Abstract
Somatic mutations of Kit have been found in leukemias and gastrointestinal stromal tumors. The proto-oncogene c-Cbl negatively regulates Kit and Flt3 by its E3 ligase activity and acts as a scaffold. We recently identified the first c-Cbl mutation in human disease in an acute myeloid leukemia patient, called Cbl-R420Q. Here we analyzed the role of Cbl mutants on Kit-mediated transformation. Coexpression of Cbl-R420Q or Cbl-70Z with Kit induced cytokine-independent proliferation, survival, and clonogenic growth. Primary murine bone marrow retrovirally transduced with c-Cbl mutants and transplanted into mice led to a generalized mastocytosis, a myeloproliferative disease, and myeloid leukemia. Overexpression of these Cbl mutants inhibited stem cell factor (SCF)–induced ubiquitination and internalization of Kit. Both Cbl mutants enhanced the basal activation of Akt and prolonged the ligand-dependent activation. Importantly, transformation was observed also with kinase-dead forms of Kit and Flt3 in the presence of Cbl-70Z, but not in the absence of Kit or Flt3, suggesting a mechanism dependent on receptor tyrosine kinases, but independent of their kinase activity. Instead, transformation depends on the Src family kinase Fyn, as c-Cbl coimmunoprecipitated with Fyn and inhibition abolished transformation. These findings may explain primary resistance to tyrosine kinase inhibitors targeted at receptor tyrosine kinases.
Introduction
Receptor tyrosine kinases (RTKs) play a pivotal role in embryonic development and in hematopoiesis. Extracellular binding of a ligand to its respective RTK induces dimerization followed by intracellular signaling.1 The amplitude and duration of RTK signaling are tightly controlled; termination of RTK signaling occurs by receptor ubiquitination, internalization, and degradation.2-4 Ubiquitination requires an E3 ubiquitin ligase, which leads to the covalent attachment of the ubiquitin molecule to the target protein.5,6 Cbl proteins are RING domain-based E3 ligases with 3 mammalian homologs; c-Cbl, Cbl-b,7 and Cbl-c.8 c-Cbl consists of a tyrosine kinase binding domain, a linker domain, a RING finger, a proline-rich region, and finally a ubiquitin-associated domain overlapping with a leucine zipper motif.9 The highly conserved Cbl RING finger domain possesses E3 ligase activity and recruits E2 enzymes for the transfer of ubiquitin to substrates.10-12 It has been shown that Cbl directs monoubiquitination and polyubiquitination on multiple sites of RTKs, for example, for epidermal growth factor receptor (EGFR) and platelet-derived growth factor receptor (PDGFR), which then leads to internalization and degradation in the lysosomes.2,13,14
Mutations in either the tyrosine kinase binding domain, linker, or RING finger domain of Cbl result in dysregulation of RTKs.15-17 Most of these mutants are thought to act in a dominant-negative manner by competing with the wild-type receptor. V-Cbl, a mutant form of Cbl, has been found in Cas NS-1 retrovirus as a fusion protein. It lacks the C-terminus part of wild-type c-Cbl and can induce pre–B-cell lymphomas and leukemia in mice.18 Cbl-70Z is a 17-amino acid deletion mutant (at the boundary of the linker and RING finger domain) isolated from 70Z/3 mouse pre–B-cell lymphoma.17 We recently identified the first c-Cbl oncogenic mutation in human disease in an acute myeloid leukemia (AML) patient, called Cbl-R420Q, and analyzed the role of this Cbl mutant in Flt3 signaling.19 Cbl-R420Q and Cbl-70Z were able to confer interleukin-3 (IL-3)–independent growth to the 32Dcl3 cell line in the presence of wild-type Flt3 receptor.19 Subsequently, several other Cbl mutations were found in AML patients.20-24 Cbl proteins are not only important for RTK signal termination; they also mediate positive RTK signaling events to downstream effectors. Cbl has been shown to bind to signaling molecules, including the Src family kinases.25,26
Cbl has also been shown to bind to several RTKs, including the Kit receptor. Kit is a member of class III RTK family, structurally related to PDGFR, having 5 immunoglobulin-like domains, a single transmembrane helix, a cytoplasmic juxtamembrane domain, and split kinase domain.27-29 Gain-of-function mutations in Kit can lead to a malignant disease, including gastrointestinal stromal tumors and leukemia.27,30,31 In AML, Kit involves mutations at residue 816, which is encoded by exon 17.32-34 Cbl binds to Kit directly35 and indirectly via Grb2.36 Cbl is known to be phosphorylated by Src family kinases (SFKs)37,38 and mediates ubiquitination35 of the Kit receptor on activation. The role of Cbl in Kit signaling in the context of leukemia is not well understood.
Here, we found that mutant Cbl proteins, Cbl-R420Q and Cbl-70Z, displayed in vitro transforming activity for myeloid cells that was dependent on the presence of Kit. These Cbl mutants altered internalization, ubiquitination, and downstream signaling of the Kit receptor. Surprisingly, we found that kinase activity of Kit and Flt3 was not required for the transformation of Cbl-70Z. We were able to show the contribution of Src family kinases for this synergistic transformation of Cbl-70Z and kinase-dead (KD) forms of Kit and Flt3. These data suggest a kinase-independent role of Kit and Flt3 for Cbl-dependent cellular functions. Retroviral expression of Cbl mutants in transplanted bone marrow induced a generalized mastocytosis, a myeloproliferative disease and, in rare care cases, myeloid leukemia. To our knowledge, this is the first report of an in vivo model investigating the role of Cbl mutations in malignant transformation. These results point toward their critical role in preventing RTK signaling termination.
Methods
Reagents and antibodies
Recombinant human Flt3 ligand (FL), murine stem cell factor (SCF), and murine IL-3 were purchased from PeproTech. Polyclonal rabbit anti–phospho-Erk-1/2, anti–phospho-Kit (Tyr-719), anti–phospho-Src family (Tyr-416), anti–phospho-c-Cbl (Tyr-731), and anti–phospho-Akt antibodies were obtained from Cell Signaling Technology. The antibodies for Kit, Flt3, HA-tag, signal transducer, and activator of transcription 5a/b (anti-STAT5a/b), Src, Yes, Fyn, Lyn, Hck, and Lck were purchased from Santa Cruz Biotechnology. The mouse monoclonal anti–c-Cbl antibody was obtained from BD Biosciences. Antibodies for anti–Erk-1/2 and anti–phospho-STAT5a/b were purchased from Upstate Biotechnology. The mouse monoclonal antiactin antibody was purchased from Sigma-Aldrich. Phycoerythrin (PE)–labeled murine anti-Kit antibody was obtained from BD Biosciences PharMingen. The Src family inhibitors PP-1 and PP-2 and the Akt inhibitor 124005 were purchased from Calbiochem. Dasatinib was obtained from Bristol-Myers Squibb and imatinib from L C Laboratories. The PI3-kinase (PI3K) inhibitor LY294002 was purchased from Cell Signaling Technology.
Cell lines
The 32Dcl3 and COS-7 cell lines were cultured as described previously.19 To generate stable cell lines, 32D cells were electroporated with 10 μg of pAL Kit-WT, pAL Kit-D790N (Kit-KD), or pAL Flt3-K644A (Flt3-KD) together with 1 μg of pMAM/BSD (Kaken Pharmaceutical) as selection marker in 0.4-cm cuvettes at 280 V and 975 μF. Cells were selected with 15 μg/mL blasticidin (Invitrogen) in IL-3–supplemented culture. Later these cells were infected with retroviral supernatants of pMY Cbl-R420Q, pMY Cbl-70Z, or pMY (empty vector). Transduced cells were sorted with the help of green fluorescent protein marker using flow cytometry. To avoid possible clonal selection, at least 2 bulk cultures were generated for each cell line. All cell lines were maintained as previously described.19
Retrovirus preparation
Platinum-E ecotropic packaging cells39 and NIH3T3 cells were maintained in Dulbecco modified Eagle medium (Invitrogen) supplemented with 10% fetal calf serum (FCS). Platinum-E cells were transiently transfected using Lipofectamine 2000 (Invitrogen), and retroviral stocks were collected twice at 12-hour intervals beginning 24 hours after transfection. Retrovirus was titered by transduction of 5 × 104 NIH3T3 cells with serial dilutions of retrovirus in the presence of 4 μg/mL polybrene (Sigma-Aldrich). Percentage of infected cells was determined by flow cytometric analysis of enhanced green fluorescent protein (EGFP) expression after 48 hours. The titer was calculated and tested for the presence of replication-competent viral particles by transferring supernatant onto NIH3T3 cells. The NIH3T3 cells were subsequently analyzed for EGFP expression by fluorescence-activated cell scanner (FACS) analysis.
Transduction and transplantation of murine bone marrow
Murine bone marrow was harvested from female Balb/C donor mice 4 days after injection of 150 mg/kg 5-fluorouracil (Ribosepharm) and prestimulated overnight in Iscove modified Dulbecco medium/20% FCS supplemented with growth factors (10 ng/mL murine IL-3 [mIL-3], 10 ng/mL mIL-6, 50 ng/mL mSCF). Cells were transduced by 4 rounds of spin infection (1200g, 32°C, 90 minutes) every 12 hours in retroviral supernatant supplemented with growth factors and 4 μg/mL polybrene (Sigma-Aldrich). Retroviral experiments were performed using the murine stem cell virus-based retroviral construct pMY.40 Subsequently, cells were resuspended in Hanks balanced salt solution (Sigma-Aldrich) and injected into the tail vein of lethally irradiated (8 Gy) female Balb/C recipient mice. Transplanted animals were monitored for signs of disease development and by serial measurement of peripheral blood counts. All procedures were reviewed and approved by the University of Frankfurt supervisory animal care committee.
Flow cytometric immunophenotyping (FACS analysis)
Single-cell suspensions of indicated tissue samples were prepared, and red blood cells of peripheral blood were lysed before analysis. Cells were subsequently stained with PE-conjugated anti-CD117 (Kit), anti-CD11b (Mac-1), anti–Gr-1, anti-CD45R/B220, or anti-CD3 antibodies. All antibodies were purchased from BD Biosciences PharMingen.
Ubiquitination assays and immunoprecipitations
COS-7 cells were transfected transiently with the indicated constructs together with a plasmid for HA-tagged ubiquitin (Ubq) using Nanofectin according to the manufacturer's protocol. At 48 hours after transfection, cells were serum starved for 12 hours and then stimulated with 50 ng/mL SCF for 10 minutes. Cells were lysed as described before.19 For immunoprecipitation, cell lysates were incubated with goat polyclonal antibody against murine Kit and with protein A/G-Plus-Sepharose. The immunoprecipitates were washed 3 times with lysis buffer. Immunoprecipitates as well as total lysates were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoreses (SDS-PAGE) and probed with anti-HA (anti-Ubq) or anti-Kit antibodies. Immunoprecipitations and Western blot analyses were performed as described previously.19
Analysis of cell growth
The 32D cells expressing the indicated constructs were washed twice and resuspended in RPMI 1640 with 10% FCS alone or supplemented with the indicated growth factors or with inhibitors at 2 × 105 cells/mL. Viable cells were determined by trypan blue exclusion and were counted daily.
3[H]-thymidine incorporation
A total of 3 × 104 cells were starved from IL-3 in 10% FCS for 12 hours, subsequently placed in the presence of indicated cytokines, nothing, or either 15 μM of PP-1 or PP-2. After 6 hours of incubation at 37°C with 5% CO2, 0.037 MBq (1 μCi)3[H]-thymidine was added to each well, cells were harvested after incubation of 15 hours, and DNA was analyzed with scintillation counter. Experiments were repeated at least 3 times.
Site-directed mutagenesis
To generate Kit KD (Kit-D790N), aspartic acid 790 of Kit-WT was replaced with asparagine (confirmed by sequencing), using mutagenesis Kit (Stratagene).
Internalization assays
Internalization assay was as described before19 ; briefly, 32D or COS-7 (5 × 105) cells expressing indicated constructs were incubated with SCF for the indicated time points. Internalization of the receptor was stopped by adding ice-cold PBS containing 0.4% sodium azide. After washing, the cells were stained with PE-labeled anti-Kit antibody and fluorescence intensity was analyzed by flow cytometry.
Clonal growth in methylcellulose
Clonal growth was performed as described previously.41 Briefly, stably transfected 32D cells expressing Kit-WT or Kit-KD with the indicated Cbl constructs were seeded at a concentration of 103 cells/dish in the absence of any growth factors. The assays were plated as triplicates, and colonies were photographed and counted on day 6.
Results
Cbl mutants (Cbl-R420Q and Cbl-70Z) confer ligand-independent growth in cooperation with Kit
Recently, we published the role of 2 dominant-negative Cbl mutants, Cbl-R420Q and Cbl-70Z, in Flt3 signaling.19 Because early hematopoietic stem cells and leukemic progenitor cells express high levels of the Kit receptor tyrosine kinase, we were interested to analyze whether dominant-negative Cbl proteins would synergize with this receptor. First, we generated IL-3–dependent 32D cell lines stably expressing Kit (Kit-WT) and/or Cbl mutants and analyzed proliferation and viability in the presence or absence of the Kit ligand SCF.
Interestingly, cells expressing either Cbl mutant in the presence of Kit-WT rapidly proliferated in the absence of exogenous SCF or IL-3 (Figure 1A). However, in the absence of Cbl mutants, cells coexpressing Kit-WT alone proliferated only in the presence of SCF (data not shown). Proliferation was more pronounced in the cells coexpressing Cbl-70Z and Kit-WT compared with cells coexpressing Cbl-R420Q and Kit-WT. Cultures expressing one of the Cbl mutants with Kit-WT survived for extended time periods in the absence of any growth factors (Figure 1B). Importantly, 32D cells expressing either of the Cbl mutants or Kit-WT alone were not able to proliferate ligand-independently and survived for a shorter duration (Figure 1B).
Cbl mutants synergize with Kit to induce autonomous growth and in vitro colony formation. (A-B) Cbl-70Z and Cbl-R420Q induce ligand-independent proliferation and survival of 32D-Kit-WT cells. The 32D cells stably overexpressing Kit-WT and/or indicated Cbl mutants were starved from IL-3, and cells were grown in the presence of 10% FCS. Cells were counted at the indicated time points by the trypan blue exclusion method, and the data are shown as fold change of the cell number compared with the start of the experiment (A). (B) The percentage of cells in the culture that was alive at the indicated time points. (C) Ligand-independent DNA synthesis of Kit-WT cells coexpressing Cbl-70Z. The 32D cells stably expressing Kit-WT and/or Cbl constructs were starved from IL-3 for 12 hours and then cultured in the presence or absence of SCF or IL-3. Proliferation was measured by 3[H]-thymidine incorporation assays. Data are shown as percentage of thymidine incorporation relative to the thymidine incorporation of the respective cell line with IL-3 supplementation. (A-C) Data represent the average and SD of 3 independent experiments. (D-E) Cbl mutants led to cytokine-independent colony growth. The 32D cells stably overexpressing Kit-WT and/or indicated Cbl mutants were serum starved for 12 hours and then plated at a concentration of 1000 cells per dish in the absence of any growth factors. Colonies were counted on day 6. The assays were plated as triplicates. The numbers given show the results of 1 of at least 3 independent experiments per construct, which all gave similar results. (E) Photographs of the dishes were taken on day 6 and show morphologic differences in clonal growth.
Cbl mutants synergize with Kit to induce autonomous growth and in vitro colony formation. (A-B) Cbl-70Z and Cbl-R420Q induce ligand-independent proliferation and survival of 32D-Kit-WT cells. The 32D cells stably overexpressing Kit-WT and/or indicated Cbl mutants were starved from IL-3, and cells were grown in the presence of 10% FCS. Cells were counted at the indicated time points by the trypan blue exclusion method, and the data are shown as fold change of the cell number compared with the start of the experiment (A). (B) The percentage of cells in the culture that was alive at the indicated time points. (C) Ligand-independent DNA synthesis of Kit-WT cells coexpressing Cbl-70Z. The 32D cells stably expressing Kit-WT and/or Cbl constructs were starved from IL-3 for 12 hours and then cultured in the presence or absence of SCF or IL-3. Proliferation was measured by 3[H]-thymidine incorporation assays. Data are shown as percentage of thymidine incorporation relative to the thymidine incorporation of the respective cell line with IL-3 supplementation. (A-C) Data represent the average and SD of 3 independent experiments. (D-E) Cbl mutants led to cytokine-independent colony growth. The 32D cells stably overexpressing Kit-WT and/or indicated Cbl mutants were serum starved for 12 hours and then plated at a concentration of 1000 cells per dish in the absence of any growth factors. Colonies were counted on day 6. The assays were plated as triplicates. The numbers given show the results of 1 of at least 3 independent experiments per construct, which all gave similar results. (E) Photographs of the dishes were taken on day 6 and show morphologic differences in clonal growth.
Finally, we analyzed the effect of Cbl mutants on IL-3–independent and SCF-dependent proliferation in3[H]-thymidine incorporation assays. Coexpression of Cbl mutants with Kit-WT induced a pronounced proliferative growth advantage in the absence of SCF, and this growth was further enhanced after SCF stimulation (Figure 1C). Neither Cbl-R420Q– nor Kit-WT–expressing cells led to SCF-independent growth. Cells expressing Cbl-70Z alone showed a moderate growth advantage,42 which was only for a short duration (Figure 1A-B). Coexpression of Cbl-WT with Kit-WT did not show any effect on proliferation (data not shown).
To further investigate the transforming potential of the 2 mutant Cbl proteins, we examined their ability to confer clonogenic growth to 32D cells (Figure 1D-E). When expressed in 32D cells, Cbl-70Z induced the formation of very few, small colonies in the absence of growth factors (Figure 1D-E). In contrast, expression of Cbl mutants together with Kit induced the growth of numerous large and dispersed colonies (Figure 1E).
Cbl mutants induce a generalized mastocytosis and a myeloproliferative disease in a murine bone marrow transplantation model
To investigate the transforming capability of different Cbl mutants in vivo, we generated murine ecotropic retrovirus (pMY) expressing Cbl-R420Q, Cbl-70Z, or empty vector. We used a bicistronic retroviral vector coexpressing the Cbl mutants together with the EGFP via an internal ribosomal entry site.40 Bone marrow was transduced with each retrovirus at a titer of 4 × 105 retroviral particles per milliliter and analyzed by flow cytometry for EGFP expression, and the total cell number was determined. A total of 150 000 EGFP+ cells was injected into the tail vein of lethally irradiated (8 Gy) female Balb/C recipient mice. Initial control experiments showed a similar level of expression between Cbl-R420Q and Cbl-70Z (data not shown).
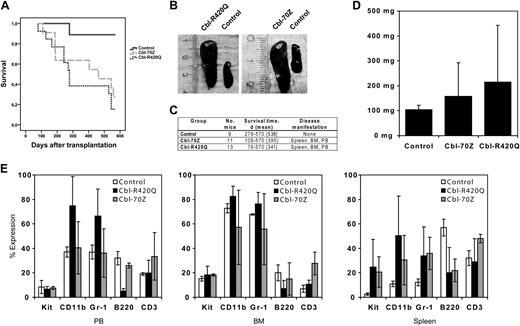
Almost all recipients of bone marrow cells transduced with Cbl mutants developed a lethal hematologic disorder with a mean latency of 341 days in the Cbl-R420Q group and 395 days in the Cbl-70Z group (Figure 2A,C). Eleven of 13 mice in the Cbl-R420Q group and 8 of 11 mice in the Cbl-70Z group died. Two animals in the Cbl-R420Q group died of a “myeloid leukemia with maturation,” matching all 5 criteria for nonlymphoid leukemia of the Bethesda proposals.43 These 2 mice showed leukocytosis of up to 140 000/μL (supplemental Figure 1B, available on the Blood website; see the Supplemental Materials link at the top of the online article) with more than 20% blasts in the peripheral blood (supplemental Figure1A) and up to 70% blast infiltration in the spleen (Figure 3). Splenomegaly (Figure 2B-D) developed with large differences in spleen weights (Figure 2D) and expansion of myeloid cells in liver and spleen were observed (Figure 3). Histology sections of spleen, liver, and bone marrow and FACS analyses of spleen, bone marrow, and peripheral blood showed extensive infiltration of blasts and mature myeloid cells (Figures 2E, 3; supplemental Figure 1D).
Expression of Cbl mutants led to a lethal mastocytosis and myeloproliferative disease in a murine bone marrow transplantation model. (A) Kaplan-Meier survival curve for recipients of bone marrow transduced with Cbl-70Z, Cbl-R420Q, or empty vector (Control). (B) Splenomegaly in mice that received transplantations of Cbl mutant-transduced bone marrow cells; empty vector (Control). (C) Analysis of mice. BM indicates bone marrow; and PB, peripheral blood. (D) Comparison of spleen weight from mice receiving bone marrow transduced with Cbl mutants. (E) Comparison of immunophenotype of peripheral blood (PB), bone marrow (BM), and spleen cells from mice receiving bone marrow transduced with Cbl mutants representing lineage-specific antigens (Kit, CD-11b, Gr-1, B220, and CD3). (D-E) Data represent the average and SD.
Expression of Cbl mutants led to a lethal mastocytosis and myeloproliferative disease in a murine bone marrow transplantation model. (A) Kaplan-Meier survival curve for recipients of bone marrow transduced with Cbl-70Z, Cbl-R420Q, or empty vector (Control). (B) Splenomegaly in mice that received transplantations of Cbl mutant-transduced bone marrow cells; empty vector (Control). (C) Analysis of mice. BM indicates bone marrow; and PB, peripheral blood. (D) Comparison of spleen weight from mice receiving bone marrow transduced with Cbl mutants. (E) Comparison of immunophenotype of peripheral blood (PB), bone marrow (BM), and spleen cells from mice receiving bone marrow transduced with Cbl mutants representing lineage-specific antigens (Kit, CD-11b, Gr-1, B220, and CD3). (D-E) Data represent the average and SD.
Cbl mutants induce a myeloid leukemia or mast cell sarcoma. Histopathology of the bone marrow (BM), liver, and spleen from mice transplanted with Cbl mutant-transduced bone marrow cells. Spleen, liver, and bone marrow sections (hematoxylin and eosin [HE], Giemsa, NACE; original magnifications, 100×-1000×) from a representative leukemic mouse (Cbl-R420Q) and a mouse with mast cell sarcoma (Cbl-70Z), which received bone marrow cells transduced with Cbl-R420Q or Cbl-70Z, respectively. Normal organ architecture is visible in a control mouse. The leukemic mouse (Cbl-R420Q) reveals a massive infiltration of immature myeloid cells with its maximum in the spleen and consecutive destruction of the normal organ structure. There are also mast cells sporadically visible in between the immature cell infiltration (arrow in the spleen pictures and Giemsa staining). In contrast, mast cells in the mice with mast-cell sarcoma have characteristic cytology with uniform, closely packed cells with round nuclei and clear, abundant cytoplasm with granules and the typical features of mast cells with numerous granules in Giemsa staining and are highly positive in NACE staining. Slides were viewed with an Olympus BX51 microscope using Olympus Soft imaging system: Cell A and the following objective lenses: 10×/0.30 UPlanFL (1st column); 40×/0.75 UPlanFL (2nd column); 100×/1.35 oil iris PlanApo (3rd and 4th columns). Images were captured on an Olympus DP70 camera and manipulated with Adobe Photoshop DP70.
Cbl mutants induce a myeloid leukemia or mast cell sarcoma. Histopathology of the bone marrow (BM), liver, and spleen from mice transplanted with Cbl mutant-transduced bone marrow cells. Spleen, liver, and bone marrow sections (hematoxylin and eosin [HE], Giemsa, NACE; original magnifications, 100×-1000×) from a representative leukemic mouse (Cbl-R420Q) and a mouse with mast cell sarcoma (Cbl-70Z), which received bone marrow cells transduced with Cbl-R420Q or Cbl-70Z, respectively. Normal organ architecture is visible in a control mouse. The leukemic mouse (Cbl-R420Q) reveals a massive infiltration of immature myeloid cells with its maximum in the spleen and consecutive destruction of the normal organ structure. There are also mast cells sporadically visible in between the immature cell infiltration (arrow in the spleen pictures and Giemsa staining). In contrast, mast cells in the mice with mast-cell sarcoma have characteristic cytology with uniform, closely packed cells with round nuclei and clear, abundant cytoplasm with granules and the typical features of mast cells with numerous granules in Giemsa staining and are highly positive in NACE staining. Slides were viewed with an Olympus BX51 microscope using Olympus Soft imaging system: Cell A and the following objective lenses: 10×/0.30 UPlanFL (1st column); 40×/0.75 UPlanFL (2nd column); 100×/1.35 oil iris PlanApo (3rd and 4th columns). Images were captured on an Olympus DP70 camera and manipulated with Adobe Photoshop DP70.
Importantly, all the animals in the Cbl-R420Q and Cbl-70Z group showed a diffuse organ infiltration (spleen, liver, bone marrow, lung, kidney, heart) of mast cells with a very variable range of mast cell infiltration (Figure 3; supplemental Figure 2A). Six mice showed massive mast cell infiltration, reminiscent of mast cell sarcoma. These cells have clear mast cell morphology with numerous granules in Giemsa staining and are highly positive in naphthol AS-D chloroacetate esterase (NACE) staining. The lesions have characteristic cytology with uniform, closely packed cells with round nuclei and clear, abundant cytoplasm with granules, which are characteristic features of mast cells (Figure 3; supplemental Figure 2B-C). One mouse had an organ infiltration of mast cells together with mature myeloid cells and 5% to 10% myeloid blast cells. Importantly, none of the control mice had histologic evidence of mastocytosis or other hematologic disease.
Thus, bone marrow transplantation of cells expressing Cbl mutants led to a generalized mastocytosis (in extreme cases mast cell sarcoma), a myeloproliferative disorder, and myeloid leukemia with a long latency and at high penetrance.
Cbl mutants inhibit ubiquitination and internalization of ligand-activated Kit
The observation that Flt3 and Kit proteins cooperated with Cbl mutants to transform myeloid cells in vitro and in vivo prompted us to study the underlying mechanisms. First, we analyzed the physical interaction between the Cbl and Kit proteins. Immunoprecipitation of overexpressed HA-tagged c-Cbl resulted in immunocomplexes that also contained Kit (Figure 4A). The association of Cbl mutants with Kit was enhanced by the presence of SCF. A consistent association of Cbl-70Z with Kit receptor was noticed in the absence of ligand (Figure 4A).
Cbl mutants bind to Kit, inhibit ubiquitination and endocytosis, and potentiate Kit-induced signaling. (A) Kit physically interacts with c-Cbl. 32D-Kit-WT cells stably transfected with HA-tagged Cbl proteins (Cbl-WT, Cbl-R420Q, and Cbl-70Z) were deprived from cytokines overnight and subsequently exposed to the indicated cytokines for 10 minutes. Cbl proteins were immunoprecipitated by anti-HA antibodies, and immunoprecipitates were resolved on SDS-PAGE. Coimmunoprecipitation of Kit was analyzed using anti-Kit antibodies. (B) Cbl mutants inhibit ubiquitination of Kit. COS-7 cells were transiently transfected with the indicated constructs together with a plasmid for HA-tagged Ubq. After 48 hours of transfection, cells were serum-starved for 12 hours and stimulated with 50 ng/mL SCF for 10 minutes or left unstimulated. Cell lysates were prepared and equal amounts of lysates were immunoprecipitated using anti-Kit antibody. The immunoprecipitates were resolved on SDS-PAGE and analyzed with anti-HA or anti-Kit antibodies. Expression of overexpressed Cbl mutants is shown in total cell lysates (TCLs) using anti-Cbl antibody. (C) Internalization of Kit is inhibited by Cbl mutants. The 32D cells stably expressing Kit in the presence or absence of different Cbl mutants were washed with PBS and then stimulated with SCF for the indicated time points. Subsequently, the amount of Kit remained on the cell surface was measured by flow cytometry after staining with a PE-labeled anti-Kit antibody. Sodium azide was used to stop the internalization. Results are expressed as mean ± SD of 3 independent experiments. (D-E) Cbl influences Kit-mediated signaling. The 32D cells with and without Kit-WT were engineered to express the indicated Cbl proteins, deprived from cytokines overnight, and subsequently exposed to the indicated cytokines for 10 minutes. Western blot analyses with the indicated antibodies were performed. (F-H) Cbl proteins change the kinetics of Kit-phosphorylation and Kit-induced Akt and Erk activity. The 32D-Kit-WT cells stably transfected with the indicated Cbl proteins were treated as in panels D and E with the exception that they were exposed to SCF for the indicated time periods. Western blot analyses using phospho-specific antibodies for Kit (F), Akt (G), and Erk1/2 (H) were performed.
Cbl mutants bind to Kit, inhibit ubiquitination and endocytosis, and potentiate Kit-induced signaling. (A) Kit physically interacts with c-Cbl. 32D-Kit-WT cells stably transfected with HA-tagged Cbl proteins (Cbl-WT, Cbl-R420Q, and Cbl-70Z) were deprived from cytokines overnight and subsequently exposed to the indicated cytokines for 10 minutes. Cbl proteins were immunoprecipitated by anti-HA antibodies, and immunoprecipitates were resolved on SDS-PAGE. Coimmunoprecipitation of Kit was analyzed using anti-Kit antibodies. (B) Cbl mutants inhibit ubiquitination of Kit. COS-7 cells were transiently transfected with the indicated constructs together with a plasmid for HA-tagged Ubq. After 48 hours of transfection, cells were serum-starved for 12 hours and stimulated with 50 ng/mL SCF for 10 minutes or left unstimulated. Cell lysates were prepared and equal amounts of lysates were immunoprecipitated using anti-Kit antibody. The immunoprecipitates were resolved on SDS-PAGE and analyzed with anti-HA or anti-Kit antibodies. Expression of overexpressed Cbl mutants is shown in total cell lysates (TCLs) using anti-Cbl antibody. (C) Internalization of Kit is inhibited by Cbl mutants. The 32D cells stably expressing Kit in the presence or absence of different Cbl mutants were washed with PBS and then stimulated with SCF for the indicated time points. Subsequently, the amount of Kit remained on the cell surface was measured by flow cytometry after staining with a PE-labeled anti-Kit antibody. Sodium azide was used to stop the internalization. Results are expressed as mean ± SD of 3 independent experiments. (D-E) Cbl influences Kit-mediated signaling. The 32D cells with and without Kit-WT were engineered to express the indicated Cbl proteins, deprived from cytokines overnight, and subsequently exposed to the indicated cytokines for 10 minutes. Western blot analyses with the indicated antibodies were performed. (F-H) Cbl proteins change the kinetics of Kit-phosphorylation and Kit-induced Akt and Erk activity. The 32D-Kit-WT cells stably transfected with the indicated Cbl proteins were treated as in panels D and E with the exception that they were exposed to SCF for the indicated time periods. Western blot analyses using phospho-specific antibodies for Kit (F), Akt (G), and Erk1/2 (H) were performed.
Under physiologic conditions, ligand activation of RTKs induces receptor internalization and degradation, which are important for RTK signal mitigation. Cbl proteins play an important role in these processes.9,44,45 Therefore, we analyzed whether Cbl mutants could abolish ubiquitination of the activated Kit receptor. COS-7 cells were transiently transfected with Kit-WT and HA-tagged Ubq in the presence or absence of Cbl mutants to study the ubiquitination. Kit was effectively ubiquitinated in the presence of endogenous Cbl after stimulation with SCF (Figure 4B), but the presence of Cbl-R420Q completely abolished ubiquitination of the activated receptor. Cbl-70Z also repressed ubiquitination, but not to the same extent as Cbl-R420Q. The expression of the Kit receptor and the Cbl mutants was similar in all the cases.
As ubiquitination often leads to endocytosis of the activated receptor,2-4 we examined the effect of Cbl mutants on endocytosis of activated Kit receptor in 32D and COS-7 cells to avoid cell type–specific effects on endocytosis. Cells transfected with Kit-WT in the presence or absence of Cbl mutants were stimulated with SCF, and the amount of Kit receptor at the surface was analyzed by FACS at the indicated time points. Cbl mutants quite effectively inhibited the initial rapid rate of ligand-induced receptor internalization (Figure 4C; supplemental Figure 3A).
Cbl mutants alter signal quality after ligand stimulation of Kit
We then analyzed the effects of Cbl mutants on the quality, amplitude, and duration of Kit signaling (Figure 4D-H). Coexpression of Cbl-70Z or Cbl-R420Q with Kit receptors induced a stronger activation of the Akt and Erk pathways as indicated by phosphorylation of the respective signaling intermediates after 10 minutes (Figure 4E). Furthermore, Akt and, to a lesser extent, Erk were phosphorylated (ie, activated) even in the absence of growth factors (Figure 4E). No activation of intracellular signaling mediators was observed when the Cbl mutants were overexpressed alone (Figure 4D). In time course experiments, we observed a prolonged activation of Kit, Akt, and Erk activity up to 240 minutes after SCF stimulation in cells expressing Cbl-70Z or Cbl-R420Q (Figure 4F-H). In addition, Cbl-70Z expression led to autophosphorylation of the Kit receptor in the absence of any ligand (Figure 4F).
Kinase activity of Kit and Flt3 is dispensable for Cbl-70Z–mediated transformation
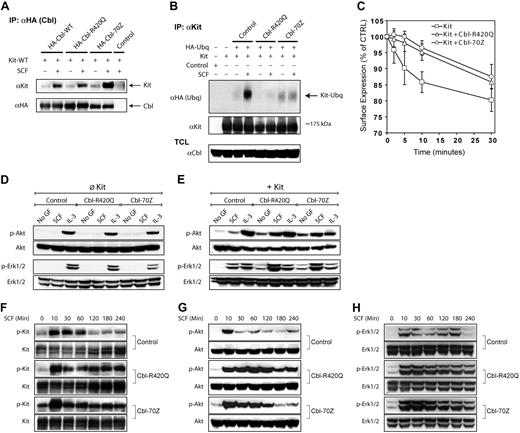
To elucidate the mechanism of transformation by Cbl mutants and Kit receptor, we generated 32D cells stably expressing KD mutants of Kit (Kit-D790N, Kit-KD) in the presence or absence of Cbl mutants. Analyzing the proliferative capacity of cells that coexpressed Kit-KD and Cbl-R420Q, we surprisingly found a subtle growth and survival advantage compared with Kit-KD alone (Figure 5A-B). This effect was more pronounced with Cbl-70Z. Interestingly, cells coexpressing Kit-KD and Cbl-70Z rapidly proliferated and survived for extended time periods in the absence of exogenous growth factors (Figure 5B-C). We generated 32D cells that expressed KD Flt3 (Flt3-K644A, Flt3-KD) in the presence and absence of Cbl mutants. Interestingly, cells coexpressing Flt3-KD and Cbl-70Z also proliferated and survived growth factor independently (Figure 5D-F). We confirmed in viability assays (data not shown) and 3[H]-thymidine incorporation assays that both KD receptors were not responsive to their respective ligand (Figure 5C,F).
Kinase activity of Kit and Flt3 is dispensable for Cbl-70Z–mediated transformation. (A-B,D-E) Cbl-70Z induces ligand-independent proliferation and survival of KD 32D-Kit (Kit-KD, A-B) or 32D-Flt3 (Flt3-KD, D-E) cells. The 32D cells stably overexpressing Kit-KD or Flt3-KD and/or indicated Cbl mutants were starved from IL-3, and cells were grown in the presence of 10% FCS. Cells were counted at the indicated time points by the trypan blue exclusion method, and the data are shown as fold change of the cell number compared with the start of the experiment (A,D). (B,E) The percentage of cells in the culture that were alive at the indicated time points. (C,F) Ligand-independent DNA synthesis of KD Kit (Kit-KD) or Flt3 (Flt3-KD) cells coexpressing Cbl mutants. 32D-Kit-KD or 32D-Flt3-KD cells stably overexpressing the indicated constructs were starved from IL-3 for 12 hours, treated with the indicated cytokines, and proliferation was measured in 3[H]-thymidine incorporation assays. Data are shown as percentage of thymidine incorporation relative to the thymidine incorporation of the respective cell line with IL-3 supplementation. (A-F) Data represent the average and SD of at least 3 independent experiments.
Kinase activity of Kit and Flt3 is dispensable for Cbl-70Z–mediated transformation. (A-B,D-E) Cbl-70Z induces ligand-independent proliferation and survival of KD 32D-Kit (Kit-KD, A-B) or 32D-Flt3 (Flt3-KD, D-E) cells. The 32D cells stably overexpressing Kit-KD or Flt3-KD and/or indicated Cbl mutants were starved from IL-3, and cells were grown in the presence of 10% FCS. Cells were counted at the indicated time points by the trypan blue exclusion method, and the data are shown as fold change of the cell number compared with the start of the experiment (A,D). (B,E) The percentage of cells in the culture that were alive at the indicated time points. (C,F) Ligand-independent DNA synthesis of KD Kit (Kit-KD) or Flt3 (Flt3-KD) cells coexpressing Cbl mutants. 32D-Kit-KD or 32D-Flt3-KD cells stably overexpressing the indicated constructs were starved from IL-3 for 12 hours, treated with the indicated cytokines, and proliferation was measured in 3[H]-thymidine incorporation assays. Data are shown as percentage of thymidine incorporation relative to the thymidine incorporation of the respective cell line with IL-3 supplementation. (A-F) Data represent the average and SD of at least 3 independent experiments.
To find out the signaling molecules supporting this transforming effect of Cbl-70Z, we examined Akt, Erk, and Stat pathways in 32D cell lines coexpressing Kit-KD or Flt3-KD and Cbl mutants. Surprisingly, we found ligand-independent activation of Akt only in the cells overexpressing Cbl-70Z in the presence of either of the KD receptors, but not in the cells overexpressing Cbl-R420Q or in the control cells (Figure 6B; supplemental Figure 3D). There were no differences in Erk or Stat5 activation (Figure 6B). Importantly, inhibition of Akt activity by its respective inhibitor abolished the cytokine-independent growth of these cells (Figure 6D). The presence of Cbl-70Z conferred ligand-independent autophosphorylation to Kit-WT (Figures 4F, 6A), but we were not able to detect autophosphorylated KD receptors in the absence or presence of any dominant-negative Cbl mutant (Figure 6A). Furthermore, we analyzed the turnover of the KD Kit receptor by ubiquitination experiments, to understand the basic mechanism of degradation of this receptor. Interestingly, we did not observe basic levels of ubiquitination of the receptor, compared with the basic level of the unstimulated Kit-WT receptor (Figure 6C). Taken together, Cbl-70Z and Cbl-R420Q not only strongly synergized with unstimulated receptor tyrosine kinases with intact kinase function, but surprisingly also induce biologically meaningful signaling events emanating from kinase-inactive class III receptor tyrosine kinases.
Cbl mutants are in part restoring KD Kit-mediated signaling. (A-B) Constitutive Akt activation in Kit-KD cells expressing Cbl-70Z. The 32D cells with Kit-WT or Kit-KD were engineered to express the indicated Cbl proteins, deprived from cytokines overnight, and subsequently exposed to the indicated cytokines for 10 minutes. Western blot analyses with the indicated antibodies were performed. (C) Kit-KD is not ubiquitylated. COS-7 cells were transiently transfected with the indicated constructs together with a plasmid for HA-tagged ubiquitin. After 48 hours of transfection, cells were serum starved for 12 hours and stimulated with 50 ng/mL SCF for 10 minutes or left unstimulated. Cell lysates were prepared, and equal amounts of lysates were immunoprecipitated using anti-Kit antibody. The immunoprecipitates were resolved on SDS-PAGE and analyzed with anti-HA or anti-Kit antibodies. (D) Proliferation of Kit-KD cells expressing Cbl-70Z is dependent on Akt activation. Kit-KD cells stably overexpressing Cbl-70Z were starved from IL-3 and cells were grown in the presence of 10% FCS and 10 to 15 μM Akt inhibitor or dimethyl sulfoxide (DMSO) as solvent. Cells were counted at the indicated time points by the trypan blue exclusion method, and the data are shown as fold change of the cell number compared with the start of the experiment.
Cbl mutants are in part restoring KD Kit-mediated signaling. (A-B) Constitutive Akt activation in Kit-KD cells expressing Cbl-70Z. The 32D cells with Kit-WT or Kit-KD were engineered to express the indicated Cbl proteins, deprived from cytokines overnight, and subsequently exposed to the indicated cytokines for 10 minutes. Western blot analyses with the indicated antibodies were performed. (C) Kit-KD is not ubiquitylated. COS-7 cells were transiently transfected with the indicated constructs together with a plasmid for HA-tagged ubiquitin. After 48 hours of transfection, cells were serum starved for 12 hours and stimulated with 50 ng/mL SCF for 10 minutes or left unstimulated. Cell lysates were prepared, and equal amounts of lysates were immunoprecipitated using anti-Kit antibody. The immunoprecipitates were resolved on SDS-PAGE and analyzed with anti-HA or anti-Kit antibodies. (D) Proliferation of Kit-KD cells expressing Cbl-70Z is dependent on Akt activation. Kit-KD cells stably overexpressing Cbl-70Z were starved from IL-3 and cells were grown in the presence of 10% FCS and 10 to 15 μM Akt inhibitor or dimethyl sulfoxide (DMSO) as solvent. Cells were counted at the indicated time points by the trypan blue exclusion method, and the data are shown as fold change of the cell number compared with the start of the experiment.
SFKs are necessary for Cbl-70Z–mediated transformation
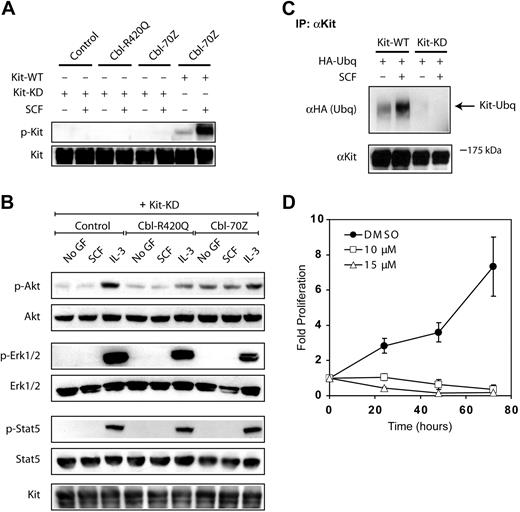
SFKs play an important role in intracellular signaling processes, for example, of cytokine receptors, which do not have tyrosine kinase activity.46,47 Because we had observed RTK activity-independent proliferation in cells overexpressing RTKs and Cbl mutants, we speculated whether SFKs could play a role in Cbl-70Z–mediated transformation. First, we analyzed the SFKs activation status in the presence of Cbl mutants. In 32D cells, we observed that some SFKs were already constitutively active in the absence of any growth factors; this activation was not further enhanced by IL-3 stimulation but was strongly enhanced in the presence of both Cbl mutants (data not shown). In addition, the overexpression of Kit in the presence of Cbl mutants showed a similar phosphorylation of SFKs irrespective of the kinase activity of the RTK (Figure 7A). Next, we examined the binding of Kit-KD, Cbl proteins, and SFKs. Immunoprecipitation experiments showed a constitutive association between phosphorylated SFKs and Cbl proteins in the absence of Kit (Figure 7B left panel). Surprisingly, immunoprecipitation of Cbl mutants resulted also in immunocomplexes with Kit-KD (Figure 7B right panel). Differences in the expression levels of Cbl mutants can be excluded as reason for the different phenotypical behavior of these mutants (Figure 7B; and data not shown). The association of Cbl mutants with Kit-KD was not enhanced by the presence of SCF. However Cbl mutants associated with KD Kit receptor, which could be a prerequisite for the transformation potential of Kit-KD together with Cbl mutants. To test this hypothesis, we used the well-established Src family inhibitors PP-1 and PP-2 and the more clinically relevant inhibitor dasatinib.48,49 Here, the phosphorylation of SFKs was completely inhibited in the presence of all Src inhibitors (supplemental Figure 3B). In addition, PP-2 completely abrogated the interaction between activated SFKs and Cbl (Figure 7C). We checked which SFKs were expressed in 32D cells and defined their binding to Cbl. By immunoprecipitation, we showed that Fyn binds to Cbl in a phosphorylation-dependent manner, but not Lck, Hck, Src, Yes, and Lyn (Figure 7D; and data not shown).
Src family tyrosine kinases play an important role in Cbl-70Z–mediated transformation. (A) Phosphorylation of SFKs is enhanced by Cbl mutants. The 32D-Kit-WT (top 2 blots) or 32D-Kit-KD (bottom 2 blots) cells stably overexpressing the indicated Cbl proteins were starved from IL-3 overnight and subsequently exposed to the indicated cytokines for 10 minutes. Western blot analyses using the phosphospecific antibody recognizing the activated form of SFK members (anti-pSrc Y416) and anti–β-actin antibody were performed as described. (B) Cbl-70Z binds stronger to p-SFKs and Cbl mutants physically interact with the KD Kit receptor. The 32D cells with or without Kit-KD stably transfected with HA-tagged Cbl proteins (Cbl-R420Q and Cbl-70Z) were treated as in panel A. Cbl proteins were immunoprecipitated by anti-HA antibodies, and immunoprecipitates were resolved on SDS-PAGE. Coimmunoprecipitation of Kit and p-SFKs was analyzed using anti-Kit or anti–phospho-SFK antibodies. (C) Interaction of SFKs and Cbl proteins is completely inhibited by the Src inhibitor PP-2. The 32D-Kit-KD cells stably overexpressing HA-tagged Cbl-70Z were starved from IL-3 in the presence of DMSO or PP-2 (15 μM) for 12 hours. Immunoprecipitation and Western blot analyses were performed as in panel B with anti–phospho-SFK and anti-HA antibodies. (D) Fyn physically binds to Cbl and its binding is phosphorylation dependent. The 32D-Kit-KD cells stably overexpressing HA-tagged Cbl-70Z were starved from IL-3 in the presence of DMSO or PP-2 (15 μM) for 12 hours. Cbl proteins were immunoprecipitated by anti-HA antibodies, and immunoprecipitates were resolved on SDS-PAGE. Coimmunoprecipitation of Fyn was analyzed using anti-Fyn antibody. (E) Src inhibitors also inhibit phosphorylation of Akt. Kit-KD cells overexpressing Cbl-70Z were treated as in panel C. In addition, cells were also starved in the presence of dasatinib (150 nM). Western blot analyses were performed with anti–phospho-Akt and anti-Akt antibodies. (F) Cbl-70Z–mediated ligand-independent DNA synthesis is blocked by Src inhibitors. The 32D cells stably expressing Kit or Flt3 and/or Cbl-70Z were starved from IL-3 for 12 hours, treated with the indicated cytokines and PP-1/PP-2 (15 μM) or DMSO for 6 hours, and incubated with 3[H]-thymidine. After 15 hours of incubation, proliferation was measured in 3[H]-thymidine incorporation assays. Data are shown as percentage of thymidine incorporation relative to the thymidine incorporation of the respective cell line with IL-3 supplementation. The data represent the average and SD of 3 independent experiments. (G) Colony growth is blocked by the Src inhibitor PP-2. The 32D cells stably overexpressing Kit or Flt3 constructs and/or Cbl-70Z were serum starved for 12 hours and then plated at a concentration of 1000 cells per dish in the presence or absence of PP-2 (15 μM) and in the absence of any growth factors. Colonies were counted on day 6. The assays were plated as triplicates. The numbers given show the results of 1 of at least 3 independent experiments per construct, which all gave similar results. (H) Imatinib and dasatinib inhibit cell proliferation substantially differently. Indicated cell lines were treated as in Figure 1A. In addition, either Kit inhibitor imatinib (2.5 μM) or dual kinase inhibitor (Kit and SFKs) dasatinib (150 nM) was added, and cells were counted at the indicated time points by the trypan blue exclusion method. The data represent the average and SD of 3 independent experiments.
Src family tyrosine kinases play an important role in Cbl-70Z–mediated transformation. (A) Phosphorylation of SFKs is enhanced by Cbl mutants. The 32D-Kit-WT (top 2 blots) or 32D-Kit-KD (bottom 2 blots) cells stably overexpressing the indicated Cbl proteins were starved from IL-3 overnight and subsequently exposed to the indicated cytokines for 10 minutes. Western blot analyses using the phosphospecific antibody recognizing the activated form of SFK members (anti-pSrc Y416) and anti–β-actin antibody were performed as described. (B) Cbl-70Z binds stronger to p-SFKs and Cbl mutants physically interact with the KD Kit receptor. The 32D cells with or without Kit-KD stably transfected with HA-tagged Cbl proteins (Cbl-R420Q and Cbl-70Z) were treated as in panel A. Cbl proteins were immunoprecipitated by anti-HA antibodies, and immunoprecipitates were resolved on SDS-PAGE. Coimmunoprecipitation of Kit and p-SFKs was analyzed using anti-Kit or anti–phospho-SFK antibodies. (C) Interaction of SFKs and Cbl proteins is completely inhibited by the Src inhibitor PP-2. The 32D-Kit-KD cells stably overexpressing HA-tagged Cbl-70Z were starved from IL-3 in the presence of DMSO or PP-2 (15 μM) for 12 hours. Immunoprecipitation and Western blot analyses were performed as in panel B with anti–phospho-SFK and anti-HA antibodies. (D) Fyn physically binds to Cbl and its binding is phosphorylation dependent. The 32D-Kit-KD cells stably overexpressing HA-tagged Cbl-70Z were starved from IL-3 in the presence of DMSO or PP-2 (15 μM) for 12 hours. Cbl proteins were immunoprecipitated by anti-HA antibodies, and immunoprecipitates were resolved on SDS-PAGE. Coimmunoprecipitation of Fyn was analyzed using anti-Fyn antibody. (E) Src inhibitors also inhibit phosphorylation of Akt. Kit-KD cells overexpressing Cbl-70Z were treated as in panel C. In addition, cells were also starved in the presence of dasatinib (150 nM). Western blot analyses were performed with anti–phospho-Akt and anti-Akt antibodies. (F) Cbl-70Z–mediated ligand-independent DNA synthesis is blocked by Src inhibitors. The 32D cells stably expressing Kit or Flt3 and/or Cbl-70Z were starved from IL-3 for 12 hours, treated with the indicated cytokines and PP-1/PP-2 (15 μM) or DMSO for 6 hours, and incubated with 3[H]-thymidine. After 15 hours of incubation, proliferation was measured in 3[H]-thymidine incorporation assays. Data are shown as percentage of thymidine incorporation relative to the thymidine incorporation of the respective cell line with IL-3 supplementation. The data represent the average and SD of 3 independent experiments. (G) Colony growth is blocked by the Src inhibitor PP-2. The 32D cells stably overexpressing Kit or Flt3 constructs and/or Cbl-70Z were serum starved for 12 hours and then plated at a concentration of 1000 cells per dish in the presence or absence of PP-2 (15 μM) and in the absence of any growth factors. Colonies were counted on day 6. The assays were plated as triplicates. The numbers given show the results of 1 of at least 3 independent experiments per construct, which all gave similar results. (H) Imatinib and dasatinib inhibit cell proliferation substantially differently. Indicated cell lines were treated as in Figure 1A. In addition, either Kit inhibitor imatinib (2.5 μM) or dual kinase inhibitor (Kit and SFKs) dasatinib (150 nM) was added, and cells were counted at the indicated time points by the trypan blue exclusion method. The data represent the average and SD of 3 independent experiments.
Consistently, we observed in proliferation assays that cells coexpressing Kit-KD or Flt3-KD together with Cbl-70Z were unable to grow in the presence of PP-1 or PP-2 (Figure 7F). Next, we investigated the effect of the Src inhibitors on the clonogenic growth mediated by Cbl mutants. Importantly, the presence of Src inhibitor PP-2 significantly inhibited the clonogenic growth in the presence of Cbl mutants and Kit or Flt3 receptor (Figure 7G). It was of interest whether the constitutive Akt activation in cells coexpressing Kit-KD and Cbl-70Z was also suppressed by Src inhibitors. We hypothesized that SFKs might be activating the PI3K pathway either directly or by phosphorylating Cbl at tyrosine 731 (Y731), which is known to be the PI3K binding site.9 Dasatinib and PP-2 completely inhibited Akt activation and Cbl phosphorylation at Y731 (Figure 7E; supplemental Figure 3C), suggesting that SFKs are upstream of Akt and are critically involved in Cbl-70Z-mediated synergistic transformation.
Finally, we used imatinib (inhibitor of Kit) and dasatinib (inhibitor of Kit and SFKs)49 to demonstrate the growth and survival mediated by Cbl mutants in the absence of Kit kinase activity. As shown in Figure 7H, dasatinib strongly inhibited the proliferation of Kit-WT (or Kit-KD) cells overexpressing Cbl-70Z, whereas imatinib reduced proliferation to a much lesser extent, most probably because imatinib does not inhibit SFKs.
Discussion
The RTKs Flt3 and Kit play an important role in leukemogenesis.50-52 Approximately 25% of all AML patients carry mutations in Flt3, whereas Kit mutations are found less frequently. Kit mutations have been preferentially associated with the core-binding factor leukemias.53 Activating mutations of Flt3 (such as Flt3-ITD and activating point mutations in the tyrosine kinase domain, Flt3-TKD) and Kit lead to aberrant signal transduction and have spurred considerable interest in the development of specific tyrosine kinase inhibitors for these RTKs.34,52,53 Recently, we identified a c-Cbl mutation in an AML patient, which leads to a defective E3 ligase activity. This was the first Cbl mutation identified in human disease.19 Several more additional mutations were subsequently found in c-Cbl and Cbl-b,20-24 suggesting that Cbl mutations are recurring events in AML. Cbl mutations probably occur in other hematologic and solid tumors as well.
Cbl has been shown to degrade several RTKs, including the EGFR and PDGFR.9,26,54,55 Recently, we showed that Flt3 is degraded in a Cbl-dependent manner.19 As shown here, the expression of Kit in myeloid 32D cells does not lead to cytokine-independent growth. However, in the presence of Cbl-R420Q or Cbl-70Z, Kit expression promotes cytokine-independent growth (Figure 1A). This effect can be enhanced after SCF stimulation. Similarly, coexpression of Kit with either Cbl-R420Q or Cbl-70Z promotes survival (Figure 1B) and proliferation (Figure 1C). In addition, coexpression of Kit and either of the Cbl mutants led to colony growth in the absence of growth factors (Figure 1D-E). We observed similar effects with Flt3,19 suggesting that this may be a general mechanism in most RTKs.
In a murine bone marrow transplantation model, we observed 2 distinct diseases with high penetrance and long latency (Figures 2–3): a generalized mastocytosis and a myeloproliferative disease. In addition, 2 mice developed a myeloid leukemia with high leukocyte counts, splenomegaly, and organ infiltration by myeloid immature cells (Figure 3). Six mice developed mast cell sarcomas. A tantalizing explanation for the massive mast cell proliferation could be the typical expression of Kit in these cells. In our opinion, the long latency is very suggestive of secondary events (mutations, epigenetic changes) occurring. These may occur in different animals, at different times with different rates, explaining the variability of disease onset and death. Postmortem analysis of Cbl expression was not performed, as endogenous Cbl is widely expressed. As the Cbl mutants affect the stability of several intracellular and surface molecules, including Flt3 and Kit in these in vivo experiments, we were unable to pinpoint the contribution of the Cbl targets to the disease phenotype. Although unproven, the similarity of Cbl-70Z and Cbl-R420Q in disease development suggests a similar mechanism for both mutants. A recent report showed that hematopoietic stem cells from Cbl−/− mice have an increased pool size, are hyperproliferative, and show enhanced long-term repopulating capacity.56 It is tempting to speculate that Cbl-inactivating mutations may promote similar function in leukemia-initiating cells. Importantly, Cbl−/− hematopoietic stem cells expanded more in the presence of SCF and thrombopoietin, which is explained by the fact that Cbl is the negative regulator of Kit and the thrombopoietin receptor (cMpl) and supports the validity of increased SCF responsiveness we observed in cells coexpressing Kit and Cbl mutants (Figure 1C). NUP98-HOXD13 (NHD13) fusion gene plays an important role in myelodysplastic syndrome and acute nonlymphocytic leukemia. Recently, an acquired Cbl mutation was shown in a Nup98-HoxD13 mouse model, which strongly supports the prominent role of Cbl mutations in vivo.57 To our knowledge, the in vivo data presented in Figures 2 and 3 are the first in vivo evidence that a Cbl mutant identified from human malignancy is involved in disease pathogenesis. The minor differences observed between Cbl-R420Q and Cbl-70Z are within the variability of animal experimentation that we and others have observed.
It is well established that an intact RING domain is required for the E3 ligase activity of Cbl.58,59 Cbl-70Z and Cbl-R420Q have a disrupted RING domain and are E3 ligase defective.19 Cbl-R420Q and Cbl-70Z therefore prevent ubiquitination and endocytosis of Kit (Figure 4B-C). We were interested to further clarify the mechanisms of transformation by Cbl mutants and performed a careful analysis of the signaling properties conferred by Kit in the presence or absence of Cbl mutants. These experiments revealed enhanced basal RTK autophosphorylation and prolonged ligand-activated MAPK and PI3K/Akt signaling in the cells expressing Cbl mutants and Kit (Figure 4F-H), which is in line with published data for EGFR,60 PDGFRα,61 and Flt3.19 However, we also observed basal activation of Akt and Erk (Figure 4E-H), which contrasts with data for Flt3, where no differences in Akt basal signaling were identified.19 The reasons for this difference are unknown but may be the result of differences in Kit and Flt3 expression. In any case, as Akt activation has been shown to cause cytokine-independent colony growth,62 this may partly explain why coexpression of Cbl mutants with Kit (in the absence of ligand) led to colony growth (Figure 1D). Coimmunoprecipitation experiments showed that Cbl mutants are part of a complex with Kit after ligand stimulation (Figure 4A). Importantly, Cbl-70Z (and to a lesser extent Cbl-R420Q) coimmunoprecipitated with Kit in the absence of ligand. This suggests a constitutive complex formation between Cbl mutants and Kit (Figure 4A), which was similarly observed for EGFR60 and Flt3.19
Our data reveal functional differences between Cbl-70Z and Cbl-R420Q. The loss of E3 ligase activity is more pronounced in cells expressing Cbl-R420Q than in those expressing Cbl-70Z (Figure 4B), whereas the consequences of expression of either Cbl mutant appear to be very similar in both the in vitro and in vivo experiments with primary murine bone marrow (Figures 2–3). However, the biologic consequences of Cbl-70Z are more dramatic in cell culture experiments performed in 32D cells (Figures 1, 5–6). Further studies are necessary to decipher these differences between Cbl mutants, including the novel mutations that have been described since our initial observation20-24 and have not yet been molecularly characterized. However, it is possible that conformational changes or differences in subcellular localization are involved.
We were interested in defining the kinase requirements of the Cbl mutants/RTK interaction. Importantly, although our data revealed that the biologic functions mediated by Kit and Cbl-70Z were in large part dependent on the receptor kinase activity, we observed a small but biologically dominant effect also of kinase-inactive Kit and Flt3 on cells expressing Cbl mutants (Figure 5). Although it was not sufficient for cytokine-independent growth that the cells expressed Cbl-70Z, KD Kit (Figure 5A-C), and Flt3 (Figure 5D-F) synergized with Cbl mutants. It could be speculated that Kit or Flt3, which can be found to be in complex with the Cbl proteins, may act as a necessary scaffold. This is in line with a recent report that survival of cancer cells was maintained by EGFR independently of its kinase activity.63 The yet unimpressive clinical outcomes of tyrosine kinase inhibitors for treatment of multiple types of cancer suggest that kinase-independent functions of RTKs may be a significant contributor for cancer progression.
Biochemical analysis demonstrated neither autophosphorylation (or ligand-dependent phosphorylation) of Kit-KD (Figure 6A) nor downstream Erk or Stat5 activation (Figure 6B). Similarly, Kit-KD was not ubiquitinated in response to SCF (Figure 6C). Finally, constitutive Akt activation seems to play an important role in cells expressing Kit-KD and Cbl-70Z (Figure 6B,D). Cbl has been shown to degrade SFKs by ubiquitination.38,64 Therefore, up-regulation of SFK activity in the presence of Cbl mutants could be explained by inhibition of their degradation (Figure 7A). In addition, we found Cbl-R420Q and Cbl-70Z in complex with Kit-KD and SFKs (Figure 7B). Inhibition of SFKs led also to Akt inhibition (Figure 7E), suggesting that SFKs act upstream of PI3K/Akt pathway as previously reported.65,66 SFKs activate Akt either directly or by phosphorylating Cbl at tyrosine 731, which is the binding site for the PI3K.9 SFK inhibition with PP-1, PP-2, or dasatinib completely inhibited proliferation and clonogenic growth (Figure 7F-H). Previously, Fyn has been shown to bind and phosphorylate Cbl and to activate PI3K.67 Our data show that the Src family member Fyn binds to Cbl (Figure 7D).
Kit inhibition with imatinib reduced the proliferation of cells overexpressing Kit-WT and Cbl-70Z stronger compared with cells expressing Kit-KD and Cbl-70Z (Figure 7H), but much lesser than dasatinib, which in addition inhibits SFKs. This indicates that Kit kinase activity is required but not essential. These findings are extending and complementing our previous findings.19 SFKs therefore are crucial components of the complex formed by KD RTKs and Cbl mutants, and their inhibition prevents transformation.
Therefore, RTKs may not be the ideal target kinases for tyrosine kinase inhibitors in a Cbl mutation–driven disease. Instead, SFKs may be interesting target kinases, for which inhibitors are readily available. Dasatinib, for example, is a multitargeted kinase inhibitor of Brc-Abl, SFKs, Kit, PDGFR, and ephrin A receptor kinases.68-70 It also emerged as a potent inhibitor of imanitib-resistant protein tyrosine kinase activation loop mutants of Kit, and it is able to induce apoptosis in mast cell and leukemic cell lines expressing these mutations.71
The data presented here demonstrate that SFKs play a critical role in the mechanism of transformation of Cbl mutants. AML blasts expressing Cbl mutants (or dysregulated Cbl function by other mechanisms) might be resistant to tyrosine kinase inhibitors targeted for Flt3 or Kit. Although direct Cbl mutations may be rare in AML, similar mechanisms in the RTK degradation machinery may be operational in a large proportion of AMLs. Further studies are required to determine whether kinase inhibitors targeted at both RTKs and SFKs may overcome this primary resistance to RTK inhibitors.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Stefanie Parkhof for her excellent technical assistance and Drs Wallace Y. Langdon, Hamid Band, and Klaus-Michael Debatin for their generous gifts of plasmid constructs used in this study.
This work was supported by the Deutsche Krebshilfe (10-6697) and the Medical Faculty of the University of Münster (IMF Sa 110404, IZKF Mül2 018/07).
Authorship
Contribution: B.S., S.R.B., C.B., and H.S. conceptualized the idea and designed the research; B.S., M.R., S.R.B., and R.G. performed the transplantation experiments; B.S., S.R.B., and M.R. performed most of the other work presented here; B.S., S.R.B., M.R., R.G., L.T., G.K., J.D., W.E.B., C.M.-T., C.B., and H.S. were involved in data analyses and discussions; G.K. performed the mouse histology analyses; and B.S., S.R.B., and C.B. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hubert Serve, Department of Medicine, Hematology/Oncology, Goethe University, Theodor-Stern-Kai 7, D-60590 Frankfurt, Germany; e-mail: serve@em.uni-frankfurt.de.
References
Author notes
*S.R.B. and C.B. contributed equally to this study.

![Figure 1. Cbl mutants synergize with Kit to induce autonomous growth and in vitro colony formation. (A-B) Cbl-70Z and Cbl-R420Q induce ligand-independent proliferation and survival of 32D-Kit-WT cells. The 32D cells stably overexpressing Kit-WT and/or indicated Cbl mutants were starved from IL-3, and cells were grown in the presence of 10% FCS. Cells were counted at the indicated time points by the trypan blue exclusion method, and the data are shown as fold change of the cell number compared with the start of the experiment (A). (B) The percentage of cells in the culture that was alive at the indicated time points. (C) Ligand-independent DNA synthesis of Kit-WT cells coexpressing Cbl-70Z. The 32D cells stably expressing Kit-WT and/or Cbl constructs were starved from IL-3 for 12 hours and then cultured in the presence or absence of SCF or IL-3. Proliferation was measured by 3[H]-thymidine incorporation assays. Data are shown as percentage of thymidine incorporation relative to the thymidine incorporation of the respective cell line with IL-3 supplementation. (A-C) Data represent the average and SD of 3 independent experiments. (D-E) Cbl mutants led to cytokine-independent colony growth. The 32D cells stably overexpressing Kit-WT and/or indicated Cbl mutants were serum starved for 12 hours and then plated at a concentration of 1000 cells per dish in the absence of any growth factors. Colonies were counted on day 6. The assays were plated as triplicates. The numbers given show the results of 1 of at least 3 independent experiments per construct, which all gave similar results. (E) Photographs of the dishes were taken on day 6 and show morphologic differences in clonal growth.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/19/10.1182_blood-2008-12-190934/4/m_zh89990944150001.jpeg?Expires=1769144850&Signature=iWOM6Pj~Hs7vghwUJSMdik2YS3x2sobSCHikRRTsm6JStTiMLfXORhTbLVh0fqSVC-ymz0P3aeqD3ftPNQ7v6Gyf3YNQOwYqQbpLSSdvTzwtzboP78Q5Tq7dOEf6GtP-5lQ7NBQtBZF-N-FVsVzkx9A4ZsgpTHgJMf73Pt9r8wflKhkpjS8c1FT-d4nYYZ5-dQDzRidA7YM9x3GnyHNfyNpaqEuFZUyb891ByhTUEaHFK968E1Nr-gccqobZgwH3zd0xR3XcwFD1pyu9N6NefTMXxqs0AT-NsHNsqv3vLHHClTfp9ZUhO7ZoocIPfy32xSk9JMAswnxXJC~ZHkQtXA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Cbl mutants induce a myeloid leukemia or mast cell sarcoma. Histopathology of the bone marrow (BM), liver, and spleen from mice transplanted with Cbl mutant-transduced bone marrow cells. Spleen, liver, and bone marrow sections (hematoxylin and eosin [HE], Giemsa, NACE; original magnifications, 100×-1000×) from a representative leukemic mouse (Cbl-R420Q) and a mouse with mast cell sarcoma (Cbl-70Z), which received bone marrow cells transduced with Cbl-R420Q or Cbl-70Z, respectively. Normal organ architecture is visible in a control mouse. The leukemic mouse (Cbl-R420Q) reveals a massive infiltration of immature myeloid cells with its maximum in the spleen and consecutive destruction of the normal organ structure. There are also mast cells sporadically visible in between the immature cell infiltration (arrow in the spleen pictures and Giemsa staining). In contrast, mast cells in the mice with mast-cell sarcoma have characteristic cytology with uniform, closely packed cells with round nuclei and clear, abundant cytoplasm with granules and the typical features of mast cells with numerous granules in Giemsa staining and are highly positive in NACE staining. Slides were viewed with an Olympus BX51 microscope using Olympus Soft imaging system: Cell A and the following objective lenses: 10×/0.30 UPlanFL (1st column); 40×/0.75 UPlanFL (2nd column); 100×/1.35 oil iris PlanApo (3rd and 4th columns). Images were captured on an Olympus DP70 camera and manipulated with Adobe Photoshop DP70.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/19/10.1182_blood-2008-12-190934/4/m_zh89990944150003.jpeg?Expires=1769144850&Signature=vk-QkW6Dz64Zx-vTRKnAvffIGIyTav3gdlHS4hkw23y1GGT~Bdj998u9sUX44Nu9FE4ggW59qhCqlfR9tgwKtBEmWkzTZo-JpG8hBM-l9Ax5FJm2TRxn27c26QC10sFD4sAnLOalxPFvbRsfyh3E2efiJ0HYMtHkZfVu~wd0tvwnokIG~wtfUuBgEdtTAXqIY7RXGXDj48S8tphkoeHDsxt7E2gXRo-lweIO5wOVHI~ZNDfVd-uCHCYfaUVa6FPWcjgDbmg0oxspTKbw52R5O4DR-Q6WgweTOzZdAZSavC7kNUvor4cILUHphl0cx3n-n~LwSyY3Numxg80-QoQvCw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Kinase activity of Kit and Flt3 is dispensable for Cbl-70Z–mediated transformation. (A-B,D-E) Cbl-70Z induces ligand-independent proliferation and survival of KD 32D-Kit (Kit-KD, A-B) or 32D-Flt3 (Flt3-KD, D-E) cells. The 32D cells stably overexpressing Kit-KD or Flt3-KD and/or indicated Cbl mutants were starved from IL-3, and cells were grown in the presence of 10% FCS. Cells were counted at the indicated time points by the trypan blue exclusion method, and the data are shown as fold change of the cell number compared with the start of the experiment (A,D). (B,E) The percentage of cells in the culture that were alive at the indicated time points. (C,F) Ligand-independent DNA synthesis of KD Kit (Kit-KD) or Flt3 (Flt3-KD) cells coexpressing Cbl mutants. 32D-Kit-KD or 32D-Flt3-KD cells stably overexpressing the indicated constructs were starved from IL-3 for 12 hours, treated with the indicated cytokines, and proliferation was measured in 3[H]-thymidine incorporation assays. Data are shown as percentage of thymidine incorporation relative to the thymidine incorporation of the respective cell line with IL-3 supplementation. (A-F) Data represent the average and SD of at least 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/19/10.1182_blood-2008-12-190934/4/m_zh89990944150005.jpeg?Expires=1769144850&Signature=gfzP6e2L8tiBbuWAcSYFRF6OaN9eL0QOP3IXBOgl2gf7S8t5l5rVnQ8Q3q8vgH6vYWjrL5cNnD7jBuoh3w8kaFIeQJeLD3zXFJ6sCr4kr7TqsbR8HUg-GEqC4Xi7OQvMBH46s8lgIFHvEn2O~aIN-0KQbXTv3rmzuiKQ-af68ajWppaJKooa6noTi3NVx3UdR0rQGdX-88~JmrJVBQRXb~8lzIIrTnkAJvyrJO2TBbvukvDk5cuTQDnuEDbj-oVki3pcAqhAeoUQmqXFMD-K~bkcln-fOYEpj4uUPEHSeY4Sby1eSqckZEeg3t-ugtamr7P6j5L2PBYYi~UoodhI6Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. Src family tyrosine kinases play an important role in Cbl-70Z–mediated transformation. (A) Phosphorylation of SFKs is enhanced by Cbl mutants. The 32D-Kit-WT (top 2 blots) or 32D-Kit-KD (bottom 2 blots) cells stably overexpressing the indicated Cbl proteins were starved from IL-3 overnight and subsequently exposed to the indicated cytokines for 10 minutes. Western blot analyses using the phosphospecific antibody recognizing the activated form of SFK members (anti-pSrc Y416) and anti–β-actin antibody were performed as described. (B) Cbl-70Z binds stronger to p-SFKs and Cbl mutants physically interact with the KD Kit receptor. The 32D cells with or without Kit-KD stably transfected with HA-tagged Cbl proteins (Cbl-R420Q and Cbl-70Z) were treated as in panel A. Cbl proteins were immunoprecipitated by anti-HA antibodies, and immunoprecipitates were resolved on SDS-PAGE. Coimmunoprecipitation of Kit and p-SFKs was analyzed using anti-Kit or anti–phospho-SFK antibodies. (C) Interaction of SFKs and Cbl proteins is completely inhibited by the Src inhibitor PP-2. The 32D-Kit-KD cells stably overexpressing HA-tagged Cbl-70Z were starved from IL-3 in the presence of DMSO or PP-2 (15 μM) for 12 hours. Immunoprecipitation and Western blot analyses were performed as in panel B with anti–phospho-SFK and anti-HA antibodies. (D) Fyn physically binds to Cbl and its binding is phosphorylation dependent. The 32D-Kit-KD cells stably overexpressing HA-tagged Cbl-70Z were starved from IL-3 in the presence of DMSO or PP-2 (15 μM) for 12 hours. Cbl proteins were immunoprecipitated by anti-HA antibodies, and immunoprecipitates were resolved on SDS-PAGE. Coimmunoprecipitation of Fyn was analyzed using anti-Fyn antibody. (E) Src inhibitors also inhibit phosphorylation of Akt. Kit-KD cells overexpressing Cbl-70Z were treated as in panel C. In addition, cells were also starved in the presence of dasatinib (150 nM). Western blot analyses were performed with anti–phospho-Akt and anti-Akt antibodies. (F) Cbl-70Z–mediated ligand-independent DNA synthesis is blocked by Src inhibitors. The 32D cells stably expressing Kit or Flt3 and/or Cbl-70Z were starved from IL-3 for 12 hours, treated with the indicated cytokines and PP-1/PP-2 (15 μM) or DMSO for 6 hours, and incubated with 3[H]-thymidine. After 15 hours of incubation, proliferation was measured in 3[H]-thymidine incorporation assays. Data are shown as percentage of thymidine incorporation relative to the thymidine incorporation of the respective cell line with IL-3 supplementation. The data represent the average and SD of 3 independent experiments. (G) Colony growth is blocked by the Src inhibitor PP-2. The 32D cells stably overexpressing Kit or Flt3 constructs and/or Cbl-70Z were serum starved for 12 hours and then plated at a concentration of 1000 cells per dish in the presence or absence of PP-2 (15 μM) and in the absence of any growth factors. Colonies were counted on day 6. The assays were plated as triplicates. The numbers given show the results of 1 of at least 3 independent experiments per construct, which all gave similar results. (H) Imatinib and dasatinib inhibit cell proliferation substantially differently. Indicated cell lines were treated as in Figure 1A. In addition, either Kit inhibitor imatinib (2.5 μM) or dual kinase inhibitor (Kit and SFKs) dasatinib (150 nM) was added, and cells were counted at the indicated time points by the trypan blue exclusion method. The data represent the average and SD of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/19/10.1182_blood-2008-12-190934/4/m_zh89990944150007.jpeg?Expires=1769144850&Signature=s0TMjNMsuRBLQZi3f44lZCKtYejQ6HpwAsDB~MqdaNDMc0x~KFZUZOvQ~TBeuJ~7xMJnRNKWXtagqSe1fhui0DbeMwv87n1rULY5ozZ1t5Cle-XhGZMytjR9bjf-aNmN77c~pnf8R1hbO~Sn4nIEptcxOvDlgV3KmQpY1HjGZqXhlaxbVAGw5uDqB6VZ~cMHnJ8YuflrZEobijxvclnU5hdg7q8mntPjjtBIZz8xE~rpfc5O-ZSHxHktGrZRBg67aAH4csg71iwp1fwFQNqpJoZzWxmsGTxLKnsC1kea5-reInq1w9y4C-cxgOV7CzX9gzdHmLC1cG44gSUUmpsBrQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal