Abstract
Chimeric fusion genes are highly prevalent in childhood acute lymphoblastic leukemia (ALL) and are mostly prenatal, early genetic events in the evolutionary trajectory of this cancer. ETV6-RUNX1–positive ALL also has multiple (∼ 6 per case) copy number alterations (CNAs) as revealed by genome-wide single-nucleotide polymorphism arrays. Recurrent CNAs are probably “driver” events contributing critically to clonal diversification and selection, but at diagnosis, their developmental timing is “buried” in the leukemia's covert natural history. This conundrum can be resolved with twin pairs. We identified and compared CNAs in 5 pairs of monozygotic twins with concordant ETV6-RUNX1–positive ALL and 1 pair discordant for ETV6-RUNX1 positive ALL. We compared, within each pair, CNAs classified as potential “driver” or “passenger” mutations based upon recurrency and, where known, gene function. An average of 5.1 (range 3-11) CNAs (excluding immunoglobulin/T-cell receptor alterations) were identified per case. All “driver” CNAs (total of 32) were distinct within each of the 5 twin pairs with concordant ALL. “Driver” CNAs in another twin with ALL were all absent in the shared ETV6-RUNX1–positive preleukemic clone of her healthy co-twin. These data place all “driver” CNAs secondary to the prenatal gene fusion event and most probably postnatal in the sequential, molecular pathogenesis of ALL.
Introduction
The common chromosome abnormalities observed in childhood acute lymphoblastic leukemia (ALL), chimeric fusion genes generated by chromosome translocation and hyperdiploidy, are predominantly prenatal in origin but, in themselves, insufficient for leukemia to develop, suggesting a minimal 2 “hit,” pre/postnatal model for molecular pathogenesis and etiology.1 This view finds support in modeling experiments with mice2,3 and human cells.4 Recent observations suggest, however, substantially more genetic complexity in ALL than previously suspected. Data derived from high-resolution single-nucleotide polymorphism (SNP) arrays have indicated that cases of B-cell precursor ALL with the common chimeric fusion gene ETV6-RUNX1 have, in addition to the fusion gene, an average of 6 additional DNA aberrations or copy number alterations (CNAs; range 1-21), several being recurrent deletions in genes with functions impacting on B-cell lineage differentiation or cell-cycle control5-8 and therefore presumed “driver” mutations.9 This provides a challenge to the presumption that the fusion gene itself is necessary and sufficient as a first prenatal hit generating the preleukemic phase of disease.1,4 A key question then becomes the issue of when, in relation to fusion genes, these multiple CNAs arise in multistep molecular pathogenesis and clonal evolution. This is difficult to ascertain in diagnostic samples as the most recent subclonal expansions can disguise prior historical sequences of genetic events. Identical twin pairs with concordant ETV6-RUNX1–positive ALL provide a unique and tractable approach to this problem.
ETV6-RUNX1 genes have highly variable but clone-specific breakpoints and fusion region genomic sequences and are therefore patient specific.1 The only exception to this is with monozygotic twins with concordant ALL who share the same acquired (ie, nonconstitutive) but unique fusion gene sequence reflecting monoclonal origin in 1 fetus in utero.10 Concordance of ALL then arises as a consequence of intraplacental spread of an initiated preleukemic clone from one twin to the other. The ALL concordance rate of 10% to 15% in such a twin context10 suggests the necessity of additional genetic hits for which there is significant discordance (ie, ∼ 85%-90%). The most common additional genetic abnormality in ETV6-RUNX1–positive ALL is deletion of the normal ETV6 allele,11 which can be subclonal (by fluorescent in situ hybridization [FISH]) and therefore secondary to ETV6-RUNX1.11,12 ETV6 deletions can also be discordant in twin pairs, indicating they are probably postnatal in origin.12-15 If ETV6 deletions and all other recurrent and presumed “driver” CNAs identified more recently in ALL5-8 are consistently secondary to ETV6-RUNX1 and postnatal, then a straightforward and testable prediction is that they should be distinct or different within twin pairs with concordant ALL. Contrariwise, if one or all of such genetic alterations precede or occur simultaneously with gene fusion, then they should be identical in twin pairs with ALL.
Methods
Patients
Five pairs of identical (monozygotic, monochorionic) twins with concordant ETV6-RUNX1 fusion gene positive ALL and a further twin pair, who were discordant for ETV6-RUNX1 fusion gene positive ALL, were accrued from several centers for the study. The clinical, immunophenotypic, cytogenetic, and FISH data are given in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Ethical review committee approval was obtained for the study from the Royal Marsden NHS Trust. We used high-resolution (500 K) SNP mapping arrays, and by comparing with matched normal or remission blood DNA, we explored the clonal relationships of their respective “within pair” genetic abnormalities.
Genome-wide copy number analyses
DNA was extracted according to standard methods from leukemic bone marrow acquired at diagnosis and peripheral blood taken during clinical remission. The diagnostic leukemic DNA was derived from bone marrow samples with a median leukemic blast cell count of 90% (supplemental Table 1) SNP array analysis was carried out using GeneChip Human Mapping 250K Nsp and 250K Sty arrays according to the manufacturer's instructions (Affymetrix). This accounts for 500 000 SNPs with a mean physical distance between SNPs of 2.5 kb. GTYPE was used for analysis of signal intensity and for genotype calling (Affymetrix; full SNP genotype data are available at www.icr.ac.uk/array/array.html). The mean call rate was 95% (range 81.25%-98.84%; supplemental Table 5). For copy number estimates, the Genome Orientated Laboratory File (GOLF Version 2.2.9) software package was used.16,17 Hybridization values were normalized to the median value of each array. Copy number was determined based on the log2 ratio of the signal intensity from the leukemia sample versus matched peripheral blood DNA taken during clinical remission, in all but 1 twin pair, and was done by visual inspection; potential copy number changes included at least 3 SNPs. In this case (twin pair 3), no remission sample was available, and the pooled signal intensity of 15 unrelated ALL remission samples was used.
FISH
Interphase FISH was performed on methanol-acetic acid fixed cells, which were only available for twin pairs 5 and 6, using standard methods. Bacterial artificial chromosome (BAC) or fosmid probes for the region of interest were obtained from the BACPAC Resource Center (Children's Hospital, Oakland Research Institute, Oakland, CA; http://bacpac.chori.org), labeled with biotin-16-dUTP or digoxigenin-11-dUTP, hybridized, and detected with streptavidin Cy5 (for biotin-labeled probes) and (1) monoclonal anti-digoxigenin (Sigma-Aldrich), (2) horse anti–mouseimmunoglobulin (Ig)–Texas Red (Vector Laboratories), and (3) goat anti–horse Ig–Texas Red (Jackson Immunochemicals; for digoxigenin-labeled probes). Fluorescent signals were viewed using a Zeiss Axioskop fluorescence microscope equipped with epifluorescence and filters for DAPI (4′,6-diamidino-2-phenylindole), FITC (fluorescein isothiocyanate), Texas Red, and Cy5, and images were captured and analyzed using a charge-coupled device (Photometrics) and SmartCapture software (Digital Scientific). The fosmid and BAC probes used were tested on normal metaphase smears to confirm correct chromosome localization. No metaphase cells were available for twin pairs 5 and 6.
Results
Monozygotic twins concordant for ETV6-RUNX1–positive ALL
Table 1 summarizes the data, and Figure 1A through C provide illustrative examples of some key “paired” SNP array data in the monozygotic twins concordant for ETV6-RUNX1 ALL. The total number of CNAs (including TCR [T-cell receptor] and Ig rearrangements) in the 5 twin pair samples averaged 7.9 aberrations, with a range of 5 to 15. To facilitate evaluation of the complex datasets generated, we elected, before data acquisition, to divide CNAs into potential “drivers,” potential neutral or nonfunctional “passengers,” and “physiological” CNAs, while recognizing that these distinctions are not entirely unambiguous. Potentially functional or driver CNAs (“drivers”) were defined as those that were observed to be recurrent in prior series of SNP or comparative genomic hybridization arrays screens of ETV6-RUNX1 ALL5-8 and, when deletions were small, then occurring in or close to genes with functions known to be relevant to leukemogenesis or B-cell regulation5 (see supplemental Table 2 for list of recurrent CNAs). It is accepted that driver status ultimately requires confirmation by functional studies.9 CNAs that were nonrecurrent (this study and prior series5-8 ) in gene poor regions or in genes of no known functional relevance to lymphocyte biology or leukemia were considered as likely “passengers.” We recognize that anonymous and sporadic CNAs could occasionally transpire to be true “drivers.” As in prior studies,5 we classify gene deletions in IGH or TCR loci as most probably “physiological” and associated with the developmental rearrangements of these loci associated with diversification of antigen receptors in the lymphoid lineages (supplemental Table 3).
CNAs identified by SNP array
| Twin sets . | Number of CNAs (identical) . | Total (identical) . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Twin set 1 . | Twin set 2 . | Twin set 3 . | Twin set 4 . | Twin set 5 . | ||||||||||||
| 1a . | . | 1b . | 2a . | . | 2b . | 3a . | . | 3b . | 4a . | . | 4b . | 5a . | . | 5b . | ||
| Total | 7 | 6 | 9 | 5 | 9 | 15 | 8 | 7 | 5 | 8 | 79 | |||||
| “Drivers” | 3 | (0) | 3 | 5 | (0) | 2 | 2 | (0) | 7 | 2 | (0) | 3 | 2 | (0) | 3 | 32 (0) |
| “Passengers” | 1 | (1) | 1 | 2 | (0) | 1 | 3 | (3) | 4 | 2 | (0) | 1 | 2 | (0) | 2 | 19 (4) |
| “Physiological” | 3 | (2) | 2 | 2 | (1) | 2 | 4 | (1) | 4 | 4 | (0) | 3 | 1 | (0) | 3 | 28 (4) |
| Twin sets . | Number of CNAs (identical) . | Total (identical) . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Twin set 1 . | Twin set 2 . | Twin set 3 . | Twin set 4 . | Twin set 5 . | ||||||||||||
| 1a . | . | 1b . | 2a . | . | 2b . | 3a . | . | 3b . | 4a . | . | 4b . | 5a . | . | 5b . | ||
| Total | 7 | 6 | 9 | 5 | 9 | 15 | 8 | 7 | 5 | 8 | 79 | |||||
| “Drivers” | 3 | (0) | 3 | 5 | (0) | 2 | 2 | (0) | 7 | 2 | (0) | 3 | 2 | (0) | 3 | 32 (0) |
| “Passengers” | 1 | (1) | 1 | 2 | (0) | 1 | 3 | (3) | 4 | 2 | (0) | 1 | 2 | (0) | 2 | 19 (4) |
| “Physiological” | 3 | (2) | 2 | 2 | (1) | 2 | 4 | (1) | 4 | 4 | (0) | 3 | 1 | (0) | 3 | 28 (4) |
CNA indicates copy number alteration.
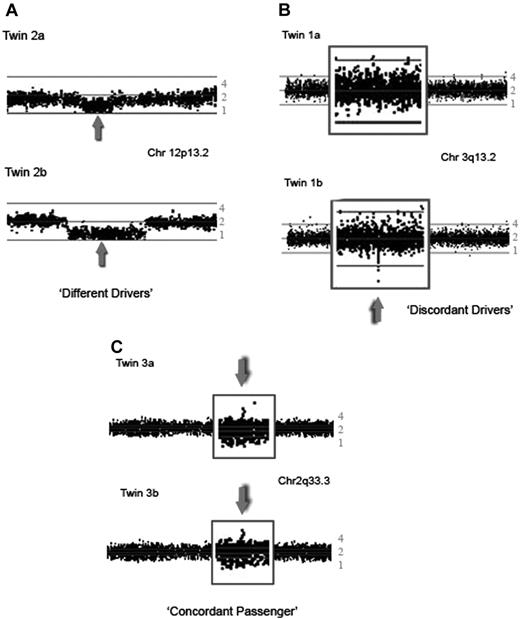
SNP array plots of CNAs detected in the leukemic bone marrow of twins with concordant ETV6-RUNX1 fusion gene positive ALL. The ratio of loss of heterozygosity of the leukemic versus the matched remission sample is plotted on a logarithmic scale to estimate the copy number. Each dot represents the log2 ratio of 1 SNP, with a moving average of 3 SNPs. The middle line represents a copy number of 2, the lower line a copy number of 1, and the top a copy number of 4. (A) 250K Sty SNP array analysis showing the short arm of chromosome 12 of twin set 2. Both twins have a deletion of 12p13.2, including the ETV6 gene (gray arrow); however, the boundaries and size of the deletion are different in each twin. (B) 250K Nsp plot of part of the long arm of chromosome 3 of twin set 1 showing a discordant deletion of 147 kb in region 3q13.2, which includes the BTLA and CD200 genes, seen in twin 1b, but not in twin 1a. (C) 250K Sty plot of part of the long arm of chromosome 2 of twin set 3 showing an identical gain of a 208-kb region at 2q33.3.
SNP array plots of CNAs detected in the leukemic bone marrow of twins with concordant ETV6-RUNX1 fusion gene positive ALL. The ratio of loss of heterozygosity of the leukemic versus the matched remission sample is plotted on a logarithmic scale to estimate the copy number. Each dot represents the log2 ratio of 1 SNP, with a moving average of 3 SNPs. The middle line represents a copy number of 2, the lower line a copy number of 1, and the top a copy number of 4. (A) 250K Sty SNP array analysis showing the short arm of chromosome 12 of twin set 2. Both twins have a deletion of 12p13.2, including the ETV6 gene (gray arrow); however, the boundaries and size of the deletion are different in each twin. (B) 250K Nsp plot of part of the long arm of chromosome 3 of twin set 1 showing a discordant deletion of 147 kb in region 3q13.2, which includes the BTLA and CD200 genes, seen in twin 1b, but not in twin 1a. (C) 250K Sty plot of part of the long arm of chromosome 2 of twin set 3 showing an identical gain of a 208-kb region at 2q33.3.
Physiological IGH or TCR CNAs were either identical (4/28) or different (24/28) in twin pairs (Table 1 and supplemental Table 3). Where such deletions are shared, we take this to indicate that the ETV6-RUNX1 fusion arose in a progenitor B cell that had either already undergone these rearrangements or simultaneously acquired them.
Sporadic CNAs classified as “passengers” (Table 1) were either identical (4/19) or distinct (15/19) in twin pairs. The most likely explanation for sharing of “passenger” CNAs is that either these preceded the ETV6-RUNX1 fusion event in the same cell or its predecessor, or that they represent collateral damage from whatever caused a simultaneous ETV6-RUNX1 fusion in the same “target” cell. Formally, it is also possible that shared passenger CNAs arose after the initiating ETV6-RUNX1 lesion. However, this would require fixation in the same subclone of both twins by subsequent (and distinct) “driver” CNAs. Passenger CNAs were all in regions with either no annotated gene or involved genes with no known or predictable role in leukemogenesis (supplemental Table 3), thus endorsing their likely passenger or neutral status.
A total of 32 “driver” mutations were identified (average 3.2 per case, range 2-7). These were all within the same chromosome regions or genes as observed in previous studies of childhood ALL, including genes encoding predominantly B-lineage transcription factors or cell-cycle regulators5-8 (Table 2). Significantly, all 5 twin pairs were discordant for all 32 CNAs (Table 1, Figure 1A-B). Discordance was reflected in either both twins sharing the same gene deletion (eg, in ETV6) but with different genomic boundaries of deletion (Figure 1A) or particular genes (or regions) being deleted in one twin and present in the other (Figure 1B). We have confirmed differences in “driver” CNAs in twin pair 5 by multicolor FISH (Figure 2). Insufficient cells were available from the other 4 twin pairs.
Presence and size of “driver” CNAs and associated genes in twin pairs
| Cytoband . | Size of CNA (Mb)* . | Gene(s) in region . | Reported in karyotype† . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Twin set 1 . | Twin set 2 . | Twin set 3 . | Twin set 4 . | Twin set 5 . | ||||||||
| 1a . | 1b . | 2a . | 2b . | 3a . | 3b . | 4a . | 4b . | 5a . | 5b . | |||
| Deletions | ||||||||||||
| 1p32.3−pter | No CNA | 55.0 | Many | No | ||||||||
| 3q13.2 | No CNA | 0.147 | CD200/BTLA | No | ||||||||
| 3q26.32 | No CNA | 0.28 | TBL1XR1 | No | ||||||||
| 5q21.1−q35.3 | No CNA | 79.1 | Many, including EBF1 | No | ||||||||
| 6p22.1 | 0.147 | No CNA | HIST1H2 | No | ||||||||
| 6q15−q27 | No CNA | 79.5 | Many | No | ||||||||
| 7q34−q35 | No CNA | 6.1 | Many | No | ||||||||
| 9p | All 9p¶ | No CNA | Whole arm | Yes | ||||||||
| 9p13.2 | 0.248 | 0.436 | PAX5 + ZCCHC7 | No | ||||||||
| 9p21.3 | 1.39¶ | No CNA | CDKN2A, CDKN2B | No | ||||||||
| 12p | 8.98‡ | 20.7‡ | 27.1¶ | No CNA | 22.3 | No CNA | 20.7 | Many, including ETV6 | Yes | |||
| 12p13 | 0.866‡§ | 0.866‡§ | 4.44¶ | 9.96 | 6.5 | 0.259 | No CNA | Many including ETV6 | Yes | |||
| 13q12.3−q34 | 83.5 | 81.2 | Many, including RB1 | No | ||||||||
| 21q22.3−qter | 4.9 | No CNA | Many (not including RUNX1) | No | ||||||||
| Gains | ||||||||||||
| 5pter−q21.1 | no CNA | 101.4 | Many | No | ||||||||
| 6pter−q15 | no CNA | 91.3 | Many | No | ||||||||
| All 10 | All 10 | No CNA | Many | Yes | ||||||||
| 15q14−q15.1 | No CNA | 4.2 | Many including LTK | No | ||||||||
| 21q | All 21q | No CNA | No CNA | 19.8 | No CNA | 20.8 | Many, including RUNX1 | Yes | ||||
| Cytoband . | Size of CNA (Mb)* . | Gene(s) in region . | Reported in karyotype† . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Twin set 1 . | Twin set 2 . | Twin set 3 . | Twin set 4 . | Twin set 5 . | ||||||||
| 1a . | 1b . | 2a . | 2b . | 3a . | 3b . | 4a . | 4b . | 5a . | 5b . | |||
| Deletions | ||||||||||||
| 1p32.3−pter | No CNA | 55.0 | Many | No | ||||||||
| 3q13.2 | No CNA | 0.147 | CD200/BTLA | No | ||||||||
| 3q26.32 | No CNA | 0.28 | TBL1XR1 | No | ||||||||
| 5q21.1−q35.3 | No CNA | 79.1 | Many, including EBF1 | No | ||||||||
| 6p22.1 | 0.147 | No CNA | HIST1H2 | No | ||||||||
| 6q15−q27 | No CNA | 79.5 | Many | No | ||||||||
| 7q34−q35 | No CNA | 6.1 | Many | No | ||||||||
| 9p | All 9p¶ | No CNA | Whole arm | Yes | ||||||||
| 9p13.2 | 0.248 | 0.436 | PAX5 + ZCCHC7 | No | ||||||||
| 9p21.3 | 1.39¶ | No CNA | CDKN2A, CDKN2B | No | ||||||||
| 12p | 8.98‡ | 20.7‡ | 27.1¶ | No CNA | 22.3 | No CNA | 20.7 | Many, including ETV6 | Yes | |||
| 12p13 | 0.866‡§ | 0.866‡§ | 4.44¶ | 9.96 | 6.5 | 0.259 | No CNA | Many including ETV6 | Yes | |||
| 13q12.3−q34 | 83.5 | 81.2 | Many, including RB1 | No | ||||||||
| 21q22.3−qter | 4.9 | No CNA | Many (not including RUNX1) | No | ||||||||
| Gains | ||||||||||||
| 5pter−q21.1 | no CNA | 101.4 | Many | No | ||||||||
| 6pter−q15 | no CNA | 91.3 | Many | No | ||||||||
| All 10 | All 10 | No CNA | Many | Yes | ||||||||
| 15q14−q15.1 | No CNA | 4.2 | Many including LTK | No | ||||||||
| 21q | All 21q | No CNA | No CNA | 19.8 | No CNA | 20.8 | Many, including RUNX1 | Yes | ||||
CNA indicates copy number alteration.
No CNA indicates no copy number alteration detected.
Abnormality reported by conventional karyotyping and/or FISH.
CNA consisting of large deletion on one chromosome homologue and a smaller deletion on the second homologue.
Identical sized focal deletion presumed to occur at the same time as the ETV6-RUNX1 rearrangement.
Deletion appeared subclonal.
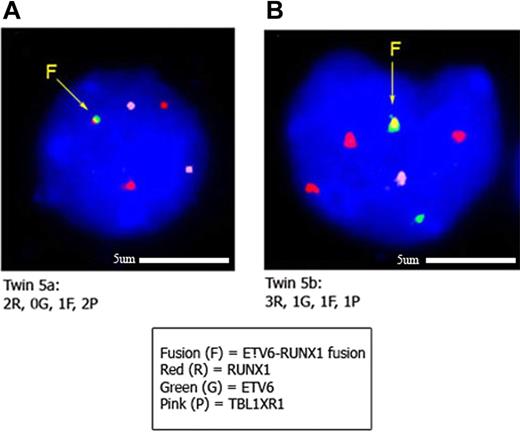
Interphase FISH to confirm CNAs identified by SNP array in twin set 5. Panels A and B show leukemic blasts from twins 5a and 5b, respectively, hybridized with the ETV6-RUNX1 ES (Vysis extra signal) probe and a fosmid probe (W12-1173N4) for the TBL1XR1 gene. The TBL1XR1 probe is labeled with biotin-16-dUTP and detected with streptavidin Cy5 (here pseudocolored pink). In panel A, the signal pattern of leukemic blasts in twin 5a is 1 yellow (corresponding to the ETV6-RUNX1 gene fusion), 2 red (1 corresponding to the normal RUNX1 gene and 1 to the residual RUNX1 gene disrupted by the translocation), and 2 pink (TBL1XR1 gene) signals. Note that the second green signal is not present, indicating loss of the normal (unrearranged) ETV6 gene. This pattern was observed in 21 of 31 cells scored. The other 10 of 31 cells had a normal pattern. In panel B, the signal pattern of leukemic blasts in twin 5b is 1 yellow (ETV6-RUNX1 gene fusion), 1 green (unrearranged ETV6), 3 red (corresponding to an additional copy of the RUNX1 gene), and 1 pink (corresponding to 1 copy of the TBL1XR1 gene). This accords with the SNP data indicating 3 discordant “driver” CNAs in this twin pair involving the ETV6, RUNX1, and TBL1XR1 genes. This pattern was observed in 31 of 32 cells scored.
Interphase FISH to confirm CNAs identified by SNP array in twin set 5. Panels A and B show leukemic blasts from twins 5a and 5b, respectively, hybridized with the ETV6-RUNX1 ES (Vysis extra signal) probe and a fosmid probe (W12-1173N4) for the TBL1XR1 gene. The TBL1XR1 probe is labeled with biotin-16-dUTP and detected with streptavidin Cy5 (here pseudocolored pink). In panel A, the signal pattern of leukemic blasts in twin 5a is 1 yellow (corresponding to the ETV6-RUNX1 gene fusion), 2 red (1 corresponding to the normal RUNX1 gene and 1 to the residual RUNX1 gene disrupted by the translocation), and 2 pink (TBL1XR1 gene) signals. Note that the second green signal is not present, indicating loss of the normal (unrearranged) ETV6 gene. This pattern was observed in 21 of 31 cells scored. The other 10 of 31 cells had a normal pattern. In panel B, the signal pattern of leukemic blasts in twin 5b is 1 yellow (ETV6-RUNX1 gene fusion), 1 green (unrearranged ETV6), 3 red (corresponding to an additional copy of the RUNX1 gene), and 1 pink (corresponding to 1 copy of the TBL1XR1 gene). This accords with the SNP data indicating 3 discordant “driver” CNAs in this twin pair involving the ETV6, RUNX1, and TBL1XR1 genes. This pattern was observed in 31 of 32 cells scored.
Some prior data have indicated that deletion of the untranslocated ETV6 can be subclonal or secondary to ETV6-RUNX1 fusion.11,12 The present study suggests that this is both a consistent feature of ETV6 deletions (9/9 deletions detected here) and, more significantly, a feature of all other CNAs in ALL (23/23 deletions or gains detected here in the 5 pairs of twins). Some discordance of genetics in leukemias can be deduced from chromosome karyotype alone.13-15 However, in the present series, 18 of the 23 non-ETV6 “driver” CNAs were invisible by karyotype (Table 2 and supplemental Table 1).
Monozygotic twins discordant for ETV6-RUNX1–positive ALL
We sought to further confirm these findings with a twin pair, previously reported,4 who were discordant for ALL (Figure 3). Twin 6a had ALL with ETV6-RUNX1 and, as identified by SNP arrays (supplemental Table 4), multiple CNAs with potential “drivers” including ETV6, PAX5 deletion, and 10p gain (Figure 3B). Her twin sister was healthy but had a persistent (over 3 years) but clinically covert preleukemic population with ETV6-RUNX1, reflecting their common or single-cell origin in utero.1,10 When these preleukemic cells (defined by positivity for ETV6-RUNX1 using FISH) were interrogated by multicolor FISH for the status of the normal (nontranslocated) ETV6 allele, PAX5 genes, and 10p, no CNA was detected in any of the cells assessed (Figure 3C). We cannot exclude that these ETV6-RUNX1 preleukemic cells harbor these CNAs at low frequencies or indeed have other genetic abnormalities, but these data further support the contention that recurrent and presumed functional gene deletions in ALL arise secondarily to gene fusion.
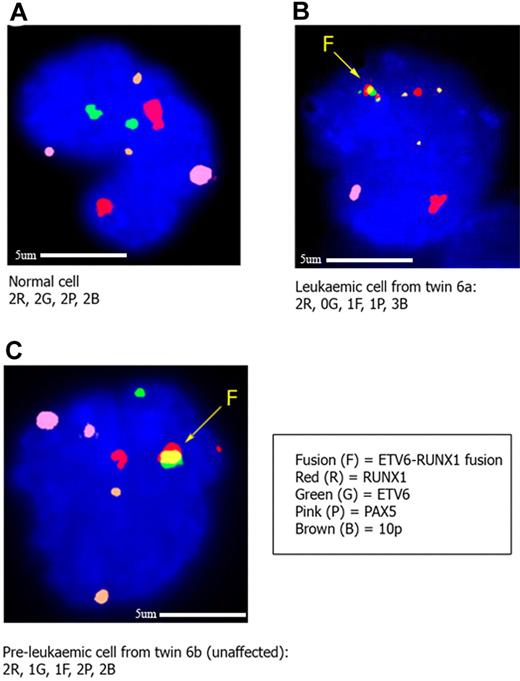
Interphase FISH to confirm CNA status in cells from twins clinically discordant for ETV6-RUNX1 positive ALL. In each case the ETV6-RUNX1 ES (Vysis extra signal) probe was cohybridized with a fosmid probe (W12-3139G6) for the PAX5 gene and a BAC probe (RP-675D23) for the region on 10p shown to be gained in the cells of the twin with leukemia by SNP array. The PAX5 probe is labeled with biotin-16-dUTP (pseudocolored pink), and the 10p probe is labeled with digoxigenin-11-dUTP (pseudocolored brown). In panel A, a normal bone marrow cell from the healthy twin (6b) shows 2 red (RUNX1 gene), 2 green (ETV6 gene), 2 pink (PAX5 gene), and 2 brown (10p) signals, respectively. In panel B, a leukemic cell from twin 6a shows 1 yellow (ETV6-RUNX1 gene fusion), 2 red (normal RUNX1 and remnant from rearranged RUNX1 gene, respectively), 1 pink (PAX5), and 3 brown (10p) signals, respectively. This confirms the findings by SNP array of a deletion of ETV6 and PAX5 and gain of 10p in this twin. In panel C, an ETV6-RUNX1 fusion gene positive cell in the unaffected twin (6b) has the following signal pattern: 1 yellow (ETV6-RUNX1 gene fusion), 2 red (normal and remnant from rearranged RUNX1), 1 green (normal ETV6), 2 pink (PAX5), and 2 brown (10p) signals, indicating that the “driver” mutations seen in the leukemic twin (loss of second ETV6, loss of PAX5, gain of 10p) are not present in the preleukemic, fusion gene positive cells of the unaffected twin. Five cells with the ETV6-RUNX1 fusion were identified in the unaffected twin (twin 6b), of a total of 4251 scored.
Interphase FISH to confirm CNA status in cells from twins clinically discordant for ETV6-RUNX1 positive ALL. In each case the ETV6-RUNX1 ES (Vysis extra signal) probe was cohybridized with a fosmid probe (W12-3139G6) for the PAX5 gene and a BAC probe (RP-675D23) for the region on 10p shown to be gained in the cells of the twin with leukemia by SNP array. The PAX5 probe is labeled with biotin-16-dUTP (pseudocolored pink), and the 10p probe is labeled with digoxigenin-11-dUTP (pseudocolored brown). In panel A, a normal bone marrow cell from the healthy twin (6b) shows 2 red (RUNX1 gene), 2 green (ETV6 gene), 2 pink (PAX5 gene), and 2 brown (10p) signals, respectively. In panel B, a leukemic cell from twin 6a shows 1 yellow (ETV6-RUNX1 gene fusion), 2 red (normal RUNX1 and remnant from rearranged RUNX1 gene, respectively), 1 pink (PAX5), and 3 brown (10p) signals, respectively. This confirms the findings by SNP array of a deletion of ETV6 and PAX5 and gain of 10p in this twin. In panel C, an ETV6-RUNX1 fusion gene positive cell in the unaffected twin (6b) has the following signal pattern: 1 yellow (ETV6-RUNX1 gene fusion), 2 red (normal and remnant from rearranged RUNX1), 1 green (normal ETV6), 2 pink (PAX5), and 2 brown (10p) signals, indicating that the “driver” mutations seen in the leukemic twin (loss of second ETV6, loss of PAX5, gain of 10p) are not present in the preleukemic, fusion gene positive cells of the unaffected twin. Five cells with the ETV6-RUNX1 fusion were identified in the unaffected twin (twin 6b), of a total of 4251 scored.
Discussion
ETV6-RUNX1 fusion is usually a prenatal, acquired genetic event in ALL. This is formally demonstrated by the sharing of identical ETV6-RUNX1 genomic sequences in all 5 monozygotic twin pairs previously assessed10,13 (2 of these pairs, numbers 1 and 3 being included in the current series), and the presence of these clonotypic sequences in the archived, neonatal blood spots of patients with ALL.1,18 This conclusion is further endorsed by the identification of fusion gene positive cells in unselected cord bloods.19 The fact that newborn cord bloods score positive for such putative preleukemic populations (with clinically undetectable frequencies in the lymphocyte fraction of 10−3 to 10−4) at rates substantially higher (∼ 100×) than the cumulative risk of childhood ALL with ETV6-RUNX1 indicates the obligatory requirement for further, postnatal genetic changes or mutations. The concordance rate of ALL in monozygotic twin children, at 10% to 15%10 also supports this contention, as does modeling with murine and human cells.2-4
ALL occurring in the twin context is no different in terms of cellular phenotype, molecular genetics and clinical features to ALL in non-twinned singletons.10 This includes the common prenatal origin of the ETV6-RUNX1 fusion1,18 and patterns of CNAs detected by SNP arrays (this study vs Mullighan et al,5 Strefford et al,6 Lilljebjörn et al,7 and Tsuzuki et al8 ). It is therefore highly likely that the sequence of genetic events we observe in twins applies more generally to childhood ALL.
These data suggest the following sequence of mutational events in the covert, evolutionary history of childhood B-cell precursor ALL. First, and as previously shown,1 an early and likely initiating (or first “hit”) gene fusion (ETV6-RUNX1) occurring prenatally, being sufficient to spawn clonal progeny, including preleukemic stem cells and as endorsed by modeling experiments with human cord blood progenitor cells.4 Second, the acquisition of multiple genetic alterations including (but possibly not restricted to) the CNAs described here and in prior studies.5-8 The latter might be expected to drive the evolution of overt leukemic stem cells4 and to culminate in a clinical diagnosis of ALL. The precise timing of these secondary CNAs has not been determined. They could occur proximal to a diagnosis, perhaps linked to environmental, exposure-based promotion of disease.20 The lack of concordance of all “driver” CNAs identified in the leukemic cells of 6 pairs of twins is most compatible with these genetic events occurring postnatally. Formally, there is a possibility that one or more of them could have arisen prenatally albeit secondary to ETV6-RUNX1 fusion and therefore in a subclone. ALL clearly does have significant subclonal diversity as indicated by long-standing karyotypic observations and more recent SNP array derived data.21 However, twin blood chimaerism10 should result in all subclones generated before birth being shared also. Subsequent discordance of all CNAs in twin pairs would therefore require differential subclone selection by further, postnatal “driver” events. Retrospective screening of serial blood samples of a healthy co-twin that eventually evolves from preleukemia to overt ALL might shed light on the CNAs timing issue. That they can arise significantly before a diagnosis of ALL as evidenced by a case of ETV6-RUNX1–positive ALL with multiple CNAs present in an aplastic phase preceding overt ALL, by some 7 months.22 The conclusion remains that all potentially or functional CNAs in ALL appear to be secondary to the likely initiating lesion, ETV6-RUNX1.
Complete genomic sequencing will be required to determine whether other mutational events, such as balanced translocations or small mutations not detectable by SNP array, contribute to this pattern of clonal evolution in childhood ALL.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We would like to thank the patients and families involved in this study.
This leukemia research was funded by a Leukaemia Lymphoma Research (London) clinical fellowship for C.M.B., a program grant for M.F.G., and Cancer Research UK for funding T.C. and B.D.Y.
Authorship
Contribution: C.M.B. carried out the majority of the experimental work, compiled data, and contributed to writing the paper; S.M.C. and T.C. carried out experimental work; B.D.Y. provided expertise in SNP array data analysis; T.O.E., M.B., E.J.G., E.R.v.W., G.C., C.J.H., R.H., and P.A. provided patient samples (cell and/or DNA) and patient diagnostic information; A.M.F. carried out experimental work and provided expertise in molecular techniques; L.K. helped manage the study, supervised FISH studies, and assisted in writing the paper; and M.G. conceived the study, acquired twin leukemic cell DNA samples, helped analyze the data, and drafted the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Prof Mel Greaves, FRS, Section of Haemato-Oncology, The Institute of Cancer Research, Brookes Lawley Bldg, 15 Cotswold Rd, Belmont, Sutton, Surrey SM2 5NG, United Kingdom;e-mail: mel.greaves@icr.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal