Abstract
Self-renewal is a feature of cancer and can be assessed by cell transplantation into immune-compromised or immune-matched animals. However, studies in zebrafish have been severely limited by lack of these reagents. Here, Myc-induced T-cell acute lymphoblastic leukemias (T-ALLs) have been made in syngeneic, clonal zebrafish and can be transplanted into sibling animals without the need for immune suppression. These studies show that self-renewing cells are abundant in T-ALL and comprise 0.1% to 15.9% of the T-ALL mass. Large-scale single-cell transplantation experiments established that T-ALLs can be initiated from a single cell and that leukemias exhibit wide differences in tumor-initiating potential. T-ALLs also can be introduced into clonal-outcrossed animals, and T-ALLs arising in mixed genetic backgrounds can be transplanted into clonal recipients without the need for major histocompatibility complex matching. Finally, high-throughput imaging methods are described that allow large numbers of fluorescent transgenic animals to be imaged simultaneously, facilitating the rapid screening of engrafted animals. Our experiments highlight the large numbers of zebrafish that can be experimentally assessed by cell transplantation and establish new high-throughput methods to functionally interrogate gene pathways involved in cancer self-renewal.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is a devastating disease of childhood and is associated with transformation of thymic precursor cells. SCL- and LMO1/LMO2-expressing leukemias account for 60% of pediatric cases and are more aggressive and treatment resistant than the other 4 subtypes,1 suggesting that distinct molecular pathways associated with transformation affect both tumor aggression and response to treatment. However, shared pathways also are used to transform T cells. P16/INK4a deletion is found in a majority of T-ALLs,2 gain-of-function mutations and deletions in NOTCH1 are common,3,4 and MYC is a central regulator of T-ALL formation.5 Taken together, T-ALL comprises a group of diseases that transform T cells by common molecular pathways, but subtypes of disease have different clinical outcomes reflected by the developmental stage at which lymphocyte differentiation is arrested and by the expression of T cell–associated developmental programs.
Rare subpopulations of self-renewing cell types have been identified in mouse and human T-ALL. Cell transplantation experiments in which the authors used T-ALLs from Pten-deficient mice have identified the c-KitmidCD3+ subpopulation as the leukemia-initiating cell (LIC).6 This cell population engrafts disease into sublethally irradiated (IR) severe combined immune deficiency (SCID) mice better than either the CD3− or c-Kit−CD3+ cells and comprises less than 0.01% of the total tumor. A rare subpopulation of human T-ALL–initiating cells also has been identified, with the CD34+/CD4− and CD34+/CD7− cell types being capable of engraftment when introduced into IR nonobese diabetic (NOD)/SCID mice.7 In these experiments, 5 × 105 to 1 × 107 unsorted leukemia cells were required for engraftment, indicating that LICs also are rare in human T-ALL.
Additional xenograft transplantation studies establish that human acute leukemias engraft into NOD/SCID/interleukin-2 receptorγ-null animals but less efficiently when introduced into partially immune compromised strains of mice.8 For the 2 ALLs described in this report, 106 unsorted leukemia cells were required to engraft disease (n = 2 of 4 or 2 of 5 transplant mice developed disease, respectively), again suggesting that ALL-initiating cells are rare. By contrast, tumor-initiating cells are frequent in syngeneic mouse models of T and B lymphoma and comprise greater than 10% of the tumor mass.9,10 In these experiments, one primary Eμ-N-RAS–induced T lymphoma was assessed for tumor-initiating potential by transplantation into syngeneic recipient animals. All 3 transplant animals that received 10 lymphoma cells went on to develop disease; however, serial transplantation, a hallmark for definitively assessing long-term self-renewal potential, was not reported. Taken together, these opposing reports suggest that there is still quite a controversy in the field governing the numbers of T-ALL cells required to remake tumor and highlight the need for further experiments to accurately determine the cell types and numbers of tumor-initiating cells in human and genetic models of T-ALL.
Transgenic zebrafish models of Myc-induced T-ALL provide a unique platform to study leukemogenesis and self-renewal. Zebrafish T-ALLs develop by 30 days of life and can be induced into various genetic backgrounds by microinjecting the rag2-mouse cMyc (rag2-mMyc) transgene directly into one cell–stage zebrafish embryos. Like human T-ALL, zebrafish leukemias are oligo-clonal,11,12 and 5 × 108 cancer cells can be isolated from a single leukemic zebrafish. T-ALL progression also can be visualized in mosaic animals; however, high-throughput methods for detecting fluorescent-labeled tumors have not been described.11,12 Although the zebrafish model of T-ALL holds immense promise for defining important pathways in human cancer, the data presented here suggests that tumor-initiating cell number cannot be accurately assessed by current cell-transplantation protocols that introduce tumor cells into IR nonimmune-matched recipients.11,13-21 If immune barriers can be overcome, the zebrafish model will afford unique opportunities to uncover the mechanisms underlying self-renewal in both normal and malignant cells.
Here, sygeneic zebrafish (CG1 strain) were used to refine cell transplantation of zebrafish cancer and to accurately quantify the numbers of self-renewing cell types in T-ALL. We show that the percentage of LIC types differs between T-ALLs and comprises between 0.1% and 15.9% of the T-ALL mass. Leukemia-initiating potential also was assessed by transplantation of single T-ALL cells, establishing that quantitative differences in leukemia-initiating potential exist between leukemias. By out-crossing CG1-strain fish to different genetic strains, it is also possible to transplant CG1 tumors into mixed genetic backgrounds or to transplant T-ALLs from a mixed genetic strain into CG1 recipient animals. Finally, high-throughput imaging methods were described to score adult transplant zebrafish for T-ALL engraftment. The methods outlined here capitalize on the large numbers of zebrafish that can be transplanted. In total, more than 1453 transplant animals were used in these studies and lay the foundation for large-scale experiments to define evolutionarily conserved self-renewal pathways in cancer that are not available in more-established models of malignancy.
Methods
Animals
Zebrafish maintenance and developmental staging were conducted as described previously.11 CG1-strain zebrafish were a gift from Dr Sergei Revskoy (Feinberg School of Medicine, Northwestern University). Albino and mylz2-green fluorescent protein (GFP) fish were obtained from Dr Leonard Zon (Children's Hospital Boston). rag2-GFP transgenic fish were created previously.11 Stable transgenic mylz2-Amcyan, mylz2-mCherry, myogenin-mCherry, and creatine-kinase-zsYellow zebrafish were generated in the AB-strain background. Specifically, each promoter was amplified from genomic DNA with the use of polymerase chain reaction. The forward primer had a XhoI site and the reverse a BamHI site. DNA fragments were purified, digested with XhoI and BamHI, and inserted into the rag2-GFP vector. Next, clones were digested with BamHI and HindIII to release the GFP cassette, and either mCherry, zsYellow, or Amcyan were cloned into the modified vector. Plasmid DNA was digested with XhoI to linearize the transgene. DNA was phenol:chloroform extracted, ethanol precipitated, quantified by gel electrophoresis, and diluted to 120 ng/μL in 0.5×TE + 100mM KCl. Next, AB-strain fish were microinjected at the one-cell stage of life and allowed to grow to adulthood. F0-injected animals were incrossed and the resulting progeny scored to identify stable F1 transgenic progeny. Transgenic F1 animals were outcrossed to AB-strain animals to propagate the line. All animal experiments were approved by the MGH Subcommittee on Research Animal Care.
Generation of mosaic transgenic animals that develop T-ALL and rhabdomyosarcoma
The rag2-GFP,22 rag2-mouse cMyc,11 rag2-kRASG12D, and rag2-dsRED18 constructs have been described previously, whereas the rag2-Amcyan and rag2-zsYellow constructs were made essentially as described.11 DNA constructs were linearized with either XhoI or Not1, phenol:chloroform extracted, and ethanol precipitated. DNA was diluted in 0.5 × TE + 100mM KCl to 60 ng/μL, with coinjection of 2 transgenes having 30 ng of each construct/microliter. DNA was microinjected into the one-cell–stage animals. Tumor-bearing zebrafish were identified at 30 to 60 days of life on the basis of fluorescent protein expression and subsequently used for cell transplantation experiments when animals were 60 to 120 days of age. RNA also was extracted from total tumor and wild-type AB-strain kidney marrow, muscle, or 24-hour embryos. After Trizol extraction, RNA was treated with DNAse and then made into complementary cDNA. Samples were assessed for expression of lck, immunoglobulin M, rag2, mouse cMyc, and ef1-alpha as previously described.11
Cell transplantation and FACS
Primary T-ALLs and RAS-induced rhabdomyosarcomas were transplanted into non-IR CG1-, AB-strain, mixed CG1/albino-strain, or CG1/AB-strain recipient fish. T-ALLs also were transplanted into sublethally IR AB-strain recipients (single dose 23 Gy, 2 days before transplantation).11,13 Cells were either sorted by fluorescence-activated cell sorting (FACS) to isolate fluorescent T-ALL cells (propidium iodide was used to exclude dead cells), or unsorted cells were introduced into recipients by intraperitoneal injection (5 μL of injection volume). In some instances, fluorescent T-ALL cells were isolated from transplant recipients and used as donor cells for subsequent serial transplantation. For limiting dilution analysis, red blood cells from CG1-strain zebrafish were used as carrier cells along with T-ALL cells. Transplantation was completed essentially as described.11,13
Limiting dilution analysis
Limiting dilution analysis was completed with the use of the Bonnefoix linear regression method. Accuracy of this test is determined by correlation coefficient (R2 values). Subsequent analysis with L-calc and limdil software confirmed our limiting dilution calculations and provided 95% confidence intervals and t test calculations to compare tumor-initiating cell numbers between samples (Table 1; supplemental Tables 3 and 5, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). All 3 analyses provided similar results.
Leukemia-initiating cells are abundant in zebrafish T-ALL
| T-ALL donor . | Recipients per experiment . | LIC frequency . | 95% Confidence . | Correlation R2 value . |
|---|---|---|---|---|
| T-ALL #5-primary leukemia | 44 | 1/250 | 1/94 to 1/663 | 0.9802 |
| T-ALL #5-transplanted leukemia | 42 | 1/57 | 1/30 to 1/110 | 0.9982 |
| T-ALL #6-primary leukemia | 46 | 1/71 | 1/37 to 1/139 | 0.9917 |
| T-ALL #6-transplanted leukemia | 40 | 1/60 | 1/32 to 1/114 | 0.9112 |
| T-ALL #7-primary leukemia | 40 | 1/77 | 1/39 to 1/151 | 0.999 |
| T-ALL #7-transplanted leukemia | 41 | 1/135 | 1/59 to 1/306 | 0.9859 |
| T-ALL #8-primary leukemia | 35 | 1/845* | 1/277 to 1/1212 | 0.9852 |
| T-ALL donor . | Recipients per experiment . | LIC frequency . | 95% Confidence . | Correlation R2 value . |
|---|---|---|---|---|
| T-ALL #5-primary leukemia | 44 | 1/250 | 1/94 to 1/663 | 0.9802 |
| T-ALL #5-transplanted leukemia | 42 | 1/57 | 1/30 to 1/110 | 0.9982 |
| T-ALL #6-primary leukemia | 46 | 1/71 | 1/37 to 1/139 | 0.9917 |
| T-ALL #6-transplanted leukemia | 40 | 1/60 | 1/32 to 1/114 | 0.9112 |
| T-ALL #7-primary leukemia | 40 | 1/77 | 1/39 to 1/151 | 0.999 |
| T-ALL #7-transplanted leukemia | 41 | 1/135 | 1/59 to 1/306 | 0.9859 |
| T-ALL #8-primary leukemia | 35 | 1/845* | 1/277 to 1/1212 | 0.9852 |
LIC frequency and 95% confidence intervals were calculated by the L-calc program, and correlation coefficient R2 values show how well the data fit by use of the Bonnefoix limiting dilution method.
LIC indicates leukemia-initiating cell; and T-ALL, T-cell acute lymphoblastic leukemia.
Sample T-ALL 8 = primary leukemia is significantly different from all other samples (P < .05) except sample T-ALL 5 = primary leukemia. The raw data used to calculate LIC frequency are shown in supplemental Table 4.
Assessing tumor engraftment
Transplant recipient fish were analyzed for fluorescent tumor engraftment with (1) an Olympus SZX16 stereomicroscope outfitted with a Prior Lumen 200 epi-fluorescence illuminator and an Olympus DP-72 color camera or (2) the light-emitting diode (LED) fluorescence macroscope at 10, 20, 30, 35, 45, 60, and 85 days after transplantation. The anterior portion of leukemic fish was fixed in 4% paraformaldehyde, processed, embedded in paraffin, and sectioned. Sections were stained with hematoxylin and eosin. In addition, leukemias also were analyzed by cytospin and May-Grunwald Giemsa staining to confirm lymphoblast morphology.
LED fluorescence macroscope and image analysis
The inverted LED fluorescence macroscope is composed of digital video cameras capable of still image photography (Logitech QuickCam, model S5500) that are positioned underneath a suspended plexiglass cover (supplemental Figure 4). High-intensity LED lights illuminate samples from below (405- to 420-nm blacklight, 420- to 470-nm blue light, 500- to 570-nm green light, incandescence light for brightfield imaging, PAR20 style bulbs), and a set of filters fit over the camera lenses, allowing only emitted light of specific wavelengths to be detected. Two digital cameras interface with a Lenovo computer and images are captured with the use of the AMCap imaging software. Video exposure, gain, intensity, and capture rate are controlled within the AMCap Software package. Brightfield and fluorescent images were imported into Photoshop and merged. For dual spectrum imaging, video was simultaneously captured by the use of 2 cameras fitted with different filter sets. Digital images from each video frame were merged with use of the Osirix software.
Results
CG1-strain zebrafish
Eggs were manually extruded from a single Golden strain female, fertilized with ultraviolet-inactivated sperm, and subjected to heat shock to produce gynogenetic diploid animals as previously described.23 Gynogenetic females were raised to adulthood, and their eggs were used in a second round of heat shock. The resulting F3 progeny were incrossed to create the CG1-strain (gift from Dr Sergei Revskoy and Igor Mizgirev, Northwestern University). The CG1 clonal fish line has not been described previously and is not related to the CB1 or CW1 lines.23
To validate that CG1-strain fish are genetically similar, a genome-wide survey was performed on 281 loci in 4 CG1 animals and 4 AB-strain zebrafish. In total, 275 of 281 CA repeat polymorphisms are identical in all 4 CG1 fish. The primer sets used in this analysis are scattered throughout the genome and are present on all 25 chromosomes (supplemental Table 1). Of these primer combinations, 63 distinguished allelic differences between CG1 and AB fish, and as expected the AB-strain sibling fish were heterogeneous at many loci (n = 165 of 281). We conclude that CG1 fish are 97.9% identical.
T-ALLs arising in CG1-strain zebrafish can be transplanted into non-IR, CG1 recipients
rag2-mouse cMyc (mMyc) and rag2-GFP, rag2-dsREDexpress, rag2-zsYellow, or rag2-Amcyan constructs were coinjected into one-cell–stage CG1-strain animals. Because injection of multiple transgenes into one-cell–stage embryos leads to cosegregation and coexpression in developing tumors,20 T-ALLs that develop were fluorescently labeled. T-ALLs were first detected in the thymus of mosaic animals and disseminated widely as leukemias progressed, similar to previous reports.11 Leukemias expressed T cell–specific lck, rag1, and mouse cMyc but failed to express high levels of immunoglobulin M or the muscle-specific factor, desmin, confirming that leukemias were T cell in origin (supplemental Figure 1). T-ALL cells were disassociated from 60- to 120-day-old leukemic fish and subjected to FACS, with dead cells being excluded by propidium iodide (94.0%-97.3% pure, 97.9%-98.9% viable). Fluorescent-labeled T-ALL cells were analyzed by cytospin to confirm lymphoblast morphology (n = 12; supplemental Figure 2) or transplanted into non-IR CG1- and AB-strain animals and gamma-IR AB-strain fish at limiting dilution (23 Gy, 2 days before transplantation, 5 × 104, 1 × 104, 1 × 103, and 1 × 102 T-ALL cells supplemented up to 5 × 104 cells with red blood cells [RBCs], n = 3 tumors analyzed total).
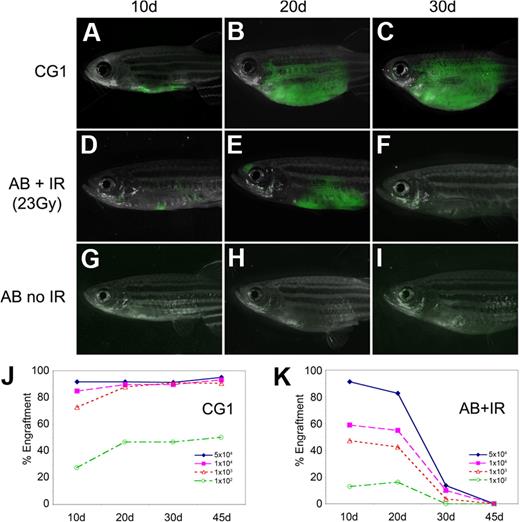
Fluorescent-labeled T-ALL cells from CG1-strain animals engrafted robustly into both non-IR CG1 recipients and IR AB-strain animals (AB + IR) by 10 days after transplantation (Figure 1A,D; supplemental Table 2); however, AB + IR recipients had fewer engrafted animals compared with CG1 recipients at both 10 and 20 days after transplantation (compare Figure 1J and 1K) and less tumor burden (Figure 1A-B and D-E). Most T-ALLs arising in AB + IR recipient fish had completely regressed by 30 days after transplantation, whereas those arising in CG1 fish continued to grow (Figure 1C,F,J-K). Sublethal irradiation of AB-strain recipient animals also led to increased death of recipient animals, with 21 of 124 AB + IR recipients dying by 45 days after transplantation compared with 5 of 114 CG1 recipients (P = .003, Fisher exact test). This difference could not be accounted for by death as the result of tumor burden (supplemental Table 2). Limiting dilution experiments establish that leukemia-initiating frequency was underestimated by 20- to 33-fold when primary leukemias were introduced into AB-IR recipient animals (supplemental Table 3). As expected, T-ALL cells failed to engraft into non-IR AB-strain recipients (Figure 1G-I).11 Taken together, our data show that limiting dilution analysis and cell transplantation into IR AB-strain animals will severely underestimate tumor-initiating cell number due to inefficient ablation of the immune system and irradiation-induced lethality of recipient fish.
T-ALLs from CG1-strain zebrafish engraft into non-IR CG1-strain recipients. GFP-labeled T-ALLs were isolated from primary leukemic fish, and 103 FACS-sorted GFP-labeled leukemia cells were transplanted into non-IR CG1- and AB-strain animals (A-C and G-I, respectively) or IR AB-strain fish (D-F). Transplant fish were scored for engraftment at 10, 20, and 30 days after transplantation. Panels are merged images of fluorescent and brightfield photographs. The engraftment kinetics differ greatly when T-ALLs are introduced into CG1 (J) or IR AB-strain zebrafish (AB + IR; K). Percent engraftment is the percentage of recipient animals that have visibly engrafted T-ALL at each time point. Panels J and K are combined data from 3 independent experiments (n = 238 transplants total). The raw data for this experiment are available in supplemental Table 2.
T-ALLs from CG1-strain zebrafish engraft into non-IR CG1-strain recipients. GFP-labeled T-ALLs were isolated from primary leukemic fish, and 103 FACS-sorted GFP-labeled leukemia cells were transplanted into non-IR CG1- and AB-strain animals (A-C and G-I, respectively) or IR AB-strain fish (D-F). Transplant fish were scored for engraftment at 10, 20, and 30 days after transplantation. Panels are merged images of fluorescent and brightfield photographs. The engraftment kinetics differ greatly when T-ALLs are introduced into CG1 (J) or IR AB-strain zebrafish (AB + IR; K). Percent engraftment is the percentage of recipient animals that have visibly engrafted T-ALL at each time point. Panels J and K are combined data from 3 independent experiments (n = 238 transplants total). The raw data for this experiment are available in supplemental Table 2.
LICs comprise 0.1% to 1.4% of the primary T-ALL
To assess the number of self-renewing cells contained within the bulk of the primary T-ALL tumor mass, fluorescent-labeled T-ALL cells were isolated from CG1-strain zebrafish and introduced into CG1-strain recipients at limiting dilution (1 × 104, 1 × 103, 1 × 102, 1 × 101 T-ALL cells supplemented up to 2 × 104 cells with RBC carrier cells, purity 95%-99%, viability 94%-99.9%, n = 4 T-ALLs analyzed). Engraftment was assessed at 20, 30, 45, and 85 days after transplantation by epi-fluorescence stereomicroscopy (raw data are presented in supplemental Table 4) and LIC number was calculated after 85 days after transplantation by the use of 3 methods: the Bonnefoix limiting dilution method,24 the L-calc statistical software (StemCell Technologies), and the web-based limdil program (http://bioinf.wehi.edu.au/software/elda/index.html). All 3 analyses gave similar results (Table 1; supplemental Table 5). In total, these experiments suggest that LICs are abundant in T-ALL and comprise between 0.1% and 1.4% of the primary tumor mass. Moreover, these results show that leukemia-initiating frequency can differ between T-ALLs (compare #6 and #8, P < .001).
The ability to induce disease after serial transplantation is a hallmark of self-renewing cancer cells. To determine whether leukemia-initiating potential is retained after serial transplantation, fluorescent-labeled leukemias were isolated from primary transplant animals that received 1 × 104 T-ALL cells and introduced at limiting dilution into CG1 secondary recipients (purity 93.9%-95.7%, viability 94.3%-97.2%). Most T-ALLs retain similar proportions of LICs compared with primary leukemias (n = 3; Table 1; supplemental Table 5). Serial transplantation confirms that leukemia-initiating potential is found in a large portion of T-ALL cells.
Single-cell transplantation confirms that tumor-initiating potential differs between T-ALLs
Although LICs are abundant in zebrafish T-ALL, their number differs between leukemias (Table 1). To further validate that T-ALLs can exhibit marked differences in tumor-initiating potential, single-cell transplants were completed with the use of either primary leukemias (n = 2), a single-passage T-ALL (n = 1), or a serially passaged T-ALL that had high leukemia-initiating capacity (T-ALL #9, serial passage 3). For single-cell transplants, cells were sorted into 96-well plates with each well supplemented with 2 × 104 RBCs. Test sorts verified that single cells were found in most wells (n = 45 of 48; supplemental Figure 3). One GFP-labeled primary leukemia engrafted 2 of 105 transplant animals by 60 days after transplantation, whereas single cells from the other primary and one-time passaged T-ALL failed to engraft disease by 120 days after transplantation (n = 0 of 210), indicating that tumor-initiating cell number was low in these T-ALLs.
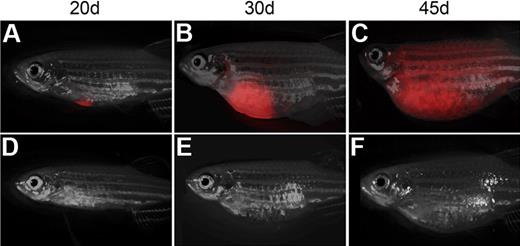
Tumor purity and viability were very high in all 3 samples after FACS (> 95% viability and > 97% purity), and 1 × 104 cells from these same leukemias lead to engraftment in all recipient animals (n = 6 per tumor). One CG1-strain T-ALL was serially transplanted 3 times (T-ALL #9). This leukemia had high leukemia initiating frequency and engrafted robustly into recipient animals (n = 7 of 44, viability 97.9% and purity 94.1%; Figure 2). These results highlight the large-scale single-cell transplantation experiments that are now possible in zebrafish (n = 359 animals) and confirm that leukemias can exhibit wide variation in leukemia-initiating potential with 0.1% to 15.9% or less of all leukemia cells having the capacity to remake tumor (P < .003; Fisher exact test).
Single T-ALL cells can be efficiently transplanted into non-IR CG1 recipient. Transplant recipient fish receiving 1 FACS-sorted dsRED+ T-ALL cell at 20 (A,D), 30 (B,E), and 45 days after transplantation (C,F). One representative animal engrafted T-ALL by 20 days (A-C), whereas one never developed leukemia (D-F). Images photographed at 7.0×. All panels are merged images of fluorescent and brightfield photographs.
Single T-ALL cells can be efficiently transplanted into non-IR CG1 recipient. Transplant recipient fish receiving 1 FACS-sorted dsRED+ T-ALL cell at 20 (A,D), 30 (B,E), and 45 days after transplantation (C,F). One representative animal engrafted T-ALL by 20 days (A-C), whereas one never developed leukemia (D-F). Images photographed at 7.0×. All panels are merged images of fluorescent and brightfield photographs.
T-ALLs can be transplanted into CG1-outcrossed animals
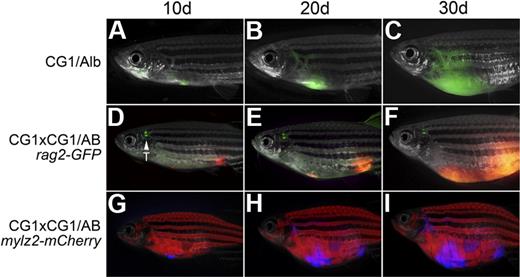
Nontumorigenic somatic cells can alter the tumor microenvironment and can affect recruitment of tumor vasculature, invasion, and metastasis. These processes can be assessed experimentally by transplanting tumor cells into syngeneic recipients that harbor specific genetic lesions. Moreover, introduction of tumors into various fluorescent transgenic zebrafish lines will allow these processes to be visualized in vivo. To determine whether CG1-strain zebrafish are amenable to such analysis, T-ALLs from CG1 fish were transplanted into the progeny of CG1-strain zebrafish crossed to albino homozygous animals. Remarkably, T-ALLs engrafted robustly into CG1/albino recipients (n = 24 of 31; Figure 3A-C; Table 2), less efficiently, however, than pure CG1 recipients (n = 27 of 27, P = .01, Fisher exact test). T-ALLs from CG1-strain zebrafish were introduced into the progeny of CG1 fish crossed with (1) wild-type AB-strain, (2) rag2-GFP (AB-strain,22 ), or (3) mylz2-mCherry (AB-strain) transgenic zebrafish. In contrast to experiments using albino animals, T-ALLs did not engraft efficiently into F1 AB-strain outcrossed recipients (n = 1 of 23 CG1/AB recipient animals developed T-ALL, P < .001, Fisher exact test). However, T-ALLs displayed increased engraftment potential when introduced into progeny, resulting from 2 rounds of outcrossing to CG1 fish—a mating of CG1 fish to CG1/AB fish (P = .001, Fisher exact test). In total, 8 of 19 CG1 × CG1/AB animals engrafted T-ALL (Figure 3D-I; Table 2), and by the third out-cross to CG1-strain fish, all animals were able to engraft T-ALLs arising from CG1-strain fish (n = 18 of 18). Taken together, our experiments show that it will now be possible to outcross CG1-strain zebrafish to specific lines of interest, allowing T-ALLs from CG1-strain fish to be efficiently transplanted into mixed mutant and/or transgenic backgrounds without the need for major histocompatibility complex matching or immune ablation by gamma-irradiation. Our experiments also indicate that albino-strain fish are more closely related to CG1 fish and that strain differences will have major impacts on the number of outcrosses required to create syngeneic recipient animals.
T-ALLs from CG1 fish are capable of engraftment into CG1 outcrossed animals. A GFP-labeled T-ALL engrafted into the progeny of CG1 by albino fish (CG1/Alb; A-C). Engraftment of T-ALLs into AB-strain zebrafish required 2 rounds of outcrossing to CG1 fish (D-I). A dsRED-labeled T-ALL engrafted into the progeny of a CG1 by CG1/AB rag2-GFP transgenic animal (D-F) or an Amcyan-labeled T-ALL engrafted into the progeny of a CG1 by CG1/AB mylz2-mCherry (G-I). Images photographed at 7.0×. All panels are merged images of fluorescent and brightfield photographs. GFP-labeled thymus is marked by T.
T-ALLs from CG1 fish are capable of engraftment into CG1 outcrossed animals. A GFP-labeled T-ALL engrafted into the progeny of CG1 by albino fish (CG1/Alb; A-C). Engraftment of T-ALLs into AB-strain zebrafish required 2 rounds of outcrossing to CG1 fish (D-I). A dsRED-labeled T-ALL engrafted into the progeny of a CG1 by CG1/AB rag2-GFP transgenic animal (D-F) or an Amcyan-labeled T-ALL engrafted into the progeny of a CG1 by CG1/AB mylz2-mCherry (G-I). Images photographed at 7.0×. All panels are merged images of fluorescent and brightfield photographs. GFP-labeled thymus is marked by T.
T-ALLs from mixed CG1 backgrounds can be introduced into nonirradiated CG1 recipients
| Tumor ID . | Donor background . | No. of transplanted cells . | Recipient background . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| AB + IR . | CG1 . | CG1 × Alb . | AB noIR . | AB/CG1 . | AB/CG1 × CG1 . | [AB/CGi × CG1] × CG1 . | |||
| 1 | CG1 | 5 × 105 | 5 of 5 | 7 of 7 | 7 of 7 | 0 of 6 | NA | NA | NA |
| 3 | CG1 | 5 × 104 | 3 of 4 | 6 of 6 | 3 of 9 | 0 of 7 | NA | NA | NA |
| 4 | CG1 | 5 × 105 | 2 of 3 | 14 of 14 | 14 of 15 | 0 of 2 | NA | NA | NA |
| Mixed* | CG1 | 5 × 105 | NA | NA | NA | NA | 1 of 23 | 8 of 19 | 18 of 18 |
| A | CG1/Alb | 5 × 105 | 1 of 1 | 0 of 7 | 0 of 9 | 0 of 6 | NA | NA | NA |
| B | CG1/Alb | 5 × 105 | 2 of 5 | 0 of 7 | 0 of 6 | 0 of 7 | NA | NA | NA |
| C | CG1/Alb | 5 × 105 | 2 of 2 | 0 of 4 | 0 of 6 | 0 of 5 | NA | NA | NA |
| D.1 | CG1/Alb × CG1 | 5 × 105 | NA | 0 of 13 | NA | NA | NA | NA | NA |
| D.2 | CG1/Alb × CG1 | 5 × 105 | NA | 0 of 13 | NA | NA | NA | NA | NA |
| D.3 | CG1/AIb × CG1 | 5 × 105 | NA | 0 of 11 | NA | NA | NA | NA | NA |
| D.4 | CG1/AIb × CG1 | 5 × 105 | NA | 0 of 11 | NA | NA | NA | NA | NA |
| D.5 | CG1/AIb × CG1 | 5 × 105 | NA | 11 of 12 | NA | NA | NA | NA | NA |
| D.6 | CG1/Alb × CG1 | 5 × 105 | NA | 11 of 11 | NA | NA | NA | NA | NA |
| Tumor ID . | Donor background . | No. of transplanted cells . | Recipient background . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| AB + IR . | CG1 . | CG1 × Alb . | AB noIR . | AB/CG1 . | AB/CG1 × CG1 . | [AB/CGi × CG1] × CG1 . | |||
| 1 | CG1 | 5 × 105 | 5 of 5 | 7 of 7 | 7 of 7 | 0 of 6 | NA | NA | NA |
| 3 | CG1 | 5 × 104 | 3 of 4 | 6 of 6 | 3 of 9 | 0 of 7 | NA | NA | NA |
| 4 | CG1 | 5 × 105 | 2 of 3 | 14 of 14 | 14 of 15 | 0 of 2 | NA | NA | NA |
| Mixed* | CG1 | 5 × 105 | NA | NA | NA | NA | 1 of 23 | 8 of 19 | 18 of 18 |
| A | CG1/Alb | 5 × 105 | 1 of 1 | 0 of 7 | 0 of 9 | 0 of 6 | NA | NA | NA |
| B | CG1/Alb | 5 × 105 | 2 of 5 | 0 of 7 | 0 of 6 | 0 of 7 | NA | NA | NA |
| C | CG1/Alb | 5 × 105 | 2 of 2 | 0 of 4 | 0 of 6 | 0 of 5 | NA | NA | NA |
| D.1 | CG1/Alb × CG1 | 5 × 105 | NA | 0 of 13 | NA | NA | NA | NA | NA |
| D.2 | CG1/Alb × CG1 | 5 × 105 | NA | 0 of 13 | NA | NA | NA | NA | NA |
| D.3 | CG1/AIb × CG1 | 5 × 105 | NA | 0 of 11 | NA | NA | NA | NA | NA |
| D.4 | CG1/AIb × CG1 | 5 × 105 | NA | 0 of 11 | NA | NA | NA | NA | NA |
| D.5 | CG1/AIb × CG1 | 5 × 105 | NA | 11 of 12 | NA | NA | NA | NA | NA |
| D.6 | CG1/Alb × CG1 | 5 × 105 | NA | 11 of 11 | NA | NA | NA | NA | NA |
The rag2-mMyc and rag2-GFP, rag2-dsREDexpress, or rag2-zsYellow transgenes were coinjected into various genetic backgrounds at the one-cell stage of life, and T-ALLs that developed were introduced into recipient animals by intraperitoneal injection. Transplanted animals were scored for T-ALL engraftment at 10, 20, and 30 days. Table is a compilation of the total number of animals that had engrafted disease by 30 days.
AB indicates AB strain; Alb, albino; IR, irradiation; and NA, not assayed.
Mixed is a combination of 3 independent experiments in which T-ALLs arising in CG1 animals were introduced into AB-strain fish that were once outcrossed to CG1-strain animals (AB/CG1), twice outcrossed (AB/CG1 × CG1), or 3 times outcrossed ([AB/CG1 × CG1] × CG1).
T-ALLs from a mixed CG1genetic background can be transplanted into CG1 recipients
Generating zebrafish tumors that have heritable genetic lesions will be important for identifying genes that modulate tumor engraftment, metastatic potential, and self-renewal. As such, T-ALLs were generated in zebrafish of mixed genetic backgrounds by microinjecting rag2-mMyc and rag2-GFP or rag2-dsREDexpress into one-cell–stage (1) CG1 embryos, (2) the offspring of a CG1 by albino homozygous cross (CG1/Alb), or (3) the progeny of a CG1/Alb by CG1 cross (CG1/Alb × CG1). T-ALLs arising in mosaic transgenic animals were transplanted into non-IR CG1-strain recipient animals (Table 2). As expected, T-ALLs from CG1-strain zebrafish engrafted robustly into CG1 recipients (n = 3 T-ALLs, 10 fish per tumor, data not shown), whereas T-ALLs from CG1/albino fish failed to engraft into CG1 recipients (n = 3 tumors assayed; Table 2). By contrast, a subset of T-ALLs arising from CG1/Alb × CG1 fish engrafted into non-IR CG1-strain recipients (n = 2 of 6 T-ALLs assayed), and in cases in which transplantation was possible, the percentage of animals with tumor engraftment was remarkably high (n = 22 of 23 transplanted zebrafish engrafted T-ALLs). Similar results also were seen when a kRASG12D-induced transgenic model of embryonal rhabdomyosarcomas was used (supplemental Table 6). Specifically, injection of rag2-kRASG12D into AB-strain mylz2-mCherry transgenic animals that had been outcrossed to CG1 fish 3 times produced transplantable rhabdomyosarcomas. All 4 rhabdomyosarcomas assayed could engraft with 100% efficiency when introduced into CG1 recipient animals (n = 25 of 25; supplemental Table 6). Taken together, these experiments provide new methodologies to rapidly introduce existing transgenic reporter lines and genetic mutations into syngeneic zebrafish.
High-throughput imaging with the LED fluorescence macroscope
Having accurately determined the LIC number in zebrafish T-ALL and developed methods to introduce tumor cells into a variety of genetic and transgenic backgrounds, we wanted to streamline the methods for detecting tumor engraftment in recipient animals. For example, screening transplant fish for engraftment with the use of a conventional fluorescence-dissecting microscope is laborious and time-consuming because screening is limited to visually inspecting one fish at a time (Figure 4Q-T). To expedite screening of both normal and tumor-bearing fluorescent transgenic zebrafish, a low-cost, high-throughput machine has been developed that can score a whole Petri dish of adult transgenic zebrafish—the LED fluorescence macroscope (supplemental Figure 4). In total, the LED fluorescence macroscope is capable of discriminating 5 colors (Figure 4), including Amcyan (blacklight, 480/30 filter), GFP (blacklight, 520/30 filter), zsYellow (blue light, 575/52 filter), dsRED express (blue light, 610/40), and mCherry (green light, 610/40 filter or 640/35). The emission/excitation spectra for each fluorescent protein are shown in supplemental Figure 5.
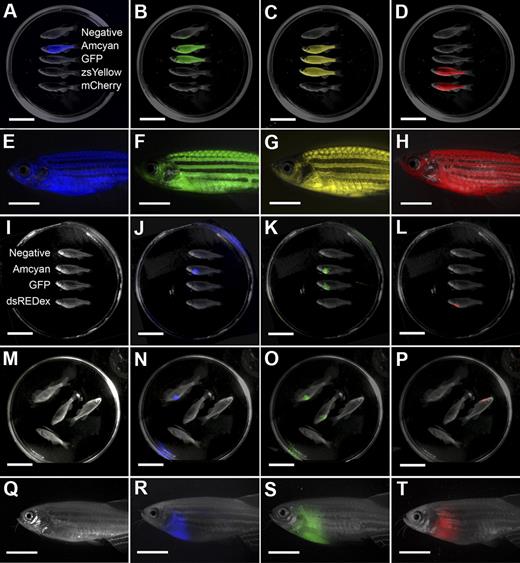
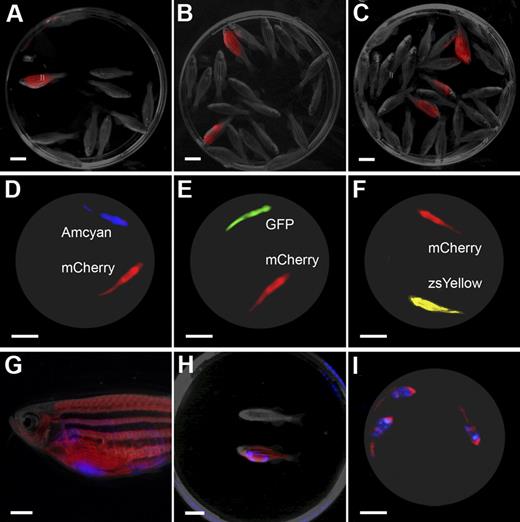
The LED fluorescence macroscope can image 5 fluorescent fluorophores in normal muscle and T-cell acute lymphoblastic leukemia (T-ALL). Whole animal imaging of wild-type (Negative), mylz2-Amcyan, mylz2-GFP, creatine kinase-zsYellow, mylz2-mCherry transgenic zebrafish (A-D). Amcyan was imaged by the use of a blacklight and 480/30 filter (A,J,N), GFP (blacklight, 520/30 filter; B,K,O), zsYellow (blue light, 575/52 filter; C), dsRED express (green light, 610/40 filter; L,P), and mCherry (green light, 610/40 filter; D). Transgenic zebrafish imaged using an epi-fluorescence stereomicroscope at the lowest magnification (7.0×; E-H). The LED fluorescence macroscope is also capable of imaging engraftment of T-cell acute lymphoblastic leukemia in recipient animals. Whole-animal imaging of recipient fish transplanted with 1.5 × 104 Amcyan-, GFP-, or dsRED express–labeled leukemia cells at 30 days after transplantation (I-L). Animals from panels I through L were mixed to demonstrate that fluorescent-labeled animals can be easily delineated (M-P). Transplant recipients imaged by the use of an epi-fluorescence stereomicroscope at low magnification (7.0×, Q-T). Scale bars are 2 cm in panels A through D and I through L, and 5-mm in panels E through H and Q through T.
The LED fluorescence macroscope can image 5 fluorescent fluorophores in normal muscle and T-cell acute lymphoblastic leukemia (T-ALL). Whole animal imaging of wild-type (Negative), mylz2-Amcyan, mylz2-GFP, creatine kinase-zsYellow, mylz2-mCherry transgenic zebrafish (A-D). Amcyan was imaged by the use of a blacklight and 480/30 filter (A,J,N), GFP (blacklight, 520/30 filter; B,K,O), zsYellow (blue light, 575/52 filter; C), dsRED express (green light, 610/40 filter; L,P), and mCherry (green light, 610/40 filter; D). Transgenic zebrafish imaged using an epi-fluorescence stereomicroscope at the lowest magnification (7.0×; E-H). The LED fluorescence macroscope is also capable of imaging engraftment of T-cell acute lymphoblastic leukemia in recipient animals. Whole-animal imaging of recipient fish transplanted with 1.5 × 104 Amcyan-, GFP-, or dsRED express–labeled leukemia cells at 30 days after transplantation (I-L). Animals from panels I through L were mixed to demonstrate that fluorescent-labeled animals can be easily delineated (M-P). Transplant recipients imaged by the use of an epi-fluorescence stereomicroscope at low magnification (7.0×, Q-T). Scale bars are 2 cm in panels A through D and I through L, and 5-mm in panels E through H and Q through T.
The LED fluorescence macroscope can detect tumor engraftment of single T-ALL cells and is capable of multispectral, real-time imaging. Animals engrafted with a single dsRED-labeled T-ALL cell can be easily distinguished from nonengrafted animals at 45 days after transplantation (n = 1 of 10 fish, A; n = 2 of 20 fish, B; n = 3 of 29 fish panel C) by LED fluorescence macroscopy. Multispectral imaging with the LED fluorescence macroscope (D-I). Still image capture with dual-spectrum imaging of unanesthetized mylz2-Amcyan and mylz2-mCherry transgenic animals (D, blue and black light with 520/40 filter and 640/35 filter), mylz2-GFP and mylz2-mCherry (E, blue light with 520/40 filter and 640/35 filter), and creatine kinase-zsYellow and mylz2-mCherry (F, blue light with 575/52 filter and 640/35 filter). Animals were imaged from below at 30 frames per second, and a single frame is shown. Multispectral imaging also can be used to visualize fluorescent-labeled T-ALLs engrafted into fluorescent recipient animals. A mylz2-mCherry animal engrafted with an Amcyan-labeled T-ALL at 30 days after transplantation imaged by epi-fluorescence stereomicroscopy (G, 7.0× magnification) or by use of the LED fluorescence macroscope (blue light with 480/30 and 640/35 filters; H-I). Panel H shows a control animal (top) along with a dual transgenic leukemic fish (bottom). Three leukemic fish imaged from below at 30 frames per second and a single frame is shown (I). Scale bars are 1 cm in panels A through F and H through I; 2 mm in panel G.
The LED fluorescence macroscope can detect tumor engraftment of single T-ALL cells and is capable of multispectral, real-time imaging. Animals engrafted with a single dsRED-labeled T-ALL cell can be easily distinguished from nonengrafted animals at 45 days after transplantation (n = 1 of 10 fish, A; n = 2 of 20 fish, B; n = 3 of 29 fish panel C) by LED fluorescence macroscopy. Multispectral imaging with the LED fluorescence macroscope (D-I). Still image capture with dual-spectrum imaging of unanesthetized mylz2-Amcyan and mylz2-mCherry transgenic animals (D, blue and black light with 520/40 filter and 640/35 filter), mylz2-GFP and mylz2-mCherry (E, blue light with 520/40 filter and 640/35 filter), and creatine kinase-zsYellow and mylz2-mCherry (F, blue light with 575/52 filter and 640/35 filter). Animals were imaged from below at 30 frames per second, and a single frame is shown. Multispectral imaging also can be used to visualize fluorescent-labeled T-ALLs engrafted into fluorescent recipient animals. A mylz2-mCherry animal engrafted with an Amcyan-labeled T-ALL at 30 days after transplantation imaged by epi-fluorescence stereomicroscopy (G, 7.0× magnification) or by use of the LED fluorescence macroscope (blue light with 480/30 and 640/35 filters; H-I). Panel H shows a control animal (top) along with a dual transgenic leukemic fish (bottom). Three leukemic fish imaged from below at 30 frames per second and a single frame is shown (I). Scale bars are 1 cm in panels A through F and H through I; 2 mm in panel G.
To validate the sensitivity of the macroscope in detecting fluorescent transgenic zebrafish, 6 stable transgenic lines that express Amcyan, GFP, zsYellow, or mCherry were scored with the LED fluorescence macroscope (supplemental Table 7). A total of 140 of 141 fluorescent transgenic animals could be discriminated with the LED fluorescence macroscope (filter and light combinations shown in supplemental Table 8), whereas nontransgenic zebrafish also were negative by macroscope analysis (n = 53 of 53). Unanesthetized animals can be imaged in real-time by the use of either video or still image capture, further facilitating the rapid screening of transgenic zebrafish (supplemental Figure 6; supplemental Videos 1-4). Collectively, these results indicate that the LED fluorescence macroscope is exceedingly accurate at identifying stable transgenic lines, can image live fluorescent zebrafish in real-time without the need for anesthesia, and can effectively discriminate multiple fluorescent proteins.
To assess whether the LED fluorescence macroscope could be used to improve the detection of transplant engraftment, Myc-induced T-ALLs were generated that express Amcyan, GFP, zsYellow, or dsRED express and were transplanted into non-IR, syngeneic recipient animals (1.5 × 104 leukemia cells). LED fluorescence macroscope imaging reliably identified engrafted animals by 30 and 45 days after transplantation for all 4 fluorophores (n = 140 of 140; Figure 4I-P; supplemental Table 7) compared with conventional epi-fluorescence microscopic analysis (Figure 4Q-T; supplemental Figure 7). The LED fluorescence macroscope could also image fluorescent-labeled rhabdomyosarcomas that had been transplanted into CG1-strain recipients, suggesting that imaging methods are not limited to T-ALL (supplemental Figure 8). Finally, the LED fluorescence macroscope also identified animals that had engrafted T-ALL from single cells. A total of 105 animals transplanted with a single primary GFP-labeled T-ALL cell were scored by both conventional epi-fluorescence microscopy and macroscopic analysis. LED fluorescence macroscope imaging identified 2 engrafted animals that had developed prominent GFP-labeled T-ALL by 60 days after transplantation.
Similar results also were obtained with a dsRED-labeled T-ALL that had been serially passaged and contained large numbers of cells with tumor-initiating potential (Figure 5A-C). Specifically, 64 single-cell T-ALL transplants were completed, and subsequently, animals were analyzed for engraftment at 30 and 45 days after transplantation. All 9 fish that engrafted T-ALL could be scored by either an epi-fluorescence stereomicroscope or the LED fluorescence macroscope (Figure 5A-C). Together, the results indicate that it is now possible to perform high-throughput imaging of single-cell transplants, allowing the unprecedented study of self-renewal and clonal evolution within fluorescent leukemias. Moreover, a whole Petri dish of up to 30 animals can be scored simultaneously, compared with single inspection using an epi-fluorescence stereomicroscope.
The LED fluorescence microscope also allows for simultaneous imaging of multiple fluorescent proteins with the use of 2 cameras fitted with different filter sets. In total, 5 of 6 fluorescent combinations could be resolved by multispectral imaging (supplemental Table 9) and can easily delineate Amcyan from mCherry (Figure 5D), GFP from mCherry (Figure 5E), and zsYellow from mCherry (Figure 5F). Moreover, still images and videos of live fish could be captured in unanesthetized, swimming animals, indicating that multispectral analysis also can be completed in real time (Figure 5; supplemental Videos 5-7). Multispectral imaging also can be used to image T-ALL transplant recipient animals. In these experiments, 5 × 105 Amcyan-labeled T-ALL cells were introduced into twice outcrossed mylz2-mCherry transgenic zebrafish and imaged for engraftment at 30 days after transplantation with the use of an epi-fluorescence microscope (Figure 5G) or the LED fluorescence macroscope (still image in Figure 5H or single frame of supplemental Video 8 is shown in Figure 5I). Together, our work describes a novel methodology to simultaneously capture real-time, multichannel images and video of live dual fluorescent transgenic fish, a technique not available by traditional epi-fluorescence stereomicroscopy.
Discussion
Zebrafish have become a powerful tool for dissecting molecular pathways in cancer,25 but assessment of cancer cell self-renewal has been limited by lack of immune compromised zebrafish and genetically identical animals. For example, rag1-deficient zebrafish have been generated26 but have yet to be used in cell transplantation assays. In addition, immune compromised NOD/SCID zebrafish have not been reported in the literature. Instead, investigators have used sublethal γ-irradiation to ablate the immune system and subsequently introduce tumor cells into adult recipient animals.11,13-21 Pioneering work from Mizgireuv and Revskoy23 has shown that chemically induced liver cancers arising in clonally derived, isogenic zebrafish can be successfully transplanted into non-IR, sibling zebrafish. However, liver tumors were not assessed for tumor-initiating cell number, nor were fluorescent reporters used to visualize tumor formation and engraftment. Moreover, transplantation of tumors from mixed genetic backgrounds into clonal zebrafish was not fully described. No additional reports have used clonal fish lines to assess tumor engraftment or to identify self-renewing cell types in cancer.
The data presented here show that transplantation experiments that use irradiation to ablate the immune system will fail to accurately determine LIC number. Our previous work suggested that T-ALL–initiating cell number was approximately 1 in 1 × 103 to 1 in 2 × 104 cells.20 Similar leukemia-initiating frequency was reported recently for genetic models of zebrafish T-cell leukemia that used similar methodologies for introducing T-ALLs into nonimmune-matched, IR recipient animals.21 The detailed studies described here show that cell transplantation into nonimmune-matched gamma-IR animals severely underestimates true cancer-initiating cell number because of inefficient ablation of the immune system, subsequent recovery of immune responses by 20 days after irradiation, and death of animals as the result of gamma-irradiation directly. Transplantation experiments using clonal CG1-strain zebrafish reveal that LIC number is much greater than previously reported, with 0.1% to 15.9% of T-ALL cells capable of engraftment into syngeneic animals.
LIC number is quite high in primary zebrafish T-ALL, signifying that self-renewal may be a much more common attribute in malignant T-ALL cells than previously suggested.6-8,27 There are several possible reasons that could account for why LIC number differs so greatly between zebrafish and human T-ALL. Human T-ALL–initiating cell number has been assessed by the use of xenograft transplantation into partially immune compromised SCID or NOD/SCID recipient mice. Recent work from Quintana et al28 established that melanoma-initiating cell number is vastly underestimated when NOD/SCID recipient animals are used and reported a 100-fold increase in self-renewing cells after transplantation into NOD/SCID/IL2 receptor-gamma-deficient mice. Moreover, in the one report in which human ALLs were introduced into NOD/SCID/IL2 receptor-gamma-deficient animals, it is uncertain whether T- or B-ALLs were analyzed and which molecular subtype of leukemia was assessed.8 Our results in zebrafish raise the interesting possibility that human T-ALLs contain many more LICs than previously reported; however, additional experiments will be required to determine whether self-renewal potential differs across molecular subtypes and if common self-renewal mechanisms are used among all T-ALLs.
Having refined the methods for cell transplantation in zebrafish, there is a major need for high-throughput imaging modalities to identify animals engrafted with fluorescent-labeled tumors. The LED fluorescence macroscope is inexpensive, can score many fish at one time, and can detect multiple fluorescent channels simultaneously. The use of the LED fluorescence macroscope will not be limited to cancer biology. For example, the LED fluorescence macroscope will aid in the identification of fluorescent transgenic founder fish, can be used in conjunction with transplantation of quantum dots into the peritoneum or musculature to track individual fish within a tank, and can be used for real-time imaging of complex behaviors of adult fish. In our experiments, the LED fluorescence macroscope has sped the screening of stable transgenic zebrafish and transplant engraftment, paving the way for high-throughput cell transplantation experiments to assess the kinetics of tumor engraftment, cancer stem cell self-renewal, and clonal evolution.
The functional transplantation assays and new high-throughput detection methods outlined here afford unique opportunities to assess molecular pathways involved in leukemia self-renewal. For example, it is possible to deliver cancer-modifying transgenes to developing T-ALLs by merely coinjecting rag2-mMyc + rag2-dsREDexpress + rag2-gene of interest into one-cell–stage CG1 fish. Previous experiments suggest that cosegregation of 3 transgenes into microinjected zebrafish embryos results in the high frequency of coexpression within developing T-ALLs (n = 22 of 23).20 When coupled with FACS and limiting dilution cell transplantation, such approaches will allow direct assessment of transgene effects on self-renewal without the need for establishing stable transgenic animals or for complex breeding strategies to generate compound transgenic zebrafish.
These methods are not only amenable to Myc-induced T-ALL, but it will also be possible in any transgene-induced malignancy in zebrafish. It also is now feasible to introduce T-ALL into various genetic backgrounds to assess modifying gene effects on leukemia by breeding a mutant or transgenic line to CG1-strain animals and backcrossing these progeny to CG1 fish. Subsequent generations of backcrossed animals can be injected with cancer causing transgenes and functionally assessed for metastasis, recruitment of vasculature, or self-renewal by engraftment of tumors into CG1 recipient animals. Importantly, these processes can be visualized by transplanting tumors into syngeneic fluorescent reporter lines produced by multiple out-crossings to CG1-strain fish. Such experiments capitalize on functionally assessing immune competency without the need for complex immune matching. Although we have focused on using our high-throughput methods of cell transplantation and imaging to assess self-renewal in T-ALL, these methods will be broadly applicable to studies involving cell transplantation of hematopoietic stem cells, muscle stem cells, and organ-specific stem cells. Together our results highlight the emergence of the zebrafish as a new model of cancer cell self-renewal and provide new high-throughput methods for functionally assessing tumor-initiating cell number in cancer that do not exist in more established vertebrate models of disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Drs Keith Joung, David Louis, Kathleen Pfaff, and Xiuning Le for critical review of the manuscript.
D.M.L. is supported by National Institutes of Health grant K01 AR055619-01A1 and 3 K01 AR055619-03S1, a grant from the Sarcoma Foundation of America, a New Investigator Award from Alex Lemonade Stand, and a pilot project grant from the Harvard Stem Cell Institute. L.I.Z. is supported by National Institutes of Health 5 R01 CA103846-04.
National Institutes of Health
Authorship
Contribution: A.C.H.S., A.R.R., and D.M.L. designed experiments; A.C.H.S. A.R.R., C.D.S., M.S.I., J.S.B., N.Y.S, J.L.O.d.J., A.T.C., D.M.L., and Y.Z. performed experiments; I.V.M. and S.R. produced the CG1 strain zebrafish; A.C.H.S. and A.R.R. wrote portions of the manuscript; D.M.L. wrote the manuscript; and C.D.S., M.S.I., J.S.B., N.Y.S., J.L.O.d.J., A.T.C., Y.Z., I.V.M., S.R., and L.I.Z. edited the manuscript.
Conflict-of-interest disclosure: L.I.Z. is a founder and stockholder of Fate Inc and a scientific advisor for Stemgent. The remaining authors declare no competing financial interests.
Correspondence: David M. Langenau, PhD, Department of Pathology, Molecular Pathology Unit, Massachusetts General Hospital, 149 13th St, 6th Fl #6012, Charlestown, MA 02129; e-mail: dlangenau@partners.org.
References
Author notes
A.C.H.S. and A.R.R. contributed equally to this article.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal