 | 1930 ± 650 | .76 |
| 10-18 y | 36 | 1540 ± 480 | |
| WBC count at diagnosis | |||
| Less than 50.0 ×109/L (50 000/μL) | 132 | 2010 ± 620 | .50 |
| More than 50.0 ×109/L (50 000/μL) | 24 | 1020 ± 370 | |
| Immunophenotype | |||
| Pro | 1 | 1752 | |
| Common | 137 | 2000 ± 600 | .49* |
| Pre | 16 | 770 ± 340 | |
| Other† | 5 | ||
| BCR/ABL | |||
| Positive | 7 | 3420 ± 1100 | .52 |
| Negative | 152 | 1780 ± 540 | |
| MLL/AF4 | |||
| Positive | 1 | 1960 | |
| Negative | 138 | 2010 ± 590 | |
| TEL/AML1 | |||
| Positive | 43 | 260 ± 37 | .002 |
| Negative | 113 | 2500 ± 720 | |
| DNA index‡ | |||
| Less than 1.16 | 118 | 1700 ± 680 | .50 |
| Greater than 1.16 | 33 | 2600 ± 490 |
MESF indicates molecule of equivalent soluble fluorochrome; and WBC, white blood cell.
Common ALL versus pre-B-ALL.
Other indicates biphenotypic or nonspecified.
Determined by flow cytometry.
In contrast to the missing association with almost all of the initial parameters at diagnosis, we found a significant correlation of the CD11b expression with clinical response data monitored by FCM at different time points of induction therapy. Notably, expression of CD11b significantly correlated with both BRRs and blast percentages in PB at day 8 and in BM at days 15 and 33 (Table 2).
Correlation between CD11b expression quantified by flow cytometry in PB samples from patients with ALL at diagnosis and response to induction therapy
| . | No. of patients . | Spearman coefficient . | P . |
|---|---|---|---|
| BRR | |||
| Day 8 | 107 | −0.27 | .005 |
| Day 15 | 69 | −0.26 | .030 |
| Day 33 | 71 | −0.25 | .036 |
| Percentage of blasts | |||
| Day 8 | 107 | 0.32 | .001 |
| Day 15 | 78 | 0.22 | .053 |
| Day 33 | 77 | 0.28 | .014 |
| . | No. of patients . | Spearman coefficient . | P . |
|---|---|---|---|
| BRR | |||
| Day 8 | 107 | −0.27 | .005 |
| Day 15 | 69 | −0.26 | .030 |
| Day 33 | 71 | −0.25 | .036 |
| Percentage of blasts | |||
| Day 8 | 107 | 0.32 | .001 |
| Day 15 | 78 | 0.22 | .053 |
| Day 33 | 77 | 0.28 | .014 |
PB indicates peripheral blood; ALL, acute lymphoblastic leukemia; and BRR, blast reduction rate.
CD11b expression at diagnosis has independent prognostic effect
The diagnostic performance of CD11b with the dichotomous end points of remission status after induction therapy (FCM-MRD day 33) and risk stratification of patients (standard risk and high risk) after consolidation therapy (day 78) was investigated by the ROC curve analysis (Figure 2). ROC curves display the relationship between true positive (“sensitivity”) and false positive (“1-specificity”) rates across various discrimination threshold values used to diagnose the dichotomous condition. The measure for the ability of the variable to distinguish between the dichotomous end points is the relative area under the ROC curves, which may vary between 0.5 (the test is unreliable) and 1.0 (perfect separation). As Figure 2 shows, the area under the ROC curve values for the FCM-MRD day-33 and risk stratification groups were 0.69 and 0.71, respectively, thus showing that CD11b can discriminate patient groups with different clinical outcomes and prognosis. ROC curve analysis was used to define the optimal threshold value to provide the highest accuracy for the separation of positive and negative end point results (Figure 2). The CD11b value to predict FCM-MRD day-33 remission status was 515 MESFs, with the corresponding odds ratio of 5.2, that is, with the 5.2-fold higher probability of MRD positivity if the CD11b expression at diagnosis was more than 515 MESFs (P = .003). The criterion of CD11b more than 773 MESFs defined patients with a 5.5-fold higher probability to be stratified into the high-risk than into the standard-risk group (P = .004).
ROC curve analysis of discrimination of FCM-MRD remission status (FCM positive vs negative) and BFM risk stratification (high vs standard) according to CD11b expression levels at diagnosis. (A) The likelihood for a positive FCM-MRD remission status after induction is given by a CD11b expression at diagnosis of > 515 molecules of equivalent soluble fluorochrome (MESFs; 95% confidence interval, 1.7-15.5). (B) Patients with a CD11b expression > 773 MESFs at diagnosis are more likely Berlin-Frankfurt-Münster high-risk patients (95% confidence interval, 1.8-16.5). AUC indicates area under the curve; and OR, odds ratio.
ROC curve analysis of discrimination of FCM-MRD remission status (FCM positive vs negative) and BFM risk stratification (high vs standard) according to CD11b expression levels at diagnosis. (A) The likelihood for a positive FCM-MRD remission status after induction is given by a CD11b expression at diagnosis of > 515 molecules of equivalent soluble fluorochrome (MESFs; 95% confidence interval, 1.7-15.5). (B) Patients with a CD11b expression > 773 MESFs at diagnosis are more likely Berlin-Frankfurt-Münster high-risk patients (95% confidence interval, 1.8-16.5). AUC indicates area under the curve; and OR, odds ratio.
We further evaluated the effect of the CD11b cutoff values in context of other clinically relevant parameters available at diagnosis. The multivariate regression models included, in addition to the expression of CD11b, the parameters of age (cutoff, 10 years), WBC count (50.0 × 109/L), DNA index (1.16), sex, and BCR/ABL and MLL positivity/negativity. Results are depicted in Table 3. In the classification of both, MRD day 33 and risk stratification, the CD11b expression contributed independently to the overall prediction rates (82%, P < .001 and 85%, P = .004, respectively). Finally, to prove the influence of the observed correlation between TEL/AML1 positivity and CD11b expression, we performed the multivariate regression analysis in the cohort of patients without TEL/AML1 translocation. The CD11b effect on MRD day 33 and risk stratification remained an independent parameter with P values of .003 (overall prediction 78%) and .015 (overall prediction 89%), respectively (data not shown).
Logistic regression analysis with the use of CD11b expression
| Observed . | FCM-MRD day-33 prediction . | Risk group prediction . | ||||
|---|---|---|---|---|---|---|
| Negative . | Positive . | Total . | Standard . | High . | Total . | |
| Negative | 48 | 5 | 53 (91)* | — | — | — |
| Positive | 8 | 11 | 19 (58)† | — | — | — |
| Total | 56 (86)‡ | 16 (69)§ | 72 (82)‖ | — | — | — |
| Standard | — | — | — | 49 | 3 | 52 (94)* |
| High | — | — | — | 8 | 12 | 20 (60)† |
| Total | — | — | — | 57 (86)‡ | 15 (80)§ | 72 (85)‖ |
| Observed . | FCM-MRD day-33 prediction . | Risk group prediction . | ||||
|---|---|---|---|---|---|---|
| Negative . | Positive . | Total . | Standard . | High . | Total . | |
| Negative | 48 | 5 | 53 (91)* | — | — | — |
| Positive | 8 | 11 | 19 (58)† | — | — | — |
| Total | 56 (86)‡ | 16 (69)§ | 72 (82)‖ | — | — | — |
| Standard | — | — | — | 49 | 3 | 52 (94)* |
| High | — | — | — | 8 | 12 | 20 (60)† |
| Total | — | — | — | 57 (86)‡ | 15 (80)§ | 72 (85)‖ |
Analyzed with age, white blood cell count, DNA index, sex, cytogenetics (BCR/ABL and MLL/AF4). CD11b cutoffs were 515 molecules of equivalent soluble fluorochrome (MESFs; FCM-MRD day 33) and 773 MESFs (risk group prediction). Data are n or n (%).
Specificity.
Sensitivity.
Negative prediction value.
Positive prediction value.
Accuracy.
CD11b expression during therapy is consistently increased in MRD blasts
In addition to the prognostic significance of CD11b at diagnosis, we were interested in investigating the phenomenon of CD11b up-regulation during therapy. To this end, we analyzed a prospective series of patients with the use of the MRD panel complemented with the CD11b antibody. The MRD detection has been performed in PB (day 8) and in BM (days 15, 33, 78) cell samples, and the CD11b expression values in MRD blasts were compared with those at diagnosis in PB and BM, correspondingly.
In PB at d8, the increased CD11b expression has been observed in 88% of patients who were MRD-positive at this time point (42 of 48). The mean expression change of 4.7-fold (P < .001) was in accordance with the previously published data from a series of 29 patients.20
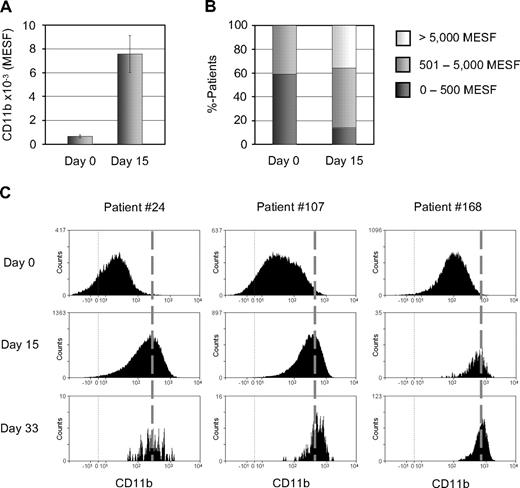
In BM at day 15, 45 of 53 patients were MRD positive, and the expression of CD11b increased by 10.5-fold from 693 to 7296 MESFs (P < .001; Figure 3A). The number of patients with a low (≤ 500 MESFs) CD11b expression changed from 59% at diagnosis to 14% at day 15. The number of cases with a medium (501-5000 MESFs) and high (> 5000 MESFs) expression increased from 41% to 50% and from 0% to 36%, respectively (Figure 3B).
CD11b protein expression is consistently increased in MRD cells. (A) Mean CD11b expression is significantly increased at day 15 in comparison with diagnosis (BM from 44 paired samples). (B) Percentages of patients with low (0-500 MESFs), medium (501-5000 MESFs) and high (> 5000 MESFs) CD11b expression at days 0 and 15. (C) Representative examples of consistently increased CD11b expression, depicted by flow cytometric histograms, under therapy.
CD11b protein expression is consistently increased in MRD cells. (A) Mean CD11b expression is significantly increased at day 15 in comparison with diagnosis (BM from 44 paired samples). (B) Percentages of patients with low (0-500 MESFs), medium (501-5000 MESFs) and high (> 5000 MESFs) CD11b expression at days 0 and 15. (C) Representative examples of consistently increased CD11b expression, depicted by flow cytometric histograms, under therapy.
At day 33, 9 of 50 patients were MRD positive. The mean CD11b expression level was 7433 MESFs and did not differ significantly from that at day 15 (7296 MESFs). Importantly, the CD11b expression, if up-regulated at day 15, remained consistently increased during further treatment (Figure 3C).
At day 78, 2 of 53 patients were MRD positive. At this time point, the CD11b expression was increased in 1 patient (day 0, 630 MESFs; day 78, 14 952 MESFs) and remained negative in the second patient.
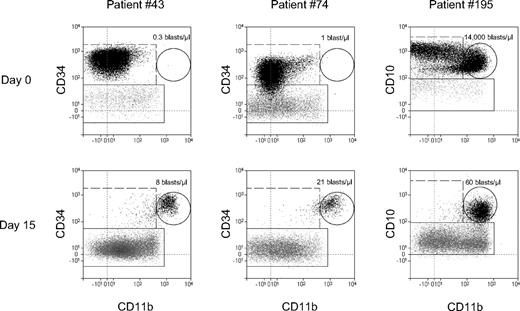
The observed increase of expression may be due either to an up-regulation of expression at the genetic level or to a therapy-specific selection of pre-existing cell subpopulations. In most cases the latter possibility cannot be excluded, because the absolute number of blasts, which is usually high at diagnosis, dramatically decreases during therapy. Nonetheless, in 5 cases (4 at day 15 and 1 at day 33) we found absolute BCs within a CD11b+ gate which were higher under therapy than initially at diagnosis, as shown in Figure 4 (left and middle cases). These calculations, however, do not exclude the principal possibility of expansion of a therapy-resistant subclone contrary to the cytoreduction of the therapy-sensitive bulk of leukemic cells. Moreover, at least in one case we observed a selection of the CD11b+ subpopulation at day 15 from the initial CD11b+ and CD11b− subpopulations at diagnosis (Figure 4 right).
CD11b expression in ALL blasts under therapy: modulation vs selection. Comparison of absolute counts of CD11b+ blasts (the numbers of blasts/μL in the dot plots) at diagnosis and day 15. In the dot plots only CD19+ cells are depicted. Leukemic cells are shown as black dots and normal B cells as gray dots. Quantification of the CD11b+ leukemic cells (gated by solid-line circles) shows 2 cases of the therapy-induced CD11b up-regulation (left and middle). The right plots indicate selection of CD11b+ cells from initial 2 CD11b+ and CD11b− subpopulations of leukemic cells.
CD11b expression in ALL blasts under therapy: modulation vs selection. Comparison of absolute counts of CD11b+ blasts (the numbers of blasts/μL in the dot plots) at diagnosis and day 15. In the dot plots only CD19+ cells are depicted. Leukemic cells are shown as black dots and normal B cells as gray dots. Quantification of the CD11b+ leukemic cells (gated by solid-line circles) shows 2 cases of the therapy-induced CD11b up-regulation (left and middle). The right plots indicate selection of CD11b+ cells from initial 2 CD11b+ and CD11b− subpopulations of leukemic cells.
CD11b as a potential MRD-specific antigen
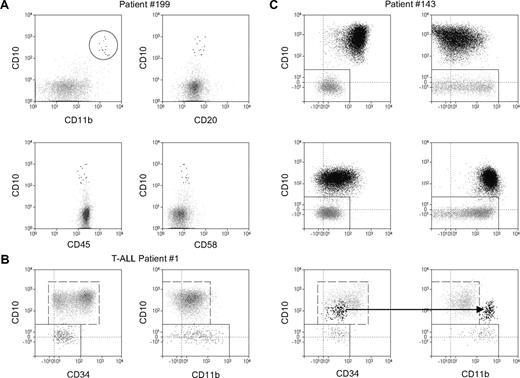
We furthermore investigated whether CD11b expression may improve discrimination of MRD cells from normal mature or immature B-lineage cells and, therefore, may serve as a MRD-specific antigen. First, we compared expression of CD11b on MRD cells with that on normal B cells. Normal B cells showed a bimodal intensity distribution of CD11b (Figures 4–5), with CD11b− and CD11b+ fractions representing presumably naive and memory B cells, respectively.31 The mean level of the CD11b expression in the memory B cells was relatively constant (5700 ± 700 MESFs). In ALL, CD11b expression levels exceeded 5700 MESFs in more than 30% of MRD-positive ALL cases (17 of 52 patients at day 8, 17 of 45 patients at day 15, 3 of 9 patients at day 33). Figure 5A shows an example of a day-15 BM sample, which contains a low number of CD19+ CD10+ cells which otherwise have a normal B-cell immunophenotype (expression of CD20, CD45, and CD58 is shown). Here, CD11b provides the CD marker that allows a reliable identification of leukemia cells.
CD11b positivity improves discrimination between ALL and normal mature and precursor B cells. Flow cytometric dot plots depict CD19+ cells from BM of ALL patients. Leukemic cells are shown as black dots and normal B cells as gray dots. (A) D15 BM sample from a patient with ALL. CD11b in combination with CD10+ allows a reliable identification of the leukemia cells, which otherwise have a normal B-cell immunophenotype. (B) Analysis of the day-78 BM sample from a patient with T-cell ALL. The dot plot CD10 versus CD34 displays immunophenotypic pattern characteristic of normal CD19+ fraction within regenerating BM. The dot plot CD10 versus CD11b shows a very low CD11b expression in normal CD19+ hematogones. (C) The same, as in panel B, dot plot combinations for a patient with PBC-ALL at day 0 (top), day 33 (middle), and day 78 (bottom) of therapy. Note the consistently increased CD11b expression and a concomitant decrease of CD34 and CD10 expression in BM during the therapy. At day 78, the CD10 versus CD11b dot plot but not the CD10 versus CD34 plot clearly discriminates leukemic cells from normal regenerating cells.
CD11b positivity improves discrimination between ALL and normal mature and precursor B cells. Flow cytometric dot plots depict CD19+ cells from BM of ALL patients. Leukemic cells are shown as black dots and normal B cells as gray dots. (A) D15 BM sample from a patient with ALL. CD11b in combination with CD10+ allows a reliable identification of the leukemia cells, which otherwise have a normal B-cell immunophenotype. (B) Analysis of the day-78 BM sample from a patient with T-cell ALL. The dot plot CD10 versus CD34 displays immunophenotypic pattern characteristic of normal CD19+ fraction within regenerating BM. The dot plot CD10 versus CD11b shows a very low CD11b expression in normal CD19+ hematogones. (C) The same, as in panel B, dot plot combinations for a patient with PBC-ALL at day 0 (top), day 33 (middle), and day 78 (bottom) of therapy. Note the consistently increased CD11b expression and a concomitant decrease of CD34 and CD10 expression in BM during the therapy. At day 78, the CD10 versus CD11b dot plot but not the CD10 versus CD34 plot clearly discriminates leukemic cells from normal regenerating cells.
Moreover, we found that the detection of CD11b can improve not only discrimination of leukemic blasts from normal B cells but also from regenerating normal precursor cells. In fact, as shown by analysis of patients with T-cell ALL with regenerating BM at day 78 (Figure 5B) and of BM samples from patients without leukemia (not shown), normal CD19+/CD10+ cells are CD11b−. Because of this fact, the application of the CD11b antibody may provide a valuable tool for a more specific and reliable identification of residual leukemic blast cells in ALL cases with regenerating BM at day 78 (Figure 5C).
Discussion
In the present study the significance of the CD11b antigen in PBC-ALL has been investigated. Its potential clinical relevance has followed from our previous study on the therapy-induced gene expression changes in leukemic blasts of patients with ALL.20 The higher mRNA level of CD11b at day 8 of induction therapy compared with initial diagnosis has been a hallmark of the genome-wide expression changes, induced by prednisone therapy.20 The observed consistent up-regulation of CD11b in therapy-resistant blasts prompted us to investigate whether CD11b+ cases may be present already at diagnosis and differ in the clinical response.
At the mRNA level, we found a considerable heterogeneity of CD11b expression in ALL cells before the therapy. Clinically, CD11b mRNA levels correlated significantly with the cytoreduction rate after the first week of the therapy. The cases with the higher CD11b expression at diagnosis showed a lower decrease of BCs. This correlation has been lost for the CD11b mRNA expression in the day-8 samples, because the consistently higher expression of CD11b has been a common feature of the blasts persisting under therapy.
Flow cytometric analysis has been applied to investigate CD11b protein expression in a large series of patients at diagnosis. This analysis confirmed the heterogeneous expression of CD11b. There was no significant correlation between CD11b expression and most of the initial laboratory, cytogenetic, and diagnostic parameters, as tested in the subgroups considered to be of clinical relevance.30 The only exception were TEL/AML1-positive ALLs which were almost totally negative for CD11b. Generally, a TEL/AML-specific immunophenotype has not been described so far.32 In an attempt to design a CD-based scoring system of TEL/AML1 rearranged cases, only one has been confirmed in a separate cohort of patients.33 By that scoring system, the presence of TEL/AML1 can be predicted by negativity or only partial expression of both CD9 and CD20.32,33 Our data suggest that CD11b may improve the immunophenotypic definition of TEL/AML1 rearrangement. Its applicability to a scoring system of TEL/AML1 warrants further investigation.
For the clinical response, the CD11b protein expression was significantly associated with therapy response parameters measured by FCM. It is remarkable that the statistically significant inverse correlation with cytoreduction was observed not only for the GC prephase (day 8) in PB but also to the later time points of induction therapy (day 15 and day 33) in BM. Moreover, the initial CD11b levels were consistently associated with percentages of blasts persisting at different time points. Of note, in the genomewide mRNA analysis reported previously,20 we also observed an increased expression of the interferon γ receptor chain 1 (IFNGR1). However, in contrast to the CD11b, IFNGR1 mRNA levels did not correlate with early therapy response. Moreover, investigation of IFNGR1 protein expression did not indicate any association with clinical parameters in the same cohorts of patients both at day 0 (n = 159) and day 8 (n = 100). This observation shows that increased gene expression in MRD cells is not necessarily indicative of clinical significance of the gene under consideration.
Optimal cutoffs for CD11b at diagnosis able to predict positive FCM-MRD status at day 33 and the high-risk assignment after consolidation therapy at day 78 were determined by ROC curve analysis. Both cutoffs showed similar values, positioned in the third quarter of the CD11b expression distribution. If the level of 773 MESFs would have been used for risk classification in the patient population analyzed herein, 21% of the standard-risk patients (12 of 56) and 35% of intermediate-risk patients (28 of 81) would have been shifted to a higher risk group. Expression levels greater than these cutoffs conferred a more than a 5-fold increased risk of MRD positivity at day 33 and assignment to the high-risk group at day 78. Moreover, the multivariate analysis indicated that CD11b expression contributes to the prognosis of therapy outcome independently from the other clinically relevant parameters at diagnosis.
CD11b is functionally described as a cell adhesion molecule that acts as a receptor for cell-surface ligands such as intracellular adhesion molecules or soluble ligands.21,34 Several studies indicated the importance of adhesion molecules in ALL, including integrins leukocyte function–associated antigen 1 (CD11a/CD18) and very late antigen 4 (CD49d/CD29).24,26,35 Notably, CD11b has not been previously reported as a therapy response-related gene. To our knowledge, it also has not been reported in the studies, which investigated genomewide expression in ALL samples at diagnosis in the context of therapy response.5-9,12 This is probably because the latter studies, in contrast to our investigation,20 have been performed in unpurified samples contaminated with mature monocytes and granulocytes, which in turn constitutively express CD11b at very high levels and as such provide a high level of noise for CD11b expression.
CD11b, which belongs to the family of leukocyte integrins as a part of the integrin Mac-1 (CD11b/CD18), is expressed mainly on cells of the myeloid lineage.24 It binds to a wide range of the ligands, including fibrinogen and complement protein iC3b, and is involved in leukocyte adhesive and migratory functions by binding to the adhesion ligands intercellular adhesion molecule-1 and -2.22,24 Because integrin-mediated signals are necessary in normal cells to block apoptosis,21 it is tempting to speculate that CD11b may functionally contribute to the survival of MRD cells and to therapy resistance. It should be considered, however, that constitutive expression of integrins has not been directly linked to their functional activity, as shown by the high expression of MAC-1 in circulating, nonadherent neutrophils.24 It is rather the conformational state of the integrin subchains which defines their activity and is regulated by the so-called inside-out signaling.21,23,25 The latter kind of signaling implies a strict regulation of the multiple conditions which should be fulfilled before the adhesive function can be activated.21
In our previous study of flow-sorted MRD blasts at the genomewide level, a number of genes have been identified, whose expression has been changed in comparison with untreated cells.20 If considered together, these changes have indicated a statistically significant shift toward more mature B cells. Given that normal cells are generally more resistant to cytotoxic treatment, this kind of systemic gene expression shift may provide a general escape mechanism in malignant precursor B cells. Within this context, the observed CD11b up-regulation is in line with the proposed overall expression shift. Moreover, given that in normal B cells the CD11b positivity is characteristic of the memory B cells, the CD11b up-regulation points to a shift to the memory type of normal B cells. Therefore, it would be of interest to investigate whether memory and naive normal B cells differ in their therapy resistance.
The changes of CD marker expression as detected by PBC-ALL–specific MRD panels have been reported elsewhere, and the mechanism of this phenomenon is a matter of discussion.14,17-19,36,37 In vitro studies suggest a glucocorticoid therapy–specific modulation of gene expression.19 As our analysis has shown, the increased expression of CD11b may be principally due to both an up-regulation of gene expression and a selection of a more resistant, CD11b+ subclone. In addition, the possibility cannot be excluded that the therapy-resistant subclone may expand under therapy contrary to the cytoreduction in the therapy-sensitive bulk of leukemic cells. Independent of the mechanisms underlying gene expression changes, the consistently increased CD11b expression makes it an attractive candidate molecule for MRD detection by immunophenotype. Because normal B-cell progenitors are CD11b−, CD11b positivity of MRD blasts may decisively contribute to the discrimination of leukemic cells in ALL cases with regenerating BM at day 78. In addition, implication of CD11b may be useful in those cases, in which the CD11b expression exceeds that in normal memory B cells. The latter condition has been fulfilled in more than 30% of ALL cases at various time points of therapy. This may particularly improve identification of a low number of blasts against a strong background of normal mature B cells.
In conclusion, starting from a genome-wide analysis of therapy-persisting (MRD) cells we identified CD11b protein as an important molecular factor in childhood ALL. Its association with therapy response parameters described by different methods and at various time points as well as increased expression in MRD cells strongly indicates an association of CD11b expression with therapy resistance mechanisms. Because the immunophenotype data are available before the response to treatment can be evaluated, CD11b expression may serve as an additional cell-biologic parameter for treatment stratification. Together with the increasing number of the parameters obtained at molecular and functional levels which may be useful in predicting MRD status after induction therapy,5,38,39 this parameter may potentially provide a part for a future basis for more individually designed treatment strategies in upcoming clinical trials. Moreover, because of the consistently increased expression levels in leukemia cells under therapy and to the cell surface allocation, CD11b has a promising potential as an MRD-specific marker in PBC-ALL which can be easily integrated into the existing antibody combinations for flow cytometric MRD detection.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Rosemarie Hoffmann, Marianne Dunken, and Birgit Oestereich for technical assistance.
This work was supported by the Federal Ministry for Education and Research (BMBF) in the National Genome Research Network (grant 01GS0870), Wilhelm Sander Stiftung (grant 2004.072.1), Alfred & Angelika Gutermuth-Stiftung, and Deutsche José Carreras Leukämie-Stiftung (grants DJCLS F05/09 and R09/19).
Authorship
Contribution: P.R. designed and performed research, analyzed data, and assisted in writing of the manuscript; R.M. performed research; G.B., G.G., M.N.D., R.K.-S., and C.H. participated in research design and manuscript drafting; M. Stanulla provided clinical data and assisted in writing of the manuscript; M. Schrappe directed the underlying clinical study; W.-D.L. contributed to the research concept and editing of the manuscript; L.K. designed research, analyzed data, and wrote the manuscript; and R.R. made patient samples vailable, analyzed data, and assisted in the writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Leonid Karawajew, Experimental and Clinical Research Center, Charité Medical School, Lindenberger Weg 80, 13125 Berlin, Germany; e-mail: leonid.karawajew@charite.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal