Abstract
MicroRNAs (miRNAs) are associated with cytogenetics and molecular subtypes of acute myelogeneous leukemia (AML), but their impact on AML pathogenesis is poorly understood. We have previously shown that miR-29b expression is deregulated in primary AML blasts. In this work, we investigated the functional role of miR-29b in leukemogenesis. Restoration of miR-29b in AML cell lines and primary samples induces apoptosis and dramatically reduces tumorigenicity in a xenograft leukemia model. Transcriptome analysis after ectopic transfection of synthetic miR-29b into leukemia cells indicates that miR-29b target apoptosis, cell cycle, and proliferation pathways. A significant enrichment for apoptosis genes, including MCL-1, was found among the mRNAs inversely correlated with miR-29b expression in 45 primary AML samples. Together, the data support a tumor suppressor role for miR-29 and provide a rationale for the use of synthetic miR-29b oligonucleotides as a novel strategy to improve treatment response in AML.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disorder that includes many entities with diverse genetic abnormalities and clinical features.1 In addition to specific chromosomal translocations, deletions, and amplifications (eg, t(15;17), −5q, −7q, +8), the discovery of gene mutations (eg, FLT3-ITD, CEBPA, and NPM1) and oncogene deregulation (ERG and BAALC overexpression) have improved the molecular and prognostic classification of AML, in particular for patients with cytogenetically normal AML (CN-AML).2 Over the past few years, a new class of small noncoding RNAs, named microRNAs (miRNAs), has changed the landscape of human genetics. Evolutionarily conserved, miRNAs are 18 to 24 nucleotides (nt) in length that regulate gene expression, for the most part, by targeting mRNAs according to the degree of complementarity with their 3′ untranslated region (UTR).3 miRNAs are involved in the regulation of critical biologic processes, including cell cycle, differentiation, development, and apoptosis.4-7 Recent data indicate that miRNAs are deregulated in diseases, such as diabetes, heart disease, and cancer.8-10 Our group and others have reported unique miRNA profiles associated with the main cytogenetic and molecular subgroups of AML.11,12 Despite this progress, very few mechanistic studies have been carried out to dissect the functional role of miRNAs in AML.
The miR-29 family is composed of 3 isoforms arranged in 2 clusters: miR-29b-1/miR-29a located on chromosome 7q32 and miR-29b-2/miR-29c located on chromosome 1q23. Interestingly, chromosome 7q32 is a region frequently deleted in myelodysplastic syndromes (MDSs) and therapy-related AML.13 MiR-29 family members have been shown to be down-regulated in high-risk chronic lymphocytic leukemia (CLL), lung cancer, invasive breast cancer, and cholangiocarcinoma.14-17 The enforced expression of miR-29b induced apoptosis in cholangiocarcinoma cell lines and reduced tumorigenicity in a xenograft model of lung cancer and rhabdomyosarcoma.17-19 We reported that expression of miR-29 family members is down-regulated in primary AML samples with chromosome translocations involving fusion of the MLL1 gene, in particular with the t(6;11) and t(9;11) translocations.11 In addition, primary CN-AML samples with wild-type nucleophosmin gene (NPM1) have low levels of miR-29s compared with CN-AML samples with NPM1 mutations.20
In this study, we explored the functional role of miR-29b in AML. We report that overexpression of synthetic miR-29b in AML cell lines and primary AML blasts induced apoptosis. Direct inoculation of synthetic miR-29b oligonucleotides into xenograft tumors dramatically decreased tumor growth. The data support a miR-29b tumor suppressor function and provide a rationale for the use of synthetic miR-29b oligonucleotides as a novel therapeutic agent in AML.
Methods
Patient samples
Frozen diagnostic bone marrow or peripheral blood samples were obtained from 100 adult AML patients from the M. D. Anderson tissue bank. Patient samples were prepared by Ficoll-Hypaque (Nygaard) gradient centrifugation, enriched for leukemic cells by CD3/CD19 depletion using MACS (Miltenyi Biotech), and cryopreserved as previously described.11 Cytogenetic analyses of the samples were performed at diagnosis, using unstimulated short-term (24-, 48-, and 72-hour) cultures with or without a direct method and G-banding. The criteria used to describe a cytogenetic clone and description of karyotype followed the recommendations of the International System for Human Cytogenetic Nomenclature.21 At least 20 bone marrow metaphase cells were analyzed in patients designated as having a normal karyotype. FLT3-ITD, activation loop D835, and NPM1 mutations analysis was performed on most of the samples as previously described.20 All patients gave informed consent in accordance with the Declaration of Helsinki for cryopreservation and use of the samples for molecular studies. Approval was obtained from the Institutional Review Board of the M. D. Anderson Cancer Center.
Microarray experiments
RNA extraction and miRNA microchip experiments were performed as described in detail elsewhere.11 A total of 5 μg of total RNA was used for hybridization on the custom miRNA microarray chip (OSU_CCC, Version 3.0), which contains approximately 1100 miRNA probes, including 345 human and 249 mouse miRNA genes, spotted in duplicates. For gene-expression profiling, RNA samples were analyzed with the use of Affymetrix U133 plus 2.0 GeneChips.
Northern blotting
Northern blotting was performed as previously described.14 Briefly, total RNA was extracted using Trizol reagent (Invitrogen). RNA samples (10 μg) were run on 12% polyacrylamide denaturing (urea) precast gels (Bio-Rad) and then transferred onto Hybond membrane (Amersham Pharmacia Biotech). The hybridization was performed with α-32P miR-29a and -b labeled probes overnight at 42°C. The miR-29 probes sequences were the complementary one to the mature miR-29s. As a loading control, we measured U6 expression after stripping the filter as previously described.14
Real-time quantification of miRNAs
The single-tube TaqMan miRNA assays were used to detect and quantify mature miRNAs as previously described22 using PCR 9700 Thermocycler ABI Prism 7900HT and the sequence detection system (Applied Biosystems). Normalization was performed with 18S. Comparative real-time polymerase chain reaction (PCR) was performed in triplicate, including no-template controls. Relative expression was calculated using the comparative cross threshold (Ct) method.23
Data analysis
miRNA microarray images were analyzed using GENEPIX PRO. Average values of the replicate spots of each miRNA were background subtracted; log2-transformed and normalized using the global median and the BRB array tools (http://linus.nci.nih.gov/BRB-ArrayTools.html). For the affymetrix experiments, Cel files generated by the GeneChip scanner were imported to the BRB software tools. The GCRMA procedure was used for background subtraction and normalization. A filtering step was performed to remove probe sets that did not show significant variation across the samples: a probe was excluded if less than 20% of expression data have at least a 1.5-fold change in either direction from gene's median value or the percentage of data missing or filtered out exceeds 50%. Class comparison within the BRB tools was used to compare mRNA expression after transfection of synthetic miR-29 versus scrambled oligonucleotides in K562 cells. To identify mRNAs that correlated with miR-29a and -29b expression in AML patients, we performed quantitative trait analysis (Spearman correlation test) using the BRB tools. The miR-29a and -b expression values for each patient were obtained using miRNA microarrays as described in “Microarray experiments.” Only normalized, log2 values were used. All the analyses were performed using BRB-ArrayTools, Version 3.6.0 (R. Simon and A. P. Lam, National Cancer Institute, Bethesda, MD) and using the R version 2.3.1 (R Foundation for Statistical Computing). To assess whether certain biologic terms were enriched or overrepresented in a signature, we used the Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/).24 In our analysis, we used high classification stringency and considered only terms that have P values less than .05, after permutation corrections (Benjamini).
Statistical analysis
Both t test and χ2 were used to compare baseline clinical features and average miRNA expression between groups of patients. All reportedP values were 2-sided and obtained using the SPSS software package (SPSS 15.0). Some graphics were performed using GraphPad prism software (www.graphpad.com).
In vitro transfection with synthetic miRNAs
The synthetic miR-29a and -29b were purchased from Ambion. A total of 5 million of K562 cells (ATCC) were nucleoporated using Amaxa (Solution V, Program T016) with 5 μg of precursor oligonucleotide and 0.5 μg green fluorescent protein plasmid in a total volume of 10 mL. The transfection efficiency of K562 cells using this method is close to 90% (www.lonzabio.com; supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The synthetic miR-212 was used as an additional control in the K562 experiments. This miRNA is not expressed in primary AML samples or in hematopoietic stem cells.11,12 MiR-29s expression levels, before and after transfection, are shown in supplemental Figure 1. Primary bone marrow or peripheral blood samples from 3 newly diagnosed AML patients were obtained after getting informed consent according to the M. D. Anderson Cancer Center Institutional Review Board guidelines. Two patients had normal cytogenetics with wild-type NPM1 and FLT3, whereas the other patient had t(9;11). After Ficoll preparation, the samples were enriched for leukemic cells by CD3/CD19 depletion. Approximately 3 × 106 primary AML blasts were nucleoporated with synthetic miR-29b or scrambled oligonucleotides (Amaxa; program T020, solution V). The expression of the oligonucleotides after transfection was assessed by quantitative real time (RT)–PCR as described before (supplemental Figure 2).
miR-29 lentivirus infection
The miR-29a and -29b lentivirus is a feline immunodeficiency virus-based construct purchased from Systems Biosciences. This construct consists of the stem loop structure of miR-29b-1 or miR-29a and 200 bp of upstream and downstream flanking genomic sequence cloned into the pMIF-cGFP-Zeo-miR plasmid (catalog no. MIFCZ301A-1). Packaging of the miR-29 constructs in pseudoviral particles was performed using the pPACKH1 packaging plasmid mix (catalog no. LV500A-1), according to the manufacturer's instructions (SBI). Lentivirus rapid titer PCR Kit (catalog no. LV950A-1) was used to measure the copy numbers and determine the multiplicity of infection to perform experiments. Kasumi-1 cells (106) were infected with the miR-29-lentivirus with an efficiency of approximately 50% as determined by green fluorescent protein measurement by flow cytometry (supplemental Figure 3). Empty lentivirus was used as a control for the experiments.
Cell growth curve
K562 (5 × 105/mL) cells were plated in 10-mL plates and transfected with scrambled or miR-29s oligonucleotides from Ambion at a final concentration of 100nM, using nucleoporation as previously described. Kasumi-1 (8 × 105/mL) cells were plated in 10-mL plates and infected with miR-29a and -29b lentivirus or control (empty lentivirus) as previously explained. Cells were harvested and counted at 24-hour intervals using a ViCell counter (Beckman Coulter). Each sample was run in triplicate. The results were validated using the MTT test (Promega).
Apoptosis experiments
To demonstrate that miR-29 induces apoptosis, we transfected or infected miR-29a and -29b or scrambled oligonucleotide/empty vector into K562, primary AML samples (n = 2) and Kasumi-1 cells, respectively. The cells were then subsequently treated with cytarabine (Ara-C; 5μM) or control (phosphate-buffered saline; Sigma-Aldrich). Annexin V/propidium iodide (PI) stain (BD Biosciences PharMingen) was performed at 48 hours after transfection/infection. Caspase 3 and 7 activity was measured at 48 hours using the Apo-ONE Homogeneous Caspase-3/7 Assay (Promega) following the manufacturer's instructions. The assay includes a profluorescent caspase 3/caspase 7 consensus substrate, rhodamine 110 bis-(N-CBZ-L-aspartyl-L-glutamyl-L-valyl-aspartic acid amide), and an optimized bifunctional cell lysis/activity buffer. The buffer efficiently lyses cultured mammalian cells and supports optimal caspase 3/caspase 7 enzymatic activity. Pro-caspase 3 protein expression was assessed by immunoblotting using the pro-caspase 3 antibody from Abcam.
Luciferase reporter experiments
The 3′ UTR segments containing the target sites for miR-29 from the MCL-1-, CXXC6, and CDK6 gene was amplified by PCR from cDNA and inserted into the pGL3 control vector (Promega), using the XBA1 site immediately downstream from the stop codon of luciferase. The primers are available in the supporting information. We also generated an insert with deletions of 4 bp from the site of perfect complementarity using the QIAGEN XL-site directed Mutagenesis Kit (Stratagene). Wild-type and mutant inserts were confirmed by sequencing. Human cell line K562 (ATCC) was grown in 10% FBS in RPMI 1640, supplemented with 1× nonessential amino acid and 1mmol/sodium pyruvate at 37°C in a humidified atmosphere of 5% CO2. The cells were cotransfected in 10-mL plates using nucleoporation (Amaxa) according to the manufacturer's protocol using 5 μg of the firefly luciferase report vector and 0.5 μg of the control vector containing Renilla luciferase, pRL-TK (Promega). For each plate, 5 μg of the synthetic miR-29b or the scrambled oligonucleotides (negative control precursor; Ambion) was used. Firefly and Renilla luciferase activities were measured consecutively using the dual-luciferase assays (Promega) 24 hours after transfection.
Western blotting
Total protein extracts from K562 cells and primary AML blasts transfected with synthetic miR-29b and scrambled oligonucleotides were extracted using radio immunoprecipitation assay (RIPA) buffer (Sigma-Aldrich). Protein expression was analyzed by Western blotting using Mcl-1, Cdk6, and GAPDH (Santa Cruz Biotechnology). The phosphorylation status of retinoblastoma (Rb) protein was explored using total Rb and Ser780 Rb antibody (Cell Signaling Technology).
In vivo studies
Animal studies were performed according to Ohio State University institutional guidelines. A total of 10 million viable K562 cells were injected subcutaneously into both flanks of 5-week-old female nude mice (Charles River Breeding Laboratories). The tumors size was measured daily until the tumor reached 50 mm3. Then, 5 μg of synthetic miR-29b or scrambled oligonucleotides diluted in Lipofectamine (Invitrogen) solution (100 μL total volume) were injected directly into the tumors (left flank = miR-29b; right flank = scrambled) at baseline and after 3, 7, and 10 days. Tumors were measured on the day of the injections and 4 days after the last injection. At that time, the mice were killed, necropsies were performed, and tumors were weighted. Tumor volumes were calculated using the equation V (in mm3) = A × B2/2, where A is the largest diameter and B is the perpendicular diameter.
Results
MiR-29 reduces cell growth and induces apoptosis in cell lines and primary AML samples
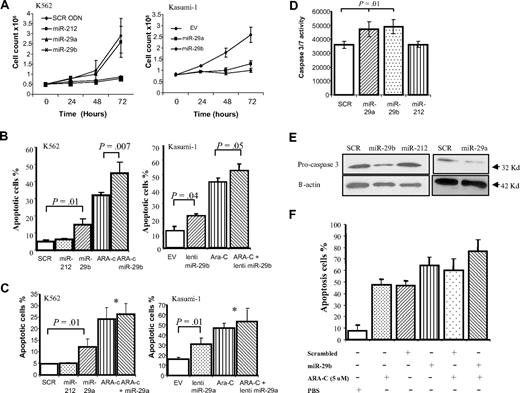
To examine the biologic effects of miR-29a and -29b in AML, we overexpressed both miRNAs in 2 myeloid leukemia cell lines (K562 and Kasumi-1) and examined cell growth curves and apoptosis. Both cell lines were chosen because they expressed low miR-29a and -29b levels (supplemental Figure 1). Synthetic miR-29a and -29b oligonucleotides as well as 2 controls (scrambled oligonucleotides and miR-212) were transfected into K562 cells using nucleoporation. Because of low efficiency and high mortality of Kasumi-1 cells with nucleoporation methods, we used a lentivirus construct to overexpress miR-29s. The empty lentivirus was used as a control (supplemental Figure 3). As shown in Figure 1A, overexpressing both miRNAs inhibited the cell growth of K562 and Kasumi-1 cell lines with respect to their respective controls. These results were validated using the MTT test (supplemental Figure 4). Next, we examined the effects of these miRNAs on apoptosis. In Figure 1B, we showed that miR-29b overexpression increased apoptosis at 48 hours (no effects were observed at 24 hours) by 3.4- and 2-fold in K562 and Kasumi-1 cells, respectively, as measured by the annexin V assay (P = .01). Further, we determined whether miR-29b transfection enhances apoptosis after treatment with Ara-C (5μM), a drug commonly used in the treatment of AML. In this experiment, K562 and Kasumi-1 cells were transfected or infected with synthetic miR-29b/lentivirus or control oligonucleotides/empty vector and immediately cultured with 5μM of Ara-C for 48 hours. Although miR-29b increased Ara-C-induced apoptosis by 1.4-fold in K562 cells (P = .007), this effect was less strong for Kasumi-1 cells (< 1-fold). Similar apoptosis results were observed by overexpressing miR-29a in both cell lines (Figure 1C). Apoptosis induction by miR-29a and -29b was associated with caspase 3 and 7 activation (Figure 1D-E). Last, we confirmed these results in 2 primary AML samples that were transfected with synthetic miR-29b or scrambled oligonucleotides and treated with Ara-C (5μM) or control (phosphate-buffered saline; Figure 1F). We found a 1.4-fold increase in apoptosis after synthetic miR-29b transfection, with respect to scrambled oligonucleotides. The antiapoptotic effects of miR-29b in primary AML samples are not as strong as in the one observed in the cell lines, in particular after Ara-C treatment. This could be explained by the high nucleoporation toxicity observed in primary AML samples.
MiR-29b reduces cell growth and induces apoptosis in cell lines and primary samples. (A) Cell growth curves of K562 cells (left panel) or Kasumi-1 cells (right panel) transfected or infected in vitro with miR-29a, -29b, and their respective controls (miR-212, scrambled oligonucleotides, or empty vector). (B) Annexin V/PI assays in K562 (left panel) and Kasumi-1 (right panel) cells after 48 hours of transfection with synthetic miR-29b/lentivirus miR-29b or controls (scrambled oligonucleotides, miR-212, or empty vector) in the presence or absence of 5μM of Ara-C. The results are shown as percentage of apoptotic cells. Data are the average of 3 independent experiments ± SD. P values were obtained using t test. *P > .05. (C) Annexin V/PI assays in K562 (left panel) and Kasumi-1 (right panel) cells after 48 hours of transfection with synthetic miR-29a/lentivirus miR-29a or controls (scrambled oligonucleotides, miR-212, or empty vector) in the presence or absence of 5μM of Ara-C. (D) The regulation of apoptosis by miR-29a and -29b was confirmed by measuring caspase 3 or caspase 7 activities. K562 cells were transfected with synthetic miR-29a and -29b or scrambled oligonucleotides and grown in 96-well microplates. After 48 hours, luminescence was measured using the caspase 3 or caspase 7 Glo Assay (Promega). P values were obtained using t test. Values are mean ± SD; n = 3. (E) Pro-caspase 3 protein expression level was measured in K562 by immunoblotting after 48 hours of transfection with synthetic miR-29a and -29b or scrambled oligonucleotides. Loading control was performed using β-actin. (F) Annexin V/PI assays in 2 primary AML samples after 48 hours of transfection with synthetic miR-29b or control (scrambled oligonucleotides) in the presence or absence of 5μM of Ara-C. The results are shown as percentage of apoptotic cells. Bars represent range.
MiR-29b reduces cell growth and induces apoptosis in cell lines and primary samples. (A) Cell growth curves of K562 cells (left panel) or Kasumi-1 cells (right panel) transfected or infected in vitro with miR-29a, -29b, and their respective controls (miR-212, scrambled oligonucleotides, or empty vector). (B) Annexin V/PI assays in K562 (left panel) and Kasumi-1 (right panel) cells after 48 hours of transfection with synthetic miR-29b/lentivirus miR-29b or controls (scrambled oligonucleotides, miR-212, or empty vector) in the presence or absence of 5μM of Ara-C. The results are shown as percentage of apoptotic cells. Data are the average of 3 independent experiments ± SD. P values were obtained using t test. *P > .05. (C) Annexin V/PI assays in K562 (left panel) and Kasumi-1 (right panel) cells after 48 hours of transfection with synthetic miR-29a/lentivirus miR-29a or controls (scrambled oligonucleotides, miR-212, or empty vector) in the presence or absence of 5μM of Ara-C. (D) The regulation of apoptosis by miR-29a and -29b was confirmed by measuring caspase 3 or caspase 7 activities. K562 cells were transfected with synthetic miR-29a and -29b or scrambled oligonucleotides and grown in 96-well microplates. After 48 hours, luminescence was measured using the caspase 3 or caspase 7 Glo Assay (Promega). P values were obtained using t test. Values are mean ± SD; n = 3. (E) Pro-caspase 3 protein expression level was measured in K562 by immunoblotting after 48 hours of transfection with synthetic miR-29a and -29b or scrambled oligonucleotides. Loading control was performed using β-actin. (F) Annexin V/PI assays in 2 primary AML samples after 48 hours of transfection with synthetic miR-29b or control (scrambled oligonucleotides) in the presence or absence of 5μM of Ara-C. The results are shown as percentage of apoptotic cells. Bars represent range.
Effects of miR-29b overexpression in a xenograft model
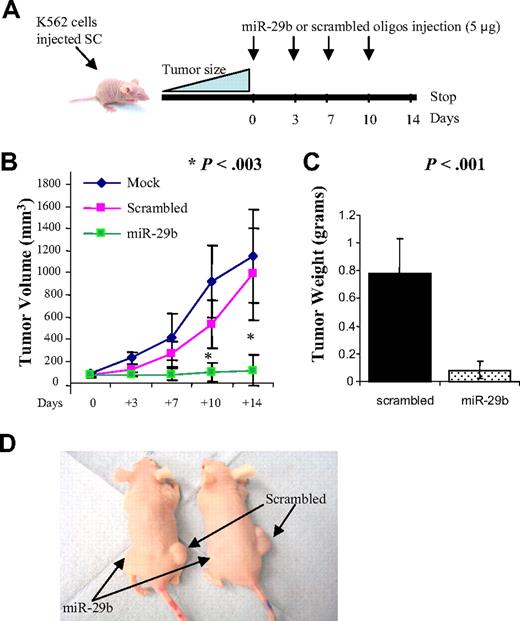
As further proof that miR-29b functions as a tumor suppressor, we tested whether miR-29b could reduce tumorigenicity in a xenograft model; approximately 10 million viable K562 cells were inoculated subcutaneously in both flanks of immunocompromised “nude” mice (12 mice per group). When tumors reached 50 mm3, synthetic miR-29b (left side) or scrambled oligonucleotides (right side) were injected directly into the tumors. The tumors were injected every 3 or 4 days for a total of 4 times (Figure 2A). As shown in Figure 2B, at 10 and 14 days after the first injection, tumors injected with synthetic miR-29b (n = 12) were significantly smaller than the scrambled oligonucleotide (n = 12) and mock controls (n = 6) (P = .003; supplemental Figure 5). At day 14, the average tumor weights for the scrambled oligonucleotides and the synthetic miR-29b inoculated mice were 0.79 g and 0.089, respectively (P = .001; Figure 2C). Two tumors disappeared completely after synthetic miR-29b injections (Figure 2D). The results of this experiment support a tumor suppression function for miR-29b in acute leukemia.
MiR-29b inhibits leukemic growth in vivo. (A) Diagram illustrating the experimental design of the mice xenograft experiment. (B) Graphic representing tumor volumes at the indicated days during the experiment for the 3 groups: mock (n = 6), scrambled (n = 12), and synthetic miR-29b (n = 12). (C) Tumor weight averages between scrambled and synthetic miR-29b-treated mice groups at the end of the experiment (day 14). P values were obtained using t test. Bars represent SD. (D) Photographs of 2 mice injected with miR-29b (left flank) or scrambled (right flank).
MiR-29b inhibits leukemic growth in vivo. (A) Diagram illustrating the experimental design of the mice xenograft experiment. (B) Graphic representing tumor volumes at the indicated days during the experiment for the 3 groups: mock (n = 6), scrambled (n = 12), and synthetic miR-29b (n = 12). (C) Tumor weight averages between scrambled and synthetic miR-29b-treated mice groups at the end of the experiment (day 14). P values were obtained using t test. Bars represent SD. (D) Photographs of 2 mice injected with miR-29b (left flank) or scrambled (right flank).
Transcriptional effects of miR-29a and -29b cluster in AML
To characterize the molecular basis of miR-29b tumor suppression in leukemias, we investigated the effects of miR-29b at the transcriptome level by performing gene-expression analysis in K562 cells transfected with synthetic miR-29b or scrambled oligonucleotides. Because miR-29a is located in a cluster with miR-29b in chromosome 7q32 and both have the same antiproliferative and antiapoptotic effects in myeloid cell lines, we also investigated the effects of miR-29a on the transcriptome using the same strategy. The transfection success was assessed by measuring miR-29s by Northern blotting (supplemental Figure 1). After ectopic transfection of miR-29a, 653 probe sets (572 genes) were differentially expressed with respect to scrambled oligonucleotides (class comparison, P < .001, false discovery rate [FDR] < .01). The ectopic transfection of miR-29b induced mRNA changes in 530 probe sets (480 genes; class comparison, P < .001, FDR < 0.01). The profiles of deregulated genes were highly similar; 503 from 530 probe sets were shared between miR-29a and -29b signatures (Figure 3A). Because both miR-29a and -29b have the same seed match sequence for target recognition, share predicted targets25 and have very similar effects on the transcriptome, we performed a supervised analysis where we pooled all the miR-29a and -29b transfections (arrays n = 6) and compared them with scrambled oligonucleotides (arrays, n = 4). After ectopic transfection of synthetic miR-29a and -29b oligonucleotides, 162 probe sets (141 genes) were significantly up-regulated and 428 probe sets (360 genes) were down-regulated with respect to the controls (class comparison, P < .001, FDR < 0.01; supplemental Table 1). Genes found deregulated in K562 cells after transfection of miR-29a and -29b were analyzed with DAVID functional analysis software to identify the most represented gene ontology (GO) categories.24 These results are shown in supplemental Table 2. In particular, genes related to apoptosis (MCL-1, TRAF4, and MYBl2) and cell cycle regulation (CDK4, CDK6, and CCND2) are targets of miR-29 regulation (supplemental Table 1). Although not overrepresented in any GO class, few genes down-regulated after miR-29s transfection are genes involved in cell proliferation pathways, such as JAK2 and IGF1. Together, these data indicate that miR-29a and -29b target apoptosis, cell cycle, and cell proliferation pathways.
MiR29a and -29b targets are specifically and significantly down-regulated after miR-29s transfections. (A) Venn diagram showing common probes between miR-29a and -29b Affymetrix signatures. Microarray mRNA profiling (Affymetrix) was performed using RNA obtained from K562 cells transfected with either synthetic miR-29a (n = 2) or miR-29b (n = 4) and scrambled oligonucleotides (n = 4). Class comparison analysis (miR-29b vs scrambled and miR-29a vs scrambled) were performed using P less than .001. The number in the intersection of the Venn diagram represents the number of probes that are shared between miR-29a and -b signatures, whereas the numbers in the left and right circle are the probes that are only differentially expressed in miR-29a and -b, respectively. (B) The box plots display the median and SD of t test score distributions in the 2 groups of Affymetrix experiments: targets of miR-29 or all other nontargets genes. The open circles represent the genes with t test score higher than 95th percentile in each individual distribution. The t test scores are positive for genes up-regulated by miR-29 transfections, whereas negative scores belong to down-regulated genes.
MiR29a and -29b targets are specifically and significantly down-regulated after miR-29s transfections. (A) Venn diagram showing common probes between miR-29a and -29b Affymetrix signatures. Microarray mRNA profiling (Affymetrix) was performed using RNA obtained from K562 cells transfected with either synthetic miR-29a (n = 2) or miR-29b (n = 4) and scrambled oligonucleotides (n = 4). Class comparison analysis (miR-29b vs scrambled and miR-29a vs scrambled) were performed using P less than .001. The number in the intersection of the Venn diagram represents the number of probes that are shared between miR-29a and -b signatures, whereas the numbers in the left and right circle are the probes that are only differentially expressed in miR-29a and -b, respectively. (B) The box plots display the median and SD of t test score distributions in the 2 groups of Affymetrix experiments: targets of miR-29 or all other nontargets genes. The open circles represent the genes with t test score higher than 95th percentile in each individual distribution. The t test scores are positive for genes up-regulated by miR-29 transfections, whereas negative scores belong to down-regulated genes.
MiR29a and -29b predicted targets are specifically and significantly down-regulated after miR-29s transfections
To investigate whether miR-29-regulated predicted targets, we performed a class comparison analysis (scrambled oligonucleotides vs pooled miR-29a and -29b transfections) using, instead of the whole genome, a subset of 602 genes, which are predicted targets according to TargetScan.25 Using the same stringency of the whole genome test (P < .001), we identified up-regulation of 5 probe sets (2 genes) and concomitant reduction in expression levels of 92 probe sets (68 genes; supplemental Table 3). The box plots in supplemental Figure 3B display the median and SD of the t test scores for either all 602 targets of miR-29 and all other expressed nontarget genes. When the number of up- and down-regulated genes were compared in a 2 × 2 table, it was apparent that miR-29a and -29b transfections specifically down-regulated mRNA levels of the predicted targets (Yates χ2 = 22, P < .001). Significantly, miR-29 targets down-regulation (Yates χ2 = 22, P < .001) was also found using a different prediction algorithm, PicTar.26 Among the miR-29b predicted targets down-regulated after miR-29s transfections, COL1A2 and DNMT3B genes have been previously validated experimentally by luciferase assays and gain-of-function experiments (supplemental Table 3).9,18
Validation of the microarray analysis in cell lines and primary AML patients
To validate the microarray data, we assayed the expression of 3 genes (MCL-1, CXXC6, and CDK6) by quantitative RT-PCR and Western blotting. We chose to validate CXXC6 because it was the most down-regulated miRNA after miR-29s transfection (supplemental Table 3). CXXC6, a member of a novel protein family, is fused to mixed lineage leukemia (MLL) in AML containing the t(10;11)(q22;q23).27 MCL-1 and CDK6 were chosen because both are oncogenes up-regulated in AML and play a key role in cancer apoptosis and cell cycle, respectively.28,29 In addition, CDK6 blocks myeloid differentiation by interfering with RUNX-1 DNA binding and RUNX-1/C/EBPα interaction.30 As shown in Figure 4, ectopic transfection of synthetic miR-29a or -29b resulted in down-regulation of MCL-1 (Figure 4A), CXXC6 (Figure 4B), and CDK6 (Figure 4C) mRNA levels with respect to the controls at 24 hours. Furthermore, we observed down-regulation of Mcl-1 and Cdk6 proteins after transfection of synthetic miR-29a and -29b oligonucleotides, but not with the controls in K562 cells (Figure 4D). These results were replicated in Kasumi-1 cells using a lentivirus vector that expressed miR-29b. As shown in Figure 4E, ectopic miR-29a or -29b infection resulted in down-regulation of Mcl-1 and Cdk6 proteins with respect to the empty construct. Because CDK6 impacts the cell cycle by targeting the Rb protein by phosphorylation,31 we measured the Rb phosphorylation status after transfection of synthetic miR-29b. As shown in Figure 4F, transfection of miR-29b in K562 cells resulted in decreased Rb phosphorylation at site Ser780 after 24 hours. Thus, miR-29b indirectly impacts Rb phosphorylation status, at least in part by targeting CDK6 (Figure 4F). These results were also reproduced in 3 primary AML samples. As shown in Figure 4G, CXXC6, CDK6, and MCL-1 mRNA levels were lower in miR-29b-transfected samples with respect to the controls at 24 hours (Figure 4G; supplemental Figure 2). Furthermore, overexpressing miR-29b in primary AML blasts resulted in Mcl-1 protein down-regulation by 55% with respect to scrambled oligonucleotides (Figure 3H-I).
Validation of the microarray analysis. (A) Quantitative RT-PCR of MCL-1, CXXC6 (B) and CDK6 (C) after 24 hours of transfection with synthetic miR-29a, -29b, or scrambled oligonucleotides in K562 cells. The results are shown as average mRNA expression after normalization with 18s and 2ΔCt calculations. Data represent the average of 3 independent experiments ± SD. (D) Western blotting of Mcl-1 and Cdk6 protein expression in K562 and Kasumi-1 cells (E) after 48 hours of transfection with synthetic miR-29a, -29b, or controls (scrambled oligonucleotides [SCR] and miR-212) or lentivirus miR-29a, -29b, or empty vector (EV) for Kasumi-1 cells. The protein loading control was performed using GAPDH. (F) Rb phosphorylation at Ser780 after 24 hours of transfection with synthetic miR-29b or control (SCR). (G) Quantitative RT-PCR of MCL-1, CXXC6, and CDK6 in 3 primary AML blasts after transfection with synthetic miR-29b or controls (SCR). The results are shown as average mRNA expression after normalization with 18s and 2ΔCt calculations. Bars represent mean ± SEM. (H) Western blotting of Mcl-1 in 3 newly diagnosed AML patients after transfection with synthetic miR-29b or controls (SCR). Loading control was performed using GAPDH. Mcl-1/GAPDH ratios are shown after densitometry evaluation of the gels. (I) Normalized Mcl-1 protein expression averages relative the mock-transfected primary AML samples. Bars represent SD.
Validation of the microarray analysis. (A) Quantitative RT-PCR of MCL-1, CXXC6 (B) and CDK6 (C) after 24 hours of transfection with synthetic miR-29a, -29b, or scrambled oligonucleotides in K562 cells. The results are shown as average mRNA expression after normalization with 18s and 2ΔCt calculations. Data represent the average of 3 independent experiments ± SD. (D) Western blotting of Mcl-1 and Cdk6 protein expression in K562 and Kasumi-1 cells (E) after 48 hours of transfection with synthetic miR-29a, -29b, or controls (scrambled oligonucleotides [SCR] and miR-212) or lentivirus miR-29a, -29b, or empty vector (EV) for Kasumi-1 cells. The protein loading control was performed using GAPDH. (F) Rb phosphorylation at Ser780 after 24 hours of transfection with synthetic miR-29b or control (SCR). (G) Quantitative RT-PCR of MCL-1, CXXC6, and CDK6 in 3 primary AML blasts after transfection with synthetic miR-29b or controls (SCR). The results are shown as average mRNA expression after normalization with 18s and 2ΔCt calculations. Bars represent mean ± SEM. (H) Western blotting of Mcl-1 in 3 newly diagnosed AML patients after transfection with synthetic miR-29b or controls (SCR). Loading control was performed using GAPDH. Mcl-1/GAPDH ratios are shown after densitometry evaluation of the gels. (I) Normalized Mcl-1 protein expression averages relative the mock-transfected primary AML samples. Bars represent SD.
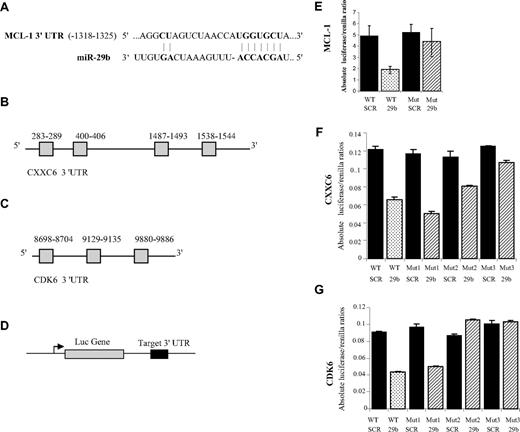
Target validation by luciferase assays
To validate these interactions, we cloned the 3′UTR of MCL-1, CXXC6, and CDK6 predicted to interact with miR-29 (Figure 5A-C) into a luciferase reporter vector and cotransfected with miR-29b into K562 cells (Figure 5D). According to TargetScan, there is only one predicted interaction site for miR-29 in the MCL-1 3′UTR (Figure 5A), whereas 4 and 3 sites are predicted to interact with miR-29 in the CXXC6 and CDK6 3′UTR, respectively (Figure 5B-C).25 A marked reduction in the luciferase/Renilla ratio was seen for MCL-1, CXXC6, and CDK6 constructs transfected with synthetic miR-29b but not with the control (scrambled oligonucleotides; Figure 5E-G). Furthermore, the observed luciferase/Renilla reduction was abrogated when we cotransfected a mutated luciferase reporter vector containing the 3′UTR of MCL-1, CXXC6, and CDK6 with 4 deleted nucleotides at the site of interactions with miR-29 (Figure 5E-G).
Mir-29b regulates MCL-1, CXXC6, and CDK6 expression through binding with their 3 UTR. (A) Predicted site of miR-29 in the MCL-1 3′ UTR mRNA (red: nucleotides involved in the target interaction). (B) Diagram illustrating the structure of the CXXC6 and CDK6 (C) 3′ UTR. There are 4 and 3 predicted miR-29 sites in the CXXC6 and CDK6 3′ UTR, respectively. (D) A wild-type (WT) luciferase reporter plasmid was generated by fusing a fragment of the MCL-1, CXXC6, and CDK6 3′UTR encompassing the miR-29 binding site downstream of the luciferase (Luc) reporter gene. (E) Luciferase activities from the MCL-1, (F) CXXC6, and (G) CDK6 constructs were determined at 24 hours and were normalized using Renilla. The mutant plasmid was generated by deleting the miR-29 binding site. The numbers next to the mutants (ie, Mut1, Mut2) represent the numbers of predicted miR-29 binding sites deleted, starting from the 5′ end of the gene. The mutations (deletions) are additive (ie, Mut2, have both the first and second site deleted). For CXXC6 we deleted up to 3 predicted sites (Mut3 includes deletions on the 3 sites: 283-9, 400-6, and 1487-93). The data represent the average of 3 independent experiments ± SD.
Mir-29b regulates MCL-1, CXXC6, and CDK6 expression through binding with their 3 UTR. (A) Predicted site of miR-29 in the MCL-1 3′ UTR mRNA (red: nucleotides involved in the target interaction). (B) Diagram illustrating the structure of the CXXC6 and CDK6 (C) 3′ UTR. There are 4 and 3 predicted miR-29 sites in the CXXC6 and CDK6 3′ UTR, respectively. (D) A wild-type (WT) luciferase reporter plasmid was generated by fusing a fragment of the MCL-1, CXXC6, and CDK6 3′UTR encompassing the miR-29 binding site downstream of the luciferase (Luc) reporter gene. (E) Luciferase activities from the MCL-1, (F) CXXC6, and (G) CDK6 constructs were determined at 24 hours and were normalized using Renilla. The mutant plasmid was generated by deleting the miR-29 binding site. The numbers next to the mutants (ie, Mut1, Mut2) represent the numbers of predicted miR-29 binding sites deleted, starting from the 5′ end of the gene. The mutations (deletions) are additive (ie, Mut2, have both the first and second site deleted). For CXXC6 we deleted up to 3 predicted sites (Mut3 includes deletions on the 3 sites: 283-9, 400-6, and 1487-93). The data represent the average of 3 independent experiments ± SD.
Correlation of miR-29a and -29b expression with mRNA expression in primary AML samples
The transient overexpression of miR-29s in K562 cells provided some important information about the global transcriptome regulation by this cluster. However, because miR-29s are overexpressed at artificially high levels in this model, it may not reflect the “steady state” gene regulation by these miRNAs. To obtain insights into the role of the miR-29s in “steady-state” target gene regulation in AML, we correlated miR-29a and 29b expression levels detected by miRNA microarray analysis (supplemental Figure 6) with mRNA expression levels detected by Affymetrix microarrays in 45 primary AML samples. Patient characteristics are described in Table 1. In particular, we included cases with wt-NPM1 CN-AML and with translocations involving the MLL gene, which are known to have low levels of miR-29b expression.11,20 We observed that 573 probe sets (484 genes) correlated inversely, whereas 716 probe sets (622 genes) correlated positively with miR-29b expression (P < .01, Spearman correlation within BRB tools; supplemental Table 4). Among the probe sets that were inversely correlated with miR-29b, 30 probes (25 genes) were miR-29b predicted targets according to TargetScan.25 Notably, MCL-1 was found inversely correlated to miR-29b (P < .001). A very similar mRNA signature was correlated with miR-29a: inverse correlation 325 probes (266 genes) and positive correlation 331 probes (281 genes) (P < .01; supplemental Table 5). Approximately 42% of the miR-29a correlated genes were also correlated with miR-29b (274 from 656 probes).
Patient characteristics
| Characteristic . | Value . |
|---|---|
| Median age, y (range) | 59.6 (23-81) |
| Sex, no. (%) | |
| Female | 14 (31) |
| Male | 31 (69) |
| Median bone marrow blasts, % (range) | 62.2 (25-91) |
| FAB, no. (%) | |
| M0-M1 | 8 (18) |
| M2 | 6 (13) |
| M3 | 4 (9) |
| M4-M5 | 24 (53) |
| Unknown | 3 (7) |
| Cytogenetics, no. (%) | |
| Normal karyotype | 31 (69) |
| FLT3-ITD | 12 |
| FLT3-TDK | 7 |
| NPM1 | 20 |
| inv16 | 2 (4) |
| t(11q23) | 6 (13) |
| t(8;21) | 1 (2) |
| t(15;17) | 4 (9) |
| del9q | 1 (2) |
| Characteristic . | Value . |
|---|---|
| Median age, y (range) | 59.6 (23-81) |
| Sex, no. (%) | |
| Female | 14 (31) |
| Male | 31 (69) |
| Median bone marrow blasts, % (range) | 62.2 (25-91) |
| FAB, no. (%) | |
| M0-M1 | 8 (18) |
| M2 | 6 (13) |
| M3 | 4 (9) |
| M4-M5 | 24 (53) |
| Unknown | 3 (7) |
| Cytogenetics, no. (%) | |
| Normal karyotype | 31 (69) |
| FLT3-ITD | 12 |
| FLT3-TDK | 7 |
| NPM1 | 20 |
| inv16 | 2 (4) |
| t(11q23) | 6 (13) |
| t(8;21) | 1 (2) |
| t(15;17) | 4 (9) |
| del9q | 1 (2) |
GO analysis of genes regulated by miR-29a and -29b in primary AML samples
Genes found negatively correlated with miR-29a and -29b in primary AML samples were analyzed with DAVID functional annotation tool to identify GO categories most represented in this subset (Table 2).24 Consistent with the cell line data, these results show that expression of miR-29a and -29b directly or indirectly affect the expression of apoptosis-related genes (Table 2). However, there were some differences between the miR-29a and -29b signatures. For example, several GO terms related to protein metabolism processes, such as posttranslational protein modification, biopolymer modification, and protein modification process, were found overrepresented only in miR-29b correlated genes (Table 2). Likewise, GO terms related to immune function, such as defense response, inflammatory response, and response to wounding were enriched only in miR-29a–correlated genes (Table 2). We also analyzed GO categories most represented in genes that were positively correlated with miR-29b. Interestingly, we found a significant enrichment of genes belonging to RNA regulation and gene transcription, including RNA metabolic process, regulation of nucleic acid metabolic process, gene expression, and regulation of transcription (Table 2). No GO categories were enriched in genes positively correlated to miR-29a.
Most significant GO categories that are correlated with miR-29 expression in primary AML samples
| . | Count . | P . | Benjamini . |
|---|---|---|---|
| GO categories enriched in genes that were negatively correlated with miR-29a and -29b expression | |||
| Annotation Cluster 1 (enrichment score 5.09) | |||
| Regulation of biologic process | 163 | <.001 | 0.009 |
| Biologic regulation | 175 | <.001 | 0.005 |
| Regulation of cellular process | 152 | <.001 | 0.005 |
| Annotation Cluster 2 (enrichment score 4.76) | |||
| Cell death | 44 | <.001 | 0.005 |
| Death | 44 | <.001 | 0.005 |
| Apoptosis | 42 | <.001 | 0.005 |
| Programmed cell death | 42 | <.001 | 0.006 |
| Cell development | 53 | <.001 | 0.066 |
| GO categories enriched in genes that were negatively correlated only with miR-29b expression | |||
| Annotation Cluster 1 (enrichment score 5.27) | |||
| Posttranslational protein modification | 69 | <.001 | 0.013 |
| Protein modification process | 77 | <.001 | 0.014 |
| Biopolymer modification | 78 | <.001 | 0.005 |
| Annotation Cluster 2 (enrichment score 3.25) | |||
| Protein metabolic process | 124 | <.001 | 0.092 |
| GO categories enriched in genes that were negatively correlated only with miR-29a expression | |||
| Annotation Cluster 1 (enrichment score 4.93) | |||
| Defense response | 25 | <.001 | 0.001 |
| Immune response | 27 | <.001 | 0.140 |
| Immune system process | 31 | <.001 | 0.120 |
| Response to stimulus | 52 | 0.016 | 0.980 |
| Annotation Cluster 2 (enrichment score 4.93) | |||
| Defense response | 25 | <.001 | 0.001 |
| Inflammatory response | 15 | <.001 | 0.045 |
| Response to wounding | 16 | <.001 | 0.230 |
| Response to external stimulus | 19 | .001 | 0.550 |
| Response to stress | 21 | .051 | 1 |
| GO categories enriched in genes that were positively correlated only with miR-29b expression | |||
| Annotation Cluster 1 (enrichment score 13.5) | |||
| Intracellular organelle | 379 | <.001 | <0.001 |
| Organelle | 379 | <.001 | <0.001 |
| Intracellular membrane-bound organelle | 339 | <.001 | <0.001 |
| Membrane-bound organelle | 339 | <.001 | <0.001 |
| Annotation Cluster 2 (enrichment score 11.9) | |||
| Regulation of cellular process | 223 | <.001 | <0.001 |
| Regulation of biologic process | 231 | <.001 | <0.001 |
| Biologic regulation | 244 | <.001 | <0.001 |
| Annotation Cluster 3 (enrichment score 9.11) | |||
| Primary metabolic process | 349 | <.001 | <0.001 |
| Macromolecule metabolic process | 311 | <.001 | <0.001 |
| Cellular metabolic process | 339 | <.001 | <0.001 |
| Metabolic process | 364 | <.001 | <0.001 |
| Annotation Cluster 4 (enrichment score 6.9) | |||
| RNA metabolic process | 157 | <.001 | <0.001 |
| Regulation of metabolic process | 152 | <.001 | <0.001 |
| Regulation of cellular metabolic process | 145 | <.001 | <0.001 |
| Regulation of nucleic acid metabolic process | 136 | <.001 | <0.001 |
| Gene expression | 175 | <.001 | <0.001 |
| Transcription | 137 | <.001 | <0.001 |
| Regulation of gene expression | 138 | <.001 | <0.001 |
| Regulation of transcription, DNA-dependent | 124 | <.001 | <0.001 |
| Regulation of transcription | 130 | <.001 | <0.001 |
| Transcription, DNA-dependent | 125 | <.001 | <0.001 |
| RNA biosynthetic process | 125 | <.001 | <0.001 |
| Annotation Cluster 5 (enrichment score 6.34) | |||
| Zinc ion binding | 124 | <.001 | <0.001 |
| Transition metal ion binding | 139 | <.001 | 0.003 |
| Annotation Cluster 6 (enrichment score 4.99) | |||
| Chromatin modification | 23 | <.001 | <0.001 |
| Establishment and/or maintenance of chromatin architecture | 27 | <.001 | 0.004 |
| Chromosome organization and biogenesis | 31 | <.001 | 0.004 |
| DNA packaging | 27 | <.001 | 0.005 |
| . | Count . | P . | Benjamini . |
|---|---|---|---|
| GO categories enriched in genes that were negatively correlated with miR-29a and -29b expression | |||
| Annotation Cluster 1 (enrichment score 5.09) | |||
| Regulation of biologic process | 163 | <.001 | 0.009 |
| Biologic regulation | 175 | <.001 | 0.005 |
| Regulation of cellular process | 152 | <.001 | 0.005 |
| Annotation Cluster 2 (enrichment score 4.76) | |||
| Cell death | 44 | <.001 | 0.005 |
| Death | 44 | <.001 | 0.005 |
| Apoptosis | 42 | <.001 | 0.005 |
| Programmed cell death | 42 | <.001 | 0.006 |
| Cell development | 53 | <.001 | 0.066 |
| GO categories enriched in genes that were negatively correlated only with miR-29b expression | |||
| Annotation Cluster 1 (enrichment score 5.27) | |||
| Posttranslational protein modification | 69 | <.001 | 0.013 |
| Protein modification process | 77 | <.001 | 0.014 |
| Biopolymer modification | 78 | <.001 | 0.005 |
| Annotation Cluster 2 (enrichment score 3.25) | |||
| Protein metabolic process | 124 | <.001 | 0.092 |
| GO categories enriched in genes that were negatively correlated only with miR-29a expression | |||
| Annotation Cluster 1 (enrichment score 4.93) | |||
| Defense response | 25 | <.001 | 0.001 |
| Immune response | 27 | <.001 | 0.140 |
| Immune system process | 31 | <.001 | 0.120 |
| Response to stimulus | 52 | 0.016 | 0.980 |
| Annotation Cluster 2 (enrichment score 4.93) | |||
| Defense response | 25 | <.001 | 0.001 |
| Inflammatory response | 15 | <.001 | 0.045 |
| Response to wounding | 16 | <.001 | 0.230 |
| Response to external stimulus | 19 | .001 | 0.550 |
| Response to stress | 21 | .051 | 1 |
| GO categories enriched in genes that were positively correlated only with miR-29b expression | |||
| Annotation Cluster 1 (enrichment score 13.5) | |||
| Intracellular organelle | 379 | <.001 | <0.001 |
| Organelle | 379 | <.001 | <0.001 |
| Intracellular membrane-bound organelle | 339 | <.001 | <0.001 |
| Membrane-bound organelle | 339 | <.001 | <0.001 |
| Annotation Cluster 2 (enrichment score 11.9) | |||
| Regulation of cellular process | 223 | <.001 | <0.001 |
| Regulation of biologic process | 231 | <.001 | <0.001 |
| Biologic regulation | 244 | <.001 | <0.001 |
| Annotation Cluster 3 (enrichment score 9.11) | |||
| Primary metabolic process | 349 | <.001 | <0.001 |
| Macromolecule metabolic process | 311 | <.001 | <0.001 |
| Cellular metabolic process | 339 | <.001 | <0.001 |
| Metabolic process | 364 | <.001 | <0.001 |
| Annotation Cluster 4 (enrichment score 6.9) | |||
| RNA metabolic process | 157 | <.001 | <0.001 |
| Regulation of metabolic process | 152 | <.001 | <0.001 |
| Regulation of cellular metabolic process | 145 | <.001 | <0.001 |
| Regulation of nucleic acid metabolic process | 136 | <.001 | <0.001 |
| Gene expression | 175 | <.001 | <0.001 |
| Transcription | 137 | <.001 | <0.001 |
| Regulation of gene expression | 138 | <.001 | <0.001 |
| Regulation of transcription, DNA-dependent | 124 | <.001 | <0.001 |
| Regulation of transcription | 130 | <.001 | <0.001 |
| Transcription, DNA-dependent | 125 | <.001 | <0.001 |
| RNA biosynthetic process | 125 | <.001 | <0.001 |
| Annotation Cluster 5 (enrichment score 6.34) | |||
| Zinc ion binding | 124 | <.001 | <0.001 |
| Transition metal ion binding | 139 | <.001 | 0.003 |
| Annotation Cluster 6 (enrichment score 4.99) | |||
| Chromatin modification | 23 | <.001 | <0.001 |
| Establishment and/or maintenance of chromatin architecture | 27 | <.001 | 0.004 |
| Chromosome organization and biogenesis | 31 | <.001 | 0.004 |
| DNA packaging | 27 | <.001 | 0.005 |
Mcl-1 protein expression is inversely expressed to miR-29b in primary AML samples
To further validate our microarray results, we correlated in primary AML samples, Mcl-1 protein, and miR-29b expression levels obtained by Western blotting and miRNA microarrays, respectively. Consistent with the mRNA microarrays data, Mcl-1 expression was opposite to that of miR-29b expression by microarrays (Figure 6A). Interestingly, 3 from 4 primary AML samples, with high level of Mcl-1 protein expression and low miR-29b levels, correspond to patient's samples characterized by heterozygous loss of chromosome 7 (Figure 6A samples 6, 8, and 9).
MiR-29 and Mcl-1 protein expression in primary AML samples with monosomy 7. (A) This panel shows the correlation between miR-29b expression by microarrays and Mcl-1 expression by Western blotting. (Top) The microarray normalized expression of miR-29b (log2 values) for 9 patients with AML (samples 1-9). Samples 6, 8, and 9 are primary AML blasts with −7 in the context of complex karyotype (> 3 cytogenetic abnormalities), whereas the others have the following karyotypes: complex karyotype (samples 1 and 4), t(9;11) (sample 2), normal karyotype (FLT-3-ITD+, NPM1 wt) (sample 3), isolated −5 (sample 5), and normal karyotype (FLT3-ITD-, NPM1 wt) (sample 7). The miR-29a microarray (log2) value for the same 1 to 9 patient samples is as follows: 8.23, 7.76. 7.36, 7.67, 8.31, 6.5, 8.1, 6.7, and 5.7. Immediately below there is the Mcl-1 protein expression by Western blotting. K562 total cell extracts were used as positive controls for Mcl-1 expression. This cell line has undetectable miR-29b expression level. GAPDH was used as a loading control for the Western blotting. (B) Chromosomal structure of the miR-29 gene family. The diagram illustrates the position and orientation of miR-29b-1 and miR-29a cluster located on human chromosome 7, and miR-29b-2 and miR-29c cluster on human chromosome 1. Note that both clusters are located in the antisense strand. (C) MiR-29a, -29b, or -29c expression by quantitative RT-PCR in 43 primary AML samples with monosomy 7 (−7) or other cytogenetic abnormalities (Other CG). The results are shown as miRNA expression after normalization with 18s and 2ΔCt calculations. Bars represent the mean.
MiR-29 and Mcl-1 protein expression in primary AML samples with monosomy 7. (A) This panel shows the correlation between miR-29b expression by microarrays and Mcl-1 expression by Western blotting. (Top) The microarray normalized expression of miR-29b (log2 values) for 9 patients with AML (samples 1-9). Samples 6, 8, and 9 are primary AML blasts with −7 in the context of complex karyotype (> 3 cytogenetic abnormalities), whereas the others have the following karyotypes: complex karyotype (samples 1 and 4), t(9;11) (sample 2), normal karyotype (FLT-3-ITD+, NPM1 wt) (sample 3), isolated −5 (sample 5), and normal karyotype (FLT3-ITD-, NPM1 wt) (sample 7). The miR-29a microarray (log2) value for the same 1 to 9 patient samples is as follows: 8.23, 7.76. 7.36, 7.67, 8.31, 6.5, 8.1, 6.7, and 5.7. Immediately below there is the Mcl-1 protein expression by Western blotting. K562 total cell extracts were used as positive controls for Mcl-1 expression. This cell line has undetectable miR-29b expression level. GAPDH was used as a loading control for the Western blotting. (B) Chromosomal structure of the miR-29 gene family. The diagram illustrates the position and orientation of miR-29b-1 and miR-29a cluster located on human chromosome 7, and miR-29b-2 and miR-29c cluster on human chromosome 1. Note that both clusters are located in the antisense strand. (C) MiR-29a, -29b, or -29c expression by quantitative RT-PCR in 43 primary AML samples with monosomy 7 (−7) or other cytogenetic abnormalities (Other CG). The results are shown as miRNA expression after normalization with 18s and 2ΔCt calculations. Bars represent the mean.
MiR-29a and -29b are down-regulated in primary AML samples with monosomy 7
Because the miR-29b-1/miR-29a cluster is located in chromosome 7 (7q32; Figure 6B), we hypothesized that primary AML samples with genomic loss of this region (eg, monosomy 7) may express low miR-29a and 29b levels. Therefore, we analyzed miR-29a, -29b, and -29c expression in primary AML samples with monosomy 7 (n = 8), either isolated or in context of other cytogenetic abnormalities, versus cases with other karyotype abnormalities (n = 35) using quantitative RT-PCR (supplemental Table 6). As shown in Figure 6C, miR-29a and -29b levels were lower in AML patient samples with monosomy 7 with respect to those with other karyotypes (miR-29a, P < .001; and miR-29b, P = .04, t test). However, miR-29c expression levels were not different between these 2 groups (P = .07, t test). MiR-29c resides on chromosome 1q32.2 (Figure 6C). These results suggest that genomic deletion of the miR-29b-1/miR-29a cluster could be responsible for the loss of miR-29a and -29b expression in this subset of AML patients. Further studies with larger numbers of patients will be needed to confirm these intriguing results.
Discussion
The data presented here indicate that forced expression of miR-29a and -29b in AML cell lines and in primary AML blasts inhibited cell growth and induced apoptosis. Interestingly, the antiapoptotic effect of miR-29s overexpression in AML cell lines was observed after 48 hours of transfection, whereas the antiproliferative effect was noticed earlier at 24 hours. These results indicate that miR-29–dependent cell proliferation effects are not the result of apoptosis and are both likely to contribute to the tumor suppressor activity of these miRNAs. The tumor suppressor effect of miR-29b in AML was further demonstrated in vivo.
To characterize the miR-29 tumor suppressor function in leukemia and identify the pathways regulated by miR-29, we analyzed the expression changes in the K562 cell line after overexpressing miR-29a or -29b. This is an effective strategy to discover target genes and identify pathways regulated by miRNAs.32,33 The results indicated that there is a significant enrichment for miR-29-predicted targets among the down-regulated genes after transfection of miR-29s. Many of the down-regulated genes are involved in critical pathways, such as apoptosis, cell cycling, and proliferation. For example, miR-29a and -29b regulate critical antiapoptotic genes, such as MCL-1. The Bcl-2 family is composed of both proapoptotic and antiapoptotic proteins, and the balance between these proteins seems to determine life or death for the cell.34 Bcl-2, Mcl-1, and Bcl-Xl are antiapoptotic proteins and promote cell survival and proliferation. In contrast, the BH3-only proteins (Bim, tBid, and Puma) are proapoptotic because of their ability to restore Bax/Bad activity and induce cell death.34 Overexpression of MCL-1 has been reported in cancer, including in AML at relapse.28-35 A recent proteomics profiling study identified that the combination of mutant P53, high levels of Mcl-1, and neurophilin-1 is associated with worse outcome in primary AML samples.36 Our results indicate that miR-29a and -29b targets Mcl-1 and induces apoptosis in AML. Both miR-29a and -29b not only directly target antiapoptotic genes but also up-regulate proapoptotic genes, such as BIM (BCL2L11) and the tumor suppressor programmed cell death-4 (PDCD4).37,38 Thus, by targeting MCL-1 and up-regulating BIM and PDCD4 transcripts, the impact of the mir-29s on the survival of AML cells could be even more robust.
We have also analyzed the correlation between miR-29a or -29b expression and mRNA expression in 45 primary AML samples. Consistent with the cell line data, apoptosis genes, such as MCL-1, were enriched in the primary blasts mRNAs that inversely correlated to miR-29a or -29b expression levels. This result was also observed at the protein level in a few primary AML samples. However, further studies using large numbers of primary AML samples will be needed to confirm this interaction.
Although there were common genes among the K562 cell line and the primary AML Affymetrix signatures, substantial differences were expected: (1) the cell line profile was obtained shortly after ectopic expression of the miR-29s, whereas the primary AML samples represent a “steady-state” balance between the miRNA and their targets; and (2) the obvious lack of stroma and immune response in the cell line system.
Another interesting finding was the enrichment for genes involved in DNA transcription and RNA metabolism among the genes positively correlated only with miR-29b but not miR-29a. Because miR-29b is also located in the nucleus,39 miR-29b might directly regulate gene transcription by a different mechanism than RNA-induced silencing complex mediated protein translation inhibition and/or mRNA cleavage.
In conclusion, we demonstrate “in vitro” and “in vivo” that miR-29b is a tumor suppressor gene by targeting multiple critical oncogenic pathways. Restoring miR-29b expression down-modulates MCL-1, induces apoptosis, and dampens cell growth in AML cells, thereby suggesting miRNA-based therapy as a novel proapoptotic approach to increase response in AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Wenjing Chen for assistance with primary AML samples handling, Prof Key Huebner for critically reviewing the manuscript, and Sharon Palko for administrative assistance.
This work was supported by the National Institutesof Health (grant P01CA055164, Lauri Strauss Discovery grant, and Kimmel translational award).
The Ohio State University has applied for a patent on clinical use of miR-29 family in cancer.
National Institutes of Health
Authorship
Contribution: C.E.A.H., V.H., M.F., and T.T. conducted functional experiments; S.V. analyzed the microarray data and performed statistical analysis of the data; N.Z., C.E.A.H., and V.H. performed the animal work; R.G., S.M.K., G.M., G.A.C., M.A., and C.M.C. designed experiments and analyzed data; R.G. wrote the manuscript; S.M.K. and M.A. obtained the patient samples; and all the authors critically reviewed and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Carlo M. Croce, The Ohio State University, Comprehensive Cancer Center, Wiseman Hall Rm 385, 400 12th Ave, Columbus, OH 43210; e-mail: carlo.croce@osumc.edu; or Ramiro Garzon, The Ohio State University, Comprehensive Cancer Center, Wiseman Hall Rm 385, 400 12th Ave, Columbus, OH 43210; e-mail: ramiro.garzon@osumc.edu.


![Figure 4. Validation of the microarray analysis. (A) Quantitative RT-PCR of MCL-1, CXXC6 (B) and CDK6 (C) after 24 hours of transfection with synthetic miR-29a, -29b, or scrambled oligonucleotides in K562 cells. The results are shown as average mRNA expression after normalization with 18s and 2ΔCt calculations. Data represent the average of 3 independent experiments ± SD. (D) Western blotting of Mcl-1 and Cdk6 protein expression in K562 and Kasumi-1 cells (E) after 48 hours of transfection with synthetic miR-29a, -29b, or controls (scrambled oligonucleotides [SCR] and miR-212) or lentivirus miR-29a, -29b, or empty vector (EV) for Kasumi-1 cells. The protein loading control was performed using GAPDH. (F) Rb phosphorylation at Ser780 after 24 hours of transfection with synthetic miR-29b or control (SCR). (G) Quantitative RT-PCR of MCL-1, CXXC6, and CDK6 in 3 primary AML blasts after transfection with synthetic miR-29b or controls (SCR). The results are shown as average mRNA expression after normalization with 18s and 2ΔCt calculations. Bars represent mean ± SEM. (H) Western blotting of Mcl-1 in 3 newly diagnosed AML patients after transfection with synthetic miR-29b or controls (SCR). Loading control was performed using GAPDH. Mcl-1/GAPDH ratios are shown after densitometry evaluation of the gels. (I) Normalized Mcl-1 protein expression averages relative the mock-transfected primary AML samples. Bars represent SD.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/26/10.1182_blood-2009-03-211938/4/m_zh89990946010004.jpeg?Expires=1765894292&Signature=yF08x29~4TsjQ6FnF5-bFS4RbReNTFgzDOGLs0MCkKX6C0YPDPoLZVL0P1ZiuDTHq0RDh2SaSqHbfC9XuUPEQFimt2DO1GOYehMZsq2y5HGXEkdo0HXCXXYTm2kRqhxX79NxM~lM40G6zg0UviaFNozEDdcnJ28Hik~ao12uk1JOiyuZF67gzX~hcxuxnfzzsN5f0UJIJ0dBAa~EsDBYaY8DWUmNXpvlrTK6xj5hX~kcQV-YE2Or0rI3-IAlw7SvO6yGGxeaWMNITnWyTvYcac260JwXZe7ttMrh67v44QDB8JRLN3JMvUlWq44Vt8G4NWTK-ei2nsbHYGPbpi-AOQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal