Abstract
Terminally mature megakaryocytes undergo dramatic cellular reorganization to produce hundreds of virtually identical platelets. A hallmark feature of this process is the generation of an elaborate system of branched protrusions called proplatelets. We recently identified RanBP10 as a tubulin-binding protein that is concentrated along polymerized microtubules in mature megakaryocytes. RanBP10 depletion in vitro caused the disturbance of polymerized filaments. Here we study the function of RanBP10 in vivo by generating deficient mice using a gene-trap approach. Mutant mice show normal platelet counts, and fetal liver-derived megakaryocytes reveal only slightly reduced proplatelet formation. However, ultrastructural analysis unveiled a significantly increased geometric axis ratio for resting platelets, and many platelets exhibited disorders in microtubule filament numbers and localization. Mutant mice showed a markedly prolonged bleeding time. Granule release, a process that depends on internal contraction of the microtubule marginal coil, also was reduced. Flow cytometry analysis revealed reduced expression of CD62P and CD63 after PAR4-peptide stimulation. These data suggest that RanBP10 plays an essential role in hemostasis and in maintaining microtubule dynamics with respect to both platelet shape and function.
Introduction
Platelets develop from megakaryocytes (MKs), large polyploid cells localized in the bone marrow. Mature MKs rearrange their entire cytoplasm into pseudopod protrusions (designated proplatelets), a process that is driven by and dependent on microtubules (MTs).1,2 MT filaments emanate from the cell cortex into the proplatelet tips, where they coil into a loop and later build the platelet marginal band. In wild-type platelets, this MT loop is normally coiled 8 to 12 times.3 Ultrastructural data previously suggested that the platelet marginal bundle contains a single coiled filament,3-6 and isolation of MT coils by platelet extraction affirmed this idea. However, the study by Patel-Hett et al7 on living platelets revealed up to 7 free plus-ends, suggesting that several shorter MT filaments associate dynamically with one longer filament, forming a bipolar array.
Each MT filament is composed of 13 protofilament heteropolymers of α- and β-tubulins. Mammalian genomes encode 6 α-tubulin and 5 β-tubulin genes. β1-tubulin (also referred to as class VI) is the most diverse isotype8 and is found exclusively in cells that harbor a marginal band: MKs, platelets, and erythroblasts.9 More than 90% of proplatelet MT filaments are composed of this β-tubulin isotype,10 whose expression depends on the MK-specific transcription factor NF-E2. Genetically engineered mice that lack a functional Tubb1 gene are thrombocytopenic, and Tubb1−/− platelets lack lentiform shape; the marginal band consists of only 2 to 3 coils, resulting in platelet spherocytosis.11,12 Although ubiquitously expressed tubulin isotypes β2 and β5 are up-regulated, the equilibrium is shifted toward monomeric tubulin. Together, these data support the notion that the marginal band is essential for maintaining platelet discoid shape.13 Platelets in β1-tubulin–nullizygous mice also show reduced reactivity to thrombin, as judged by expression of the activation marker CD62P.11 Probably, the reduced filament coiling in the platelet periphery leads to a defect in coil contraction preceding granule concentration and release.
A dynein-dynactin complex recently was implicated in MT sliding during proplatelet elongation,14 but the forces that regulate MT coiling at the proplatelet tip, influence platelet shedding, or drive the contractile process during platelet activation remain poorly defined. Because of the strong effect of β1-tubulin ablation on platelet morphology and structure,11,12 we searched for novel MT-associated proteins with a yeast 2-hybrid screen by using the C-terminal domain of β1-tubulin as the bait. This domain is the most divergent part of the protein and is known to interact with MT-associated proteins. We were able to show that secretory leukocyte protease inhibitor interacts with β1-tubulin in platelets.15 Another β1-tubulin binding factor is Ran binding protein 10 (RanBP10), a novel protein that also binds Ran. RanBP10 localizes in the cytoplasm of mature MKs and platelets, where it concentrates on polymerized noncentrosomal MTs.16 In addition, RanBP10 harbors guanine nucleotide exchange factor (GEF) activity toward Ran. For the nucleation of dynamic MT filaments in the mitotic spindle, the chromatin-associated GEF RCC1 loads Ran with guanosine triphosphate (GTP), leading to activation of Tpx2, a factor that delivers positional information for MT nucleation.17-20 During interphase, a classical MT-organizing center, composed of 2 centrioles and the γ-tubulin ring complex (γTuRC), is responsible for MT assembly. In mature proplatelet-forming MKs, however, it is difficult to detect centrioles or significant amounts of γ-tubulin,16,21,22 suggesting that other mechanisms are responsible for organization and nucleation of MTs.
Depletion of RanBP10 protein in MKs leads to the disruption of MT filaments.16 This result, together with its high GEF activity for Ran, indicates a potential role for RanBP10 as a regulator of MT organization and stabilization in mature MKs and prompted us to examine RanBP10-null platelets in a mouse model. Here, we present evidence that RanBP10, despite being dispensable for platelet biogenesis in vivo, is essential for maintaining platelet isocytosis and function. Ultrastructural analysis of mutant platelets by electron microscopy and deconvolution immunofluorescence microscopy revealed a structural defect in the platelet marginal band, resulting in a more spherocytic shape. Furthermore, RanBP10-knockout mice exhibit a markedly prolonged bleeding time and a defect in granule secretion. These results show that RanBP10 is essential for structural organization of the platelet marginal band and in hemostasis.
Methods
Mouse husbandry, MK culture, and 5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside (X-Gal) staining
RanBP10-knockout mice were generated by a gene trap approach by use of the embryonic stem cell clone RRZ226 provided from BayGenomics. After germline transmission heterozygous mice were back-bred to either C57/BL6 or 129/Sv strains, respectively. Animal care and all experiments were performed according to institutional guidelines with the approval of the ethical review board at Charité-Universitätsmedizin. Fetal liver–derived MKs were isolated and cultured as described.10 To determine genotypes by expression of the transgenic fusion protein, fetuses were stained with X-Gal and were incubated for 30 minutes in fixation buffer (100mM phosphate buffer containing 5mM EGTA (ethyleneglycoltetraacetic acid), 0.2% glutaraldehyde, and 2mM MgCl2) and washed in 100mM phosphate buffer (pH 7.3) and 2mM MgCl2. Staining was performed for 6 hours or overnight at 37°C in staining buffer (5mM K4[Fe(CN)6]–3H2O, 5mM K3[Fe(CN)6]) containing 1 mg/mL X-Gal.
Reverse transcription–polymerase chain reaction, polymerase chain reaction, cloning, and sequencing
Total RNA from bone marrow or cultured MKs was isolated by the use of Trizol (Invitrogen) according to the manufacturer's instructions. mRNA was reverse-transcribed with oligo-(dT) primers and amplified by the use of a multiplex polymerase chain reaction (PCR): forward primer 5′GCTTGTACCCTGCGGTTAATC, reverse primer (RanBP10) 5′AAGCGCTGGGTTGTCTCAATG, and reverse primer (β-geo cassette) 5′ATTCAGGCTGCGCAACTGTTGGG.
Genomic DNA was isolated from tail or ear biopsies according to standard protocols. The supernatant is used for allele-specific PCR with 2 primer sets: wild-type forward 5′-CAGAGCTATCACATTGATAGAAC, wild-type reverse 5′-TAGTCCCAGCTGGCCTAAAC, mutant forward 5′-TTATCGATGAGCGTGGTGGTTATG, mutant reverse 5′-GCGCGTACATCGGGCAAATAATATC. Primers used for breakpoint analysis PCR were as follows: forward (common) 5′GACAAGAATTTCTCTGTGTAG, reverse (wild-type) 5′-TAGTCCCAGCTGGCCTAAAC, and reverse (mutant) 5′-GTGTGGGAAAGCCTTCAAAG before cloning into the TOPO-TA-Cloning-Kit (Invitrogen).
Antiserum, platelet activation, and immunoblot analysis
Antiserum directed against mouse RanBP10 protein was recently described16 and additionally affinity purified by the use of peptide-coupled NHS-columns (GE Healthcare). A total of 10 to 30 μg of protein lysates was applied per lane before standard sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was conducted and immunoblotted onto polyvinylidenefluoride membranes, incubated with primary antibodies as indicated, and detected by the use of horseradish peroxidase–conjugated secondary antibodies (Santa Cruz Biotechnology) before standard enhanced chemoluminescence detection (GE Healthcare). For activation studies, platelets were washed and resuspended in HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid)-Tyrode buffer containing CaCl2 (1mM). Thrombin or PAR-4 peptide was added with the indicated concentrations and stimulation stopped by centrifugation. One-third volume of 4× sample buffer was added to the supernatant while the pellet was dissolved in one volume of 1× sample buffer and both fractions subjected to SDS-PAGE and blotted against platelet factor 4 (Peprotech). For granule protein studies, membranes were additionally probed against von Willebrand factor (clone H-300; Santa Cruz Biotechnology) and P-selectin (clone CTB; Santa Cruz Biotechnology).
Proplatelet and platelet studies
The percentage of proplatelet-forming MKs was determined on day 5 of culture.23 Peripheral blood was withdrawn retroorbitally into EDTA (ethylenediaminetetraacetic acid)–containing tubes, and cell counts were analyzed with mouse blood settings (Synlab). Platelet-rich plasma (PRP) was isolated from citrated blood after differential centrifugation. Platelet ultrastructure was analyzed by the use of transmission electron microscopy as described.24 In brief, blood was directly withdrawn into one volume of fixative (4% glutaraldehyde in 100mM cacodylate buffer) by heart puncture. Platelets were enriched through centrifugation at 150g for 3 minutes, pelleted at 230g for 6 minutes, and resuspended in 2% glutaraldehyde in 0.1M cacodylate buffer. Postfixation was performed with 2% OsO4 (ChemPur) for 2 hours. Dehydration was achieved by an ascending series of ethanol wash steps (50% to 100%). Ultrathin sections (70 nm) of epoxy-embedded resin were cut with an Ultracut S (Leica) and stained with uranyl acetate and lead citrate. Examination was conducted by the use of an EM 906 transmission electron microscope (Zeiss).
Flow cytometry
A total of 100 μL of fetal liver-derived cultures was stained on day 5 of culture with phycoerythrin-conjugated antibody against mouse CD61 (BD Biosciences) for 20 minutes. Incubation was stopped by adding 1 mL of 0.5% bovine serum albumin in phosphate-buffered saline and analyzed by flow cytometry (FACSCan, Becton Dickinson). For platelet activation studies, PRP was stimulated with indicated concentrations of protease-activated receptor 4 peptide (PAR4p, Bachem) or collagen (Probes & go; Labordiagnostica). After 5 minutes, FITC-conjugated anti-CD62P antibody (BD Biosciences) or unconjugated anti-CD63 (MoBiTec, MBL) followed by a secondary Alexa 488–conjugated donkey anti–rat IgG (Invitrogen) was added, and staining continued as indicated previously.
Immunofluorescence microscopy
Immunostaining was performed as described.15 Cultured MKs and platelets were cytocentrifuged onto poly-L-lysine–coated coverslips (Becton Dickinson), fixed in 4% formaldehyde, permeabilized with 0.5% Triton X-100, and blocked with 3% goat serum. Primary antibodies as indicated were incubated for 30 minutes at room temperature. Cells were washed thoroughly with phosphate-buffered saline and fluorophore-conjugated secondary antibodies added for 30 minutes. Cells were washed 3 times and nuclei counterstained by the use of Hoechst 33 258 or DAPI. Coverslips were mounted with Fluoromount-G (SouthernBiotech) and examined on an Olympus IX71 inverted fluorescence microscope by the use of 60× or 100× oil objectives. Images were acquired with a Photometrics CoolSNAP HQ ICX285 camera. Forty cross-sections were taken at 0.2-μm spacing, and images were deconvolved by the use of DeltaVision software (Applied Precision). Platelet images were acquired by an A1 laser scanning microscope (Eclipse) by the use of NIS-Elements software (Nikon).
Tail bleeding time
Six- to 12-week-old sex- and age-matched mice were anesthetized, and 5 mm of the tail tip was cut. Bleeding was monitored into 37°C warm saline with an end point set to 360 seconds. In case of continuous bleeding, the vessel was occluded by mechanical force to exclude exsanguination. Bleeding time was then indicated as prolonged.11
Aggregometry
PRP and washed platelets were prepared as recently described.12 Platelet counts were adjusted to a final concentration of 200 nL, stimulated with agonists as indicated, and light transmission was recorded over time in a standard aggregometer (möLab).
Results
RanBP10 overexpression in MKs increases MT numbers and thickness and displaces Ran
Elaboration and growth of MT filaments in mature MKs are preconditions for proplatelet formation.2 We recently analyzed the consequences of RNAi-mediated RanBP10 depletion and demonstrated that MT filaments were shortened or interrupted.16 Because of the obvious influence of RanBP10 on MT filaments, we examined the effects of overexpressing EGFP-tagged RanBP10 by retroviral infection of cultured MKs.15 Infected cells expressed EGFP (Figure 1A), and individual cells showing fluorescence markedly over the background were analyzed in detail. MKs with excess RanBP10 commonly displayed an abundance of exceptionally thick MTs that either coiled up to fill the cytoplasm or extended a significant distance from the cell. Figure 1B through D (representative examples) shows single z-sections of MKs after deconvolution, in which proplatelets (arrows in Figure 1B) and abnormal MTs induced by excess RanBP10 can be readily distinguished. We observed atypical bundled fibers in 37% of RanBP10-overexpressing cells and did not see similar MT morphologies in normal MKs after many other manipulations. Superficially, these structures resemble the cortical MTs that accumulate before proplatelet morphogenesis (Figure 1C) and probably represent thick bundles of parallel fibers. The need to assess MTs in isolated cells precludes making a biochemical distinction between the potential targets of Ran–GTP regulation: MT nucleation, dynamics, or stabilization.25,26
RanBP10 overexpression in MKs increases MT numbers and thickness and displaces Ran. (A-D) MKs that overexpress RanBP10 show many partially coiled internal MT fibers that are substantially thicker and longer than those in control MKs. MKs infected with GFP-RanBP10 retrovirus were recognized by virtue of green fluorescence (A) and their MT structure examined by β-tubulin IF (B-D; single z-sections are displayed after deconvolution). Panels A and B show the same cell, and nuclei are seen by DAPI staining (B,D). Arrows in panel B point to a bona fide proplatelet, whereas the rest of the cell is filled with thicker MT bundles. (E) Indirect Ran IF (red), coupled with DAPI nuclear stain (blue), contrasts the nuclear confinement of Ran in control EGFP virus-infected MKs (top) with extra-nuclear Ran accumulation in cells that overexpress RanBP10 (bottom). (F,G) The merged fluorescence images were rotated 90° along the axis shown in panel E to visualize x/z planes after 3-dimensional reconstruction. Ran and nuclear material overlap in distribution in control cells (F), whereas in RanBP10-overexpressing MKs, a substantial fraction of Ran is present outside the nucleus. Cross-sectional intensity profiles are displayed immediately below, with pixel position on the abscissa and fluorescence intensity on the ordinate (scale for DAPI signal on the left and for Texas Red on the right); blue and red arrows mark boundaries of the respective fluorescent signals. Scale bars: B-D, 15 μm; E, 10 μm.
RanBP10 overexpression in MKs increases MT numbers and thickness and displaces Ran. (A-D) MKs that overexpress RanBP10 show many partially coiled internal MT fibers that are substantially thicker and longer than those in control MKs. MKs infected with GFP-RanBP10 retrovirus were recognized by virtue of green fluorescence (A) and their MT structure examined by β-tubulin IF (B-D; single z-sections are displayed after deconvolution). Panels A and B show the same cell, and nuclei are seen by DAPI staining (B,D). Arrows in panel B point to a bona fide proplatelet, whereas the rest of the cell is filled with thicker MT bundles. (E) Indirect Ran IF (red), coupled with DAPI nuclear stain (blue), contrasts the nuclear confinement of Ran in control EGFP virus-infected MKs (top) with extra-nuclear Ran accumulation in cells that overexpress RanBP10 (bottom). (F,G) The merged fluorescence images were rotated 90° along the axis shown in panel E to visualize x/z planes after 3-dimensional reconstruction. Ran and nuclear material overlap in distribution in control cells (F), whereas in RanBP10-overexpressing MKs, a substantial fraction of Ran is present outside the nucleus. Cross-sectional intensity profiles are displayed immediately below, with pixel position on the abscissa and fluorescence intensity on the ordinate (scale for DAPI signal on the left and for Texas Red on the right); blue and red arrows mark boundaries of the respective fluorescent signals. Scale bars: B-D, 15 μm; E, 10 μm.
The increased cytoplasmic Ran–GEF activity imparted by excess RanBP10 may be expected to counteract, to some degree, the endogenous Ran-GAP and to influence the Ran–guanosine diphosphate (GDP)/Ran–GTP equilibrium in the cytosol. Because only Ran-GDP can enter the cell nucleus, excess Ran-GTP should accumulate outside,27,28 and experimental evidence indicates that this collection is usually perinuclear.28 In control EGFP virus-infected MKs, Ran was uniformly distributed in the nucleus and colocalized with DAPI chromatin stain (Figure 1E). In contrast, and in agreement with the previous prediction, RanBP10-overexpressing MKs consistently displayed extranuclear Ran accumulation (Figure 1E). To further examine this altered distribution, we obtained perpendicular reconstructions of IF images along the axes shown in Figure 1E. Unlike the overlap between DAPI and Ran signals in control infected cells (Figure 1F), much of the Ran appeared outside the nucleus in cells overexpressing RanBP10 (Figure 1G). Quantitative profiles of the signal intensities confirmed these observations. Thus, ectopic expression of RanBP10 in MKs results in substantial disturbance of MT dynamics.
In vivo disruption of RanBP10 by gene trap
Efficient knockdown of RanBP10 protein in primary MKs is technically difficult; low transduction rates and sustained protein stability result in the need to analyze single infected cells. We sought to study the consequences of complete RanBP10 ablation in vivo and took advantage of a gene-trap approach to generate RanBP10-deficient mice by using the embryonic stem cell clone RRZ 226. The gene trap lies within intron 2 of the RanBP10 gene, introducing a β-geo exon with a strong splice acceptor site. On the affected allele, mRNA is transcribed under the endogenous RanBP10 promoter, but exon 2 splices into the gene trap exon, which is followed by a strong termination signal (Figure 2A). We intercrossed RanBP10 heterozygous mice to generate animals of all genotypes, isolated fetuses, and stained with X-Gal to visualize reporter gene activity (Figure 2B). Blue color was absent in wild-type controls (left panel) but clearly detectable in mice harboring the transgene, with increased intensity in nullizygous mice (middle and right panels). Nonuniform staining confirmed the restricted RanBP10 spatiotemporal expression observed by Northern blot.16 Nullizygous mice showed no RanBP10 protein (Figure 2C) and no RanBP10 mRNA (Figure 2D). To establish a genotyping strategy to distinguish between heterozygous and nullizygous mice, we mapped the 20-kb gene trap insertion site with a series of overlapping PCRs and identified a 4-kb fragment (between kb 9 and 13) that is absent in intron 2 of the transgene (Figure 2E). Cloning and sequencing of the breakpoint at the 5′ insertion site (Figure 2F) enabled us to genotype mice by using primers that span the breakpoint and exclusively amplify wild-type or mutant alleles (Figure 2G). In summary, we generated mice lacking functional RanBP10. These mice were viable and showed no conspicuous phenotype, hence permitting analysis of the consequences of RanBP10 loss on MKs and platelet biogenesis.
Generation of RanBP10-deficient mice by a gene trap approach. RanBP10 transgenic mice were generated by a gene trap approach. (A) The gene trap inserted into the 20-kb intron 2 of RanBP10 (wt, disrupted endogenous RanBP10 expression and uses a splice acceptor to encode for a fusion transcript with β-galactosidase activity (mut). (B) Expression of the fusion protein was determined by X-gal staining of fetuses on gestation day 13.5 to 15.5. Wild-type fetuses (+/+) show essentially no staining compared with heterozygous or homozygous fetuses (+/−, −/−). (C) Protein lysates of RanBP10−/− platelets showed no detectable RanBP10 protein compared with wild-type (+/+) controls by immunoblot analysis. (D) RNA from fetal liver-derived MKs was reverse transcribed before PCR by the use of allele-specific primers for wild-type (wt) and mutant (mut) transcripts in a multiplex reaction. Wild-type (+/+), heterozygous (+/−), and homozygous (−/−) mice could be genotyped unambiguously. (E) The gene trap insertion site on intron 2 was mapped in overlapping 1-kb steps. The comparison of RanBP10+/− (+/−) and RanBP10−/− (−/−) displays loss of a 4-kb fragment between kb 9 and 13. (F) Amplified PCR products were cloned into pCRII-TOPO vector and sequenced. Identical nucleotides are depicted in black. The sequence of the wild-type allele (blue) differs from the gene-trap derived allele (red). (G) On the basis of the sequence of the cloned breakpoint, genomic PCR was performed by the use of allele-specific primers. This enabled identification of RanBP10 wild-type (+/+), heterozygous (+/−), and knockout (−/−) mice by genomic DNA.
Generation of RanBP10-deficient mice by a gene trap approach. RanBP10 transgenic mice were generated by a gene trap approach. (A) The gene trap inserted into the 20-kb intron 2 of RanBP10 (wt, disrupted endogenous RanBP10 expression and uses a splice acceptor to encode for a fusion transcript with β-galactosidase activity (mut). (B) Expression of the fusion protein was determined by X-gal staining of fetuses on gestation day 13.5 to 15.5. Wild-type fetuses (+/+) show essentially no staining compared with heterozygous or homozygous fetuses (+/−, −/−). (C) Protein lysates of RanBP10−/− platelets showed no detectable RanBP10 protein compared with wild-type (+/+) controls by immunoblot analysis. (D) RNA from fetal liver-derived MKs was reverse transcribed before PCR by the use of allele-specific primers for wild-type (wt) and mutant (mut) transcripts in a multiplex reaction. Wild-type (+/+), heterozygous (+/−), and homozygous (−/−) mice could be genotyped unambiguously. (E) The gene trap insertion site on intron 2 was mapped in overlapping 1-kb steps. The comparison of RanBP10+/− (+/−) and RanBP10−/− (−/−) displays loss of a 4-kb fragment between kb 9 and 13. (F) Amplified PCR products were cloned into pCRII-TOPO vector and sequenced. Identical nucleotides are depicted in black. The sequence of the wild-type allele (blue) differs from the gene-trap derived allele (red). (G) On the basis of the sequence of the cloned breakpoint, genomic PCR was performed by the use of allele-specific primers. This enabled identification of RanBP10 wild-type (+/+), heterozygous (+/−), and knockout (−/−) mice by genomic DNA.
MT filaments in MKs from RanBP10-null mice
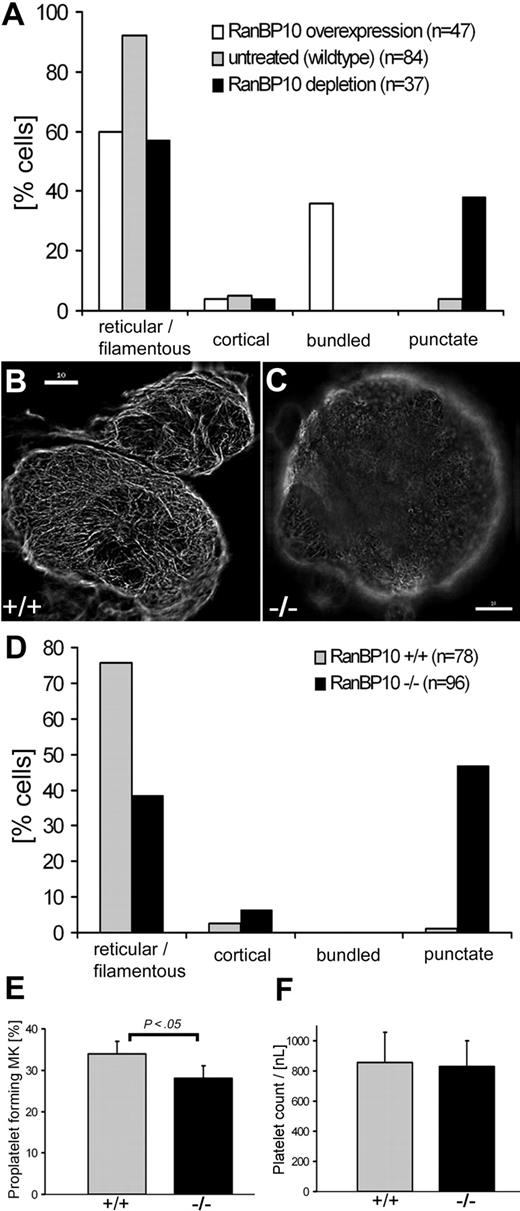
We first compared tubulin expression in mature MKs forced to overexpress EGFP–RanBP10 and those depleted for RanBP10. Whereas MKs overexpressing RanBP10 showed increased MT filament bundling, shRNA-mediated depletion of endogenous protein resulted in many short MT filaments or an absence of tubulin staining, a phenotype not detected with any other manipulation. Cortical tubulin staining is only found in cells undergoing proplatelet formation, and quantitation of the staining pattern confirmed that increasing or reducing RanBP10 expression in primary MKs affects MT filaments in opposing directions (see Figure 3A).
Punctuated MT filaments in RanBP10−/− MKs. For the quantitative evaluation of MK immunostained with an antibody against β-tubulin, 4 different MT network classifications were determined: reticular, cortical, bundled, and punctuated. The percentage of each subtype was determined individually. (A) The comparison of RanBP10-depleted, RanBP-overexpressing, and wild-type MKs indicated diverging effects on MT bundling and filament collapse. (B-C) Representative examples of immunostained MKs with reticular filamentous MT (B) and punctuated MT (C); scale bar 10 μm. (D) RanBP10-null MKs show a lack in filamentous MTs compared with wild-type cells but an increase in punctuated MT. (E) MK cultures were examined for proplatelet formation by light microscopy on day 5 of cultivation with TPO. RanBP10-null MKs have a slight decrease in proplatelet formation that is statistically significant (P < .05). Error bars represent SD. (F) Peripheral platelet counts did not differ between wild-type and mutant animals.
Punctuated MT filaments in RanBP10−/− MKs. For the quantitative evaluation of MK immunostained with an antibody against β-tubulin, 4 different MT network classifications were determined: reticular, cortical, bundled, and punctuated. The percentage of each subtype was determined individually. (A) The comparison of RanBP10-depleted, RanBP-overexpressing, and wild-type MKs indicated diverging effects on MT bundling and filament collapse. (B-C) Representative examples of immunostained MKs with reticular filamentous MT (B) and punctuated MT (C); scale bar 10 μm. (D) RanBP10-null MKs show a lack in filamentous MTs compared with wild-type cells but an increase in punctuated MT. (E) MK cultures were examined for proplatelet formation by light microscopy on day 5 of cultivation with TPO. RanBP10-null MKs have a slight decrease in proplatelet formation that is statistically significant (P < .05). Error bars represent SD. (F) Peripheral platelet counts did not differ between wild-type and mutant animals.
We next asked whether the tubulin staining pattern in RanBP10-null MKs would resemble our in vitro results and examined MT staining patterns in MKs from wild-type and knockout animals. Most MKs derived from control animals revealed the expected reticular staining pattern of MT filaments (Figure 3B), whereas only half the mutant MKs showed such staining. In 46% of the RanBP10-null cells, MT filaments were shortened and showed discontinuous MT filaments indicated by a punctate MT network (Figure 3C). Thick bundling of MTs, as observed in RanBP10-overexpressing MKs, was never seen in MKs of wild-type or RanBP10-null animals. Quantitation of the tubulin staining (Figure 3D) indicated that RanBP10-deficient mice phenocopy the reduced tubulin staining found after RNA interference and confirmed that RanBP10 plays an important role in MT filament function in MKs.
RanBP10 ablation leads to reduced proplatelet-forming MKs
Proplatelet production is impaired in β1-tubulin–deficient mice.11 Because RanBP10 was originally identified as a β1-tubulin binding protein,16 we checked for a similar influence of RanBP10. Fetal liver cells were cultivated in the presence of thrombopoietin and analyzed for occurrence and frequency of CD61+ MKs. Wild-type cultures yielded 47% CD61+ cells compared with 51% in RanBP10-knockout cultures, indicating that RanBP10 has no major influence on megakaryopoiesis per se. Cell death and proliferation rates were comparable, excluding the possibility of an indirect effect of RanBP10 on the fraction of CD61+ MKs (data not shown). Analysis of proplatelet production in culture is a bona fide technique for examining platelet biogenesis, as shown for NF-E2, GATA1 β1-tubulin, and other mouse models.11,29,30 We followed proplatelet formation by phase-contrast light microscopy. In wild-type cultures, 34% of MKs elaborated proplatelets on day 4 to 5 of culture in accordance with published data. In RanBP10-null cultures the fraction of proplatelet forming MKs was consistently less than 30% (Figure 3E). This statistically significant reduction did not, however, influence peripheral platelet counts (832 nL in mutant animals compared with 857 nL in littermate controls; see Figure 3F and Table 1); the slight reduction in proplatelet formation in vitro might be compensated in vivo. Values for hemoglobin, hematocrit, and leukocyte counts were virtually identical in the 2 groups (Table 1), but erythrocyte counts were reduced by 6% in RanBP10−/− mice and mean red cell volume was increased by 7% (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). In summary, RanBP10 is dispensable for MK formation and platelet biogenesis but affects red blood cell size and number slightly.
Blood cell counts of RanBP10-deficient mice
| . | +/+ (n = 13) . | +/− (n = 16) . | −/− (n = 17) . | P . |
|---|---|---|---|---|
| Platelets, nL | 857 ± 202 | 789 ± 217 | 832 ± 169 | — |
| Leukocytes, nL | 6.71 ± 2.53 | 8.32 ± 3.95 | 6.51 ± 2.20 | — |
| Erythrocytes, pL | 9.48 ± 0.47 | 9.58 ± 0.62 | 8.96 ± 0.35 | < .05 |
| MCV, fL | 83 ± 2.52 | 83 ± 4.08 | 89 ± 2.62 | < .05 |
| MCH, fmol/Ery | 1.01 ± 0.02 | 1.01 ± 0.05 | 1.07 ± 0.04 | < .05 |
| Hk, % | 0.79 ± 0.04 | 0.80 ± 0.05 | 0.80 ± 0.03 | — |
| Hb, mM | 9.54 ± 0.56 | 9.68 ± 0.54 | 9.54 ± 0.46 | — |
| . | +/+ (n = 13) . | +/− (n = 16) . | −/− (n = 17) . | P . |
|---|---|---|---|---|
| Platelets, nL | 857 ± 202 | 789 ± 217 | 832 ± 169 | — |
| Leukocytes, nL | 6.71 ± 2.53 | 8.32 ± 3.95 | 6.51 ± 2.20 | — |
| Erythrocytes, pL | 9.48 ± 0.47 | 9.58 ± 0.62 | 8.96 ± 0.35 | < .05 |
| MCV, fL | 83 ± 2.52 | 83 ± 4.08 | 89 ± 2.62 | < .05 |
| MCH, fmol/Ery | 1.01 ± 0.02 | 1.01 ± 0.05 | 1.07 ± 0.04 | < .05 |
| Hk, % | 0.79 ± 0.04 | 0.80 ± 0.05 | 0.80 ± 0.03 | — |
| Hb, mM | 9.54 ± 0.56 | 9.68 ± 0.54 | 9.54 ± 0.46 | — |
Ery indicates erythrocyte; Hb, hemoglobin; Hk, hematocrit; MCH, mean cellular hemoglobin; MCV, mean cellular volume; RanBP10, Ran binding protein 10; and —, not significant.
Loss of RanBP10 results in a disordered platelet marginal band
Although RanBP10 seems dispensable for megakaryopoiesis and platelet biogenesis in vivo, MT filaments play additional roles in maintaining platelet shape and function, as shown in mice lacking β1-tubulin.11 To determine whether platelet shape is affected in RanBP10-null mice, we studied their ultrastructure by using immunofluorescence and transmission electron microscopy. Wild-type platelets displayed the typical discoid appearance in electron micrographs, with abundant granules present in the center and MT coils transversely cut at both platelet poles (Figure 4A). Superficially, platelets lacking RanBP10 revealed a similar appearance, although they appeared less extended (Figure 4B). We quantified this observation and calculated a geometric axis ratio (width/length), based on a measure for platelet discoid shape,31 and found a reduction of 0.25 in wild-type compared with 0.33 in mutant platelets (Figure 4C). This difference was statistically significant (P < .05), confirming that mutant platelets have a more spherical shape. There were no obvious differences in the appearance of RanBP10-null platelets with respect to distribution, size, shape, or number of granules.
Loss of RanBP10 results in a disordered platelet marginal band. Platelets were obtained from adult mice by retro-orbital blood withdrawal, fixed, and subsequently analyzed by transmission electron microscopy. (A) The overview of wild-type platelets showed the typical discoid shape of platelets with normal granular content. (B) RanBP10−/− platelets exhibit no differences in granular content but a less discoid shape. (C) The change of the platelet shape was quantified by determination of the geometric axis ratio, which shows a statistical significant increase (P < .05; t test) between wild-type (n = 21) and RanBP10-null mice (n = 25). (D) Wild-type platelets possess a normal amount8-12 of MT coils (MT, black arrows) in the periphery. (E) In contrast some RanBP10-deficient platelets exhibit a greater amount (here up to 25) of filaments per coil coils (MT, black arrows). (F) Other platelets showed a normal number of MT filaments per coil, but an atypical appearance in profile (black arrow) and axis (black arrowhead) in one cross-section. (G) RanBP10-knockout platelets also showed additional MT coils (MT, black arrow). Immunofluorescence analysis was consistent with electron microscopy regarding the marginal band. (D) Inset shows wild-type platelets immunostained with an antibody against β-tubulin, with typical MT coils, representing the marginal band. (E) Inset shows the disordered marginal band of RanBP10−/− platelets by immunofluorescence microscopy. Despite having a normal marginal band (not shown), the platelets exhibit multiple starting points of MT coils (white asterisk) and structures that look like halved MT coils (white arrows). (H) Quantification of filaments per MT coil in wild-type (n = 17) and RanBP10 knockout platelets (n = 15) shows a small increase of MT coils for the RanBP10-deficient platelet, but the high standard deviation indicates that the number of MT coils ranges from well above to well below normal. Original magnification, ×10 000 (A-B); ×∼ 35 000 (D-G). Scale bars (insets D-E) indicate 2 μm. The resolved images and quantifications are representatives of 2 independent experiments.
Loss of RanBP10 results in a disordered platelet marginal band. Platelets were obtained from adult mice by retro-orbital blood withdrawal, fixed, and subsequently analyzed by transmission electron microscopy. (A) The overview of wild-type platelets showed the typical discoid shape of platelets with normal granular content. (B) RanBP10−/− platelets exhibit no differences in granular content but a less discoid shape. (C) The change of the platelet shape was quantified by determination of the geometric axis ratio, which shows a statistical significant increase (P < .05; t test) between wild-type (n = 21) and RanBP10-null mice (n = 25). (D) Wild-type platelets possess a normal amount8-12 of MT coils (MT, black arrows) in the periphery. (E) In contrast some RanBP10-deficient platelets exhibit a greater amount (here up to 25) of filaments per coil coils (MT, black arrows). (F) Other platelets showed a normal number of MT filaments per coil, but an atypical appearance in profile (black arrow) and axis (black arrowhead) in one cross-section. (G) RanBP10-knockout platelets also showed additional MT coils (MT, black arrow). Immunofluorescence analysis was consistent with electron microscopy regarding the marginal band. (D) Inset shows wild-type platelets immunostained with an antibody against β-tubulin, with typical MT coils, representing the marginal band. (E) Inset shows the disordered marginal band of RanBP10−/− platelets by immunofluorescence microscopy. Despite having a normal marginal band (not shown), the platelets exhibit multiple starting points of MT coils (white asterisk) and structures that look like halved MT coils (white arrows). (H) Quantification of filaments per MT coil in wild-type (n = 17) and RanBP10 knockout platelets (n = 15) shows a small increase of MT coils for the RanBP10-deficient platelet, but the high standard deviation indicates that the number of MT coils ranges from well above to well below normal. Original magnification, ×10 000 (A-B); ×∼ 35 000 (D-G). Scale bars (insets D-E) indicate 2 μm. The resolved images and quantifications are representatives of 2 independent experiments.
Platelets from β1-tubulin mice are spherical and harbor only 2 to 3 MT filaments per coil in their marginal band.11 Because RanBP10 is a β-tubulin binding protein, we asked whether lack of RanBP10 affects the composition of the marginal band. In agreement with the literature,3,11 wild-type platelets exhibited 8 to 12 MT filaments (Figure 4D). In RanBP10-null platelets we found a spectrum of reduced, normal, or highly increased filament numbers, ranging from 5 to 26 (Figure 4E,H). In single cross-sections we even found MT bundles cut in both longitudinal and transverse sections (Figure 4F), and in some platelets, coils were not restricted to the platelet poles but also located internally (Figure 4G). To verify that these atypical manifestations of MT bundles are indeed specific, we performed IF on platelets stained for β-tubulin. The majority of RanBP10-deficient platelets did not exhibit a conspicuous cortical signal but showed a disordered appearance, including open circles and multiple starting points (Figure 4E inset). In contrast, nearly all wild-type platelets revealed the expected cortical tubulin staining pattern (Figure 4D inset). These results argue strongly that RanBP10 helps keep cortical MT filaments in structurally precise configuration to avoid platelet anisocytosis (Figure 4H).
Defective hemostasis, platelet activation, and aggregation in RanBP10-deficient mice
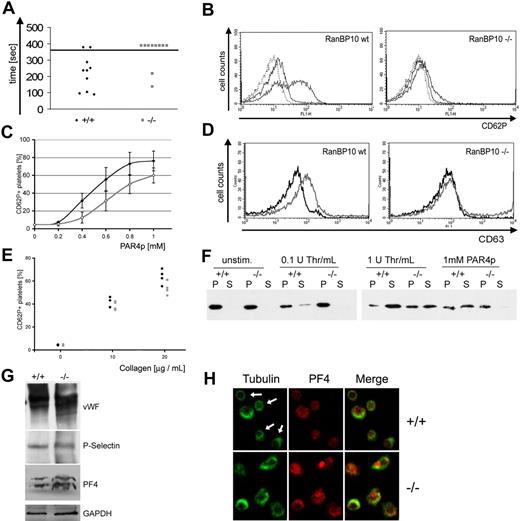
One major platelet function is to transport proteins and small molecules to sites of vascular injury. Proteins are mainly stored within α-granules, whereas nucleotides amass within electron-dense granules.32 During platelet activation, granules localize in the center before release, most likely by contraction of the cortical MT bundle.5 Having demonstrated that the MT coil is impaired in many RanBP10-null platelets, we asked next whether RanBP10-deficient mice have a bleeding defect by using a standard assay where tail bleeding is followed during the course of 6 minutes. We found that 8 of 10 RanBP10−/− mice showed a markedly prolonged bleeding time compared with only 2 of 10 wild-type animals (Figure 5A). Blood flow decreased over time but soon resumed in mutants because there was no stable plug formation. To define whether the hemostatic defect is caused by impaired platelet activation, we investigated granule release by stimulating platelets with different concentrations of PAR4p for 5 minutes, followed by flow cytometric analysis of CD62P expression. P-selectin is absent on the surface of resting platelets but highly up-regulated in response to agonist exposure, as indicated by a shift to higher fluorescence (Figure 5B left panel). RanBP10−/− platelets showed reduced CD62P release in response to PAR4p (Figure 5B right panel). We quantified this defect by measuring the fraction of CD62P+ cells compared with unstimulated platelets. In wild-type mice, 0.5mM PAR4p stimulated half-maximal activation, whereas 0.8mM was required for the same activation in mutant platelets (Figure 5C); even high concentrations of agonist did not lead to complete degranulation. Next we examined whether dense-granule release is also impaired, as manifest by up-regulation of CD63. Wild-type platelets stimulated with PAR4p showed increased CD63 expression, whereas mice lacking RanBP10 failed to release dense granules, even after exposure to 1mM PAR4p (Figure 5D). To exclude the possibility that impaired α- and dense granule release reflects diminished PAR4p reactivity, we also analyzed reactivity toward collagen. The response of RanBP10-null platelets to collagen was slightly reduced in comparison with wild-type platelets (Figure 5E). Because granule number was normal in mutant platelets (Figure 4B,E), we suspected that defective MT dynamics may prevent degranulation.
Defective hemostasis and platelet activation in RanBP10-deficient mice. (A) RanBP10 mice have a prolonged bleeding time. Tails were cut at the tip and blood flow was followed by submerging in 37°C saline until flow discontinued for at least 30 seconds. Bleeding stopped within 5 minutes in only 2 of 10 mutant mice but in 8 of 10 wild-type mice. (B) Mutant platelets fail to show full CD62P expression after agonist stimulation. Whole blood of wild-type and mutant mice was stimulated with 1mM PAR4 peptide before adding an fluorescein isothiocyanate-conjugated anti-CD62P specific antibody. Platelets were analyzed by flow cytometry showing a markedly reduced expression of CD62P. (C) Dose-response curve of PAR4p-stimulated platelets from 4 RanBP10-null (gray curve) and 5 wild-type mice (black curve), showing reduced reactivity with suboptimal concentrations. Error bars indicate 1 SD. (D) Platelets lacking RanBP10 fail to secrete the dense-granule marker CD63 after incubation with 1mM PAR4p (gray curve, right panel), compared with wild-type controls (gray curve, left panel). One representative of 3 experiments is shown. Black curves represent unstimulated controls. (E) Flow cytometric analysis from 4 wild-type (♦) or 4 RanBP10-null mice (gray circles) stimulated with 10 or 20 μg/mL collagen reveals slightly reduced CD62P expression in mutant animals. (F) Mutant mice show a reduced release of platelet granules after agonist stimulation. Washed platelets from wild-type (+/+) and knockout (−/−) mice were incubated with 1 or 0.1 U thrombin or 1mM PAR4 peptide as indicated, for 5 minutes before centrifugation. Platelet supernatant (S) was removed before adding 4× sample buffer. Pellets (P) were solubilized with 1× sample buffer overnight, and equal volumes of both fractions were subjected to SDS-PAGE. PF4 distribution was analyzed by immunoblotting. Mutant platelets failed to degranulate after low thrombin or PAR4p stimulation but showed partial response to high thrombin. (G) Normal expression of α-granule proteins in RanBP10-deficient platelets (−/−) compared with wild-type controls (+/+). Total platelet lysates were immunoblotted with antibodies against von Willebrand factor (VWF), P-selectin, and PF4. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. (H) Immunofluorescence staining against tubulin (green) showed contraction of the MT coil in most PAR4p-stimulated wild-type platelets (top left, white arrows) while staining for granule marker protein PF4 (red) was weak. In activated RanBP10−/− platelets tubulin staining revealed larger MT coils (bottom left). PF4 staining was stronger implying that granule release was incomplete, which is also shown in the merged panels. Scale bar indicates 2 μm.
Defective hemostasis and platelet activation in RanBP10-deficient mice. (A) RanBP10 mice have a prolonged bleeding time. Tails were cut at the tip and blood flow was followed by submerging in 37°C saline until flow discontinued for at least 30 seconds. Bleeding stopped within 5 minutes in only 2 of 10 mutant mice but in 8 of 10 wild-type mice. (B) Mutant platelets fail to show full CD62P expression after agonist stimulation. Whole blood of wild-type and mutant mice was stimulated with 1mM PAR4 peptide before adding an fluorescein isothiocyanate-conjugated anti-CD62P specific antibody. Platelets were analyzed by flow cytometry showing a markedly reduced expression of CD62P. (C) Dose-response curve of PAR4p-stimulated platelets from 4 RanBP10-null (gray curve) and 5 wild-type mice (black curve), showing reduced reactivity with suboptimal concentrations. Error bars indicate 1 SD. (D) Platelets lacking RanBP10 fail to secrete the dense-granule marker CD63 after incubation with 1mM PAR4p (gray curve, right panel), compared with wild-type controls (gray curve, left panel). One representative of 3 experiments is shown. Black curves represent unstimulated controls. (E) Flow cytometric analysis from 4 wild-type (♦) or 4 RanBP10-null mice (gray circles) stimulated with 10 or 20 μg/mL collagen reveals slightly reduced CD62P expression in mutant animals. (F) Mutant mice show a reduced release of platelet granules after agonist stimulation. Washed platelets from wild-type (+/+) and knockout (−/−) mice were incubated with 1 or 0.1 U thrombin or 1mM PAR4 peptide as indicated, for 5 minutes before centrifugation. Platelet supernatant (S) was removed before adding 4× sample buffer. Pellets (P) were solubilized with 1× sample buffer overnight, and equal volumes of both fractions were subjected to SDS-PAGE. PF4 distribution was analyzed by immunoblotting. Mutant platelets failed to degranulate after low thrombin or PAR4p stimulation but showed partial response to high thrombin. (G) Normal expression of α-granule proteins in RanBP10-deficient platelets (−/−) compared with wild-type controls (+/+). Total platelet lysates were immunoblotted with antibodies against von Willebrand factor (VWF), P-selectin, and PF4. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. (H) Immunofluorescence staining against tubulin (green) showed contraction of the MT coil in most PAR4p-stimulated wild-type platelets (top left, white arrows) while staining for granule marker protein PF4 (red) was weak. In activated RanBP10−/− platelets tubulin staining revealed larger MT coils (bottom left). PF4 staining was stronger implying that granule release was incomplete, which is also shown in the merged panels. Scale bar indicates 2 μm.
To test this hypothesis, we studied release of the α-granule protein platelet factor 4 (PF4) in response to agonists. In resting platelets, PF4 was solely present in pellet fractions, whereas increasing concentrations of thrombin lead to its release into supernatant fractions. In RanBP10-deficient platelets, by contrast, 1mM PAR4p or 0.1 U/mL thrombin did not result in PF4 translocation, and protein remained in the pellet. Greater thrombin concentrations (1 U/mL) led to nearly complete release of PF4 from wild-type platelets, whereas in mutants, approximately one-half of PF4 remained within the pellet fraction (Figure 5F). To exclude a reduced granule cargo load in mutant platelets, we tested for expression of von Willebrand factor, P-selectin, and PF4 by immunoblotting (Figure 5G). Glyceraldehyde-3-phosphate dehydrogenase loading control of both lanes was adjusted after densitometric analysis, showing that protein expression in RanBP10-null platelets was within 10% range of the expression found in wild-type controls (data not shown). RanBP10-null platelets thus show impaired release of a normal granule cargo load. Finally, wild-type platelets showed typical contraction of the marginal band (arrows in Figure 5H top left panel), whereas the majority of mutant platelet coils showed impaired contraction (bottom left panel). In agreement with our granule release data, immunostaining of residual PF4 in activated wild-type platelets showed only weak staining (top middle panel), whereas PF4 was readily observed in mutant platelets (bottom middle panel). Taken together, these data support the hypothesis that the MT-binding factor RanBP10 is essential for platelet function in hemostasis, most likely by modulating MT dynamics and marginal band contraction during platelet activation.
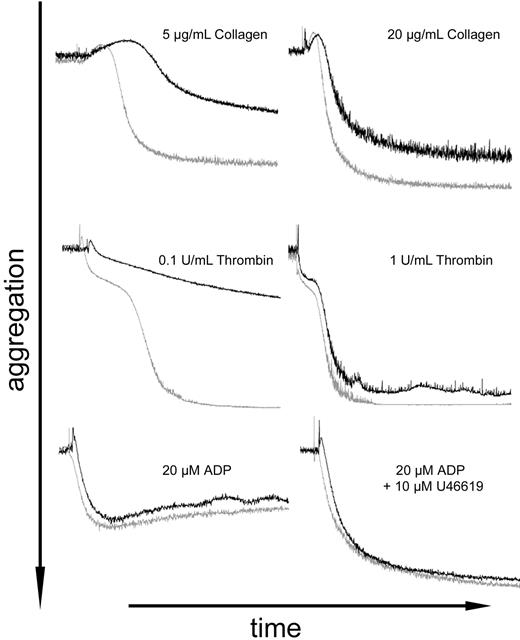
The main feature of platelet function is thrombus formation, mediated by both adhesion and aggregation. We therefore measured aggregation of wild-type and mutant platelets stimulated with collagen, thrombin, or adenosine diphosphate (ADP). As summarized in Figure 6, RanBP10−/− platelets showed reduced aggregation, especially with low concentrations of collagen or thrombin. Platelet activation with suboptimal agonist concentrations requires ADP secretion. This secretion is impaired in RanBP10−/− animals, as demonstrated by the absence of PAR4p-mediated increase on CD63 levels (Figure 5D). In contrast, aggregation in response to exogenous ADP, as well as in response to high concentrations of thrombin or collagen, which do not require dense granule-mediated ADP release, was similar in wild-type and knockout animals (Figure 6). These data strongly suggest the conclusion that the bleeding defects in RanBP10−/− mice result at least partly from diminished platelet function by reduced granule release.
Impaired platelet aggregation in RanBP10-deficient mice. Platelets from wild-type (gray curves) and RanBP10-nullizygous animals (black curves) were adjusted to 300 nL and light transmission followed by aggregometry after stimulation with collagen (5 and 20 μg/mL, top), thrombin (0.1 and 1 U/mL, middle), and 20μM ADP alone (bottom left) or together with 10μM U46619 (bottom right). Panels represent results from 2 or 3 independent experiments, depending on the agonist.
Impaired platelet aggregation in RanBP10-deficient mice. Platelets from wild-type (gray curves) and RanBP10-nullizygous animals (black curves) were adjusted to 300 nL and light transmission followed by aggregometry after stimulation with collagen (5 and 20 μg/mL, top), thrombin (0.1 and 1 U/mL, middle), and 20μM ADP alone (bottom left) or together with 10μM U46619 (bottom right). Panels represent results from 2 or 3 independent experiments, depending on the agonist.
Discussion
Cell shape and reactivity play critical roles in platelet function. Cortical MT bundles maintain a discoid shape to allow correct hemodynamic contact with vessel surfaces and contract after agonist exposure, leading to granule centralization before exocytosis. Recent work from Patel-Hett et al7 shows that the marginal band does not consist of a single coiled MT filament but that one long filament associates with several shorter ones, forming a dynamic bipolar array. The immediate platelet shape change observed after activation has been attributed to a contractile process rather than to depolymerization and repolymerization of MT filaments, because addition of the MT stabilizing agent taxol does not inhibit it.33,34 These cytoskeletal dynamics require additional proteins like dynein/dynactin, as seen in MT sliding during proplatelet elongation,14 but the mechanism is still an ill-defined one. To identify potential candidates essential for platelet biogenesis, shape, and function, we therefore searched for proteins that interact specifically with the C-terminal domain of β1-tubulin. We thus isolated secretory leukocyte protease inhibitor as a tubulin-binding partner essential for regulated proteolysis.15 A second gene highlighted in this screen is RanBP10, which encodes a previously uncharacterized protein with binding capacities for both β-tubulin and the small GTPase Ran.16 Our data on RanBP10 overexpression and RNA interference-mediated protein depletion in primary MKs reveal a dose-dependent effect on MT filament dynamics in MKs. However, its roles in platelet biogenesis and function are best studied in mouse models. RanBP10 is dispensable for MK maturation and platelet biogenesis (Figures 3) but plays an important role in maintaining platelet shape and function (Figures 4,Figure 5–6).
RanBP10 is a cytosolic protein that decorates MT filaments in MKs and platelets.16 Forced ectopic expression causes marked bundling of MT filaments, whereas RNAi-induced protein depletion leads to reduced tubulin staining. Although this appearance was phenocopied by most MKs from RanBP10-null mice, overall platelet biogenesis was hardly affected, which could be because the number of tubulin monomers is not altered and RanBP10 by itself does not influence the rate of polymerization. In addition, RanBP9, a ubiquitous homolog, might compensate for absence of RanBP10. Although we did not detect significant homology within the RhoGEF consensus domain, RanGEF activity of RanBP9 has not been excluded. Moreover, RanBP9 has been shown to bind MTs and ectopically nucleate new filaments35 and could hence obscure some of the functional effects of RanBP10 loss.
A role for RanBP10 in hemostasis
In mice lacking β1-tubulin, the dose-response curve to thrombin was reduced by approximately one order of magnitude for α-granule release.11 In contrast, serotonin release was normal and prolonged bleeding time is mainly attributed to thrombocytopenia and platelet spherocytosis rather than impaired degranulation.12 The bleeding phenotype in RanBP10-null mice, which harbor almost-normal platelet counts, could indeed be reflected in more profound malfunction of platelet reactivity compared with mice lacking β1-tubulin. To some extent, platelet reactivity in RanBP10−/− mice phenocopies defects reported in patients with TUBB1 mutations: Heterozygous carriers of the Q43P polymorphism have platelet spherocytosis, diminished dense granule release, and reduced aggregation toward low-dose collagen.36 Recently, an R318W mutation has been identified and attributed to the decreased expression of β1-tubulin and thrombocytopenia seen in some patients.37 Similarly, RanBP10-null platelets are more spherical and less uniform than wild-type platelets, and a wide range in number of MT filaments per coil (n = 5-26) suggests that RanBP10 acts to maintain marginal band integrity and prevent platelet anisocytosis. This malfunction might explain the bleeding defect observed in mutant mice and suggests that RanBP10 has vital hemostatic functions.
The cellular functions of platelets in wound healing are well characterized.38 However, the cell biologic changes underlying the process of platelet activation, namely the contraction of the peripheral MT coil preceding granule secretion, are still poorly understood. Platelet activation by stimuli like thrombin, ADP, or foreign surfaces leads to contraction of the platelet marginal band and centering of granules and organelles34,39 ; treatment with vinca alkaloids or colchicine prevents internal transformation of activated platelets.5,40,41 RanBP10-deficient platelets show reduced P-selectin expression in response to thrombin, similar to β1-tubulin knockout platelets.11 This contractile process of the marginal band is not likely the sole force responsible for the granule release reaction because in RanBP10−/− and β1-tubulin–null platelets, P-selectin is released in response to high agonist concentrations. Treatment with paclitaxel of chilled platelets results in irregular MT filaments without completely affecting granule secretion,33 but nevertheless, disordered platelet MT filaments seem to influence the activation process, most likely by impairing granule release.
Functions of RanBP10: MT bundling or Ran-GEF activity
RanBP10 also binds the small GTPase Ran and harbors Ran GEF activity.16 As RanBP10 concentrates along polymerized MT filaments in the MK periphery, it might convert cytoplasmic Ran–GDP into Ran–GTP and thus provide positional information for MT nucleation or other filament properties.25,42 In primary MKs forced to overexpress an EGFP-tagged RanBP10, we found Ran accumulating on the cytoplasmic side of the nuclear envelope, suggesting that it cannot enter the nucleus while bound to GTP (Figure 1E-F). This finding suggests that RanBP10 can indeed load Ran with GTP. It is conceivable that generation of Ran–GTP plays an essential role during platelet activation. Ran also has been detected in sciatic nerve cells, in both its GDP- and GTP-bound state, far away from the cell body.43 The authors showed that axon injury leads to importin-mediated retrograde transport mediated by the minus-end motor dynein. Because dynein also mediates sliding of MT filaments during proplatelet elongation,14 we speculate on the underlying similarities, as reviewed recently.44
Role of RanBP10 loss in other cell types
At first glance, peripheral-blood cell counts in RanBP10-null mice are comparable with wild-type controls. However, red cell counts were reduced, and the mean cellular volume was markedly increased. Both changes are statistically significant and represent a macrocytic anemia, such as found in patients with cobalamine or folate deficiency or other causes of reticulocytosis. β1-tubulin is also expressed during erythropoiesis, where yolk sac–derived fetal erythroblasts harbor a MT marginal band.9 Further studies are required to better elucidate the role of RanBP10 in both red blood cell and platelet maturation and function.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Transmission electron microscopy was generated with the help of Prof Sebastian Bachmann and Petra Schrade, core facility for electron microscopy, Charité, Berlin. We thank Prof Saumweber (Humboldt University, Berlin) for use of the deconvolution immunofluorescence microscope and Dr Renné (Karolinska Institutet, Stockholm) for setup of the gene trap approach in the initial phase of the project.
This study was supported by the Deutsche Forschungsgemeinschaft SCHU 1421/3-1 and 3-2 (to H.S.).
Authorship
Contributions: S.K., I.M., S.F., M.W., H.S., and J.F. performed experiments; R.A.S. designed certain experiments and analyzed and interpreted data; S.K. analyzed data; H.S. analyzed and interpreted data and planned the study; and S.K. and H.S. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Harald Schulze, PhD, Charité-Universitätsmedizin, Labor für Pädiatrische Molekularbiologie, Ziegelstrasse 5-9, 10098 Berlin, Germany; e-mail: harald.schulze@charite.de.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal