Abstract
Although the cancer stem cell (CSC) concept implies that CSCs are rare, recent reports suggest that CSCs may be frequent in some cancers. We hypothesized that the proportion of leukemia stem cells would vary as a function of the number of dysregulated pathways. Constitutive expression of MN1 served as a 1-oncogene model, and coexpression of MN1 and a HOX gene served as a 2-oncogene model. Leukemia-initiating cell (LIC) number and in vitro expansion potential of LICs were functionally assessed by limiting dilution analyses. LIC expansion potential was 132-fold increased in the 2- compared with the 1-oncogene model, although phenotypically, both leukemias were similar. The 2-oncogene model was characterized by granulocyte-macrophage colony-stimulating factor (GM-CSF) hypersensitivity and activated STAT/ERK signaling. GM-CSF hypersensitivity of the 2-oncogene model (MN1/HOXA9) was lost in Stat5b−/− cells, and the LIC expansion potential was reduced by 86- and 28-fold in Stat5b−/− and Stat1−/− cells, respectively. Interestingly, in 201 acute myeloid leukemia (AML) patients, coexpression of MN1 and HOXA9 was restricted to patients with the poorest prognosis and was associated with highly active STAT signaling. Our data demonstrate the functional heterogeneity of LICs and show that STAT signaling is critical for leukemia stem cell self-renewal in MN1- and HOXA9-expressing leukemias.
Introduction
Several decades ago, variability of leukemias among patients was recognized by morphologic differences and recurrent chromosomal aberrations. In the last decade, a multitude of somatic mutations has been identified in acute myeloid leukemia (AML) patients without cytogenetic aberrations, such as mutations in NPM1,1 FLT3,2,3 C/EBPα,4 or MLL.5 Many of these mutations are associated with a distinct prognosis, thus underscoring the heterogeneity of individual leukemias. Heterogeneity has been found not only between but also within individual leukemias. Moreover, in human AML and murine models only a subset of leukemic cells has shown the ability to engraft recipient mice and confer leukemia.6-9 These findings have provided support for the cancer stem cell (CSC) model, in which tumor growth relies on a subset of cancer cells at the apex of a cellular hierarchy, and in which heterogeneity within the tumor is ascribed to differentiation from the cancer stem cell.10 Enhanced self-renewal is the hallmark of CSCs and novel treatment approaches aim to specifically target the aberrant self-renewal machinery in CSCs.11 Whether the heterogeneity among leukemias is represented at the stem cell level has not been systematically investigated. However, understanding the mechanisms underlying aberrant self-renewal in individual leukemias will be of paramount importance to developing leukemia stem cell–specific therapies. To functionally measure leukemia stem cell potential, the operational assay of leukemia-initiating cells (LICs) has been developed. Cells that can initiate the leukemia upon transplantation are denoted LICs, and can be quantified by limiting dilution transplantation. As an approach to modeling leukemias with differing leukemia stem cell properties, we analyzed leukemias engineered by the potent oncogene MN1 alone or in combination with the leukemogenic oncogenes HOXA9 or NUP98-HOXD13. These combinations were chosen based on evidence that these oncogenes may impact distinct pathways in AML patients: we hypothesized that coexpression of MN1 and a HOX gene may recapitulate characteristics of poor risk AML, which presumably is driven by the most aggressive leukemia stem cells, and in which multiple pathways are activated.12
NPM1-mutated AML, accounting for 50% of patients with cytogenetically normal AML, is characterized by up-regulation of a HOX gene signature and the HOX cofactor MEIS1, whereas NPM1–wild-type AML is characterized by up-regulation of genes such as CD34, MN1, or BAALC that are also expressed in normal hematopoietic stem and progenitor cells.13-16 Several HOX genes and MN1 induce myeloid malignancies in mice that represent these NPM1-mutated AML and NPM1–wild-type AML, respectively.
The MN1 gene was first identified as a fusion partner of TEL, an ETS transcription factor, in patients with myeloid leukemia or myelodysplastic syndrome containing the translocation t(12;22)(p13;q11).17 More recent studies show that MN1 is a potent oncogene producing myeloid malignancy alone or in combination with CBFβ-MYH11.18,19 MN1 promotes proliferation and self-renewal of transduced cells and blocks their differentiation through transcriptional repression of differentiation-associated genes.18 MN1 has been implicated in transcriptional control of the RAR/RXR signaling pathway,20,21 and has been associated with induction of resistance to the differentiation-inducing agent all-trans retinoic acid both in vitro and in AML patients.18
HOXA9 was originally identified as a fusion partner of nucleoporin 98 (NUP98) in AML patients22,23 and is overexpressed in a high proportion of AML patients.24 Mice that received a transplant of HOXA9-transduced bone marrow cells survive more than 170 days without apparent disease.25 The fusion protein NUP98HOXD13 (ND13) has been found in some AML patients26 and has oncogenic potential in mice.27,28 Transgenic mice expressing ND13 have been used to identify collaborating genes by insertional mutagenesis and identified insertions in the Meis1 and Mn1 loci as the most common collaborating events.29
In the present study, constitutive expression of MN1 either alone or in combination with ND13 or HOXA9 served as a progression model of AML-expressing oncogenes from different molecular subgroups of AML. We hypothesized that different leukemias exhibit variable frequencies of LICs and distinct kinetics for disease induction as a result of the specific oncogenic pathways activated within the leukemic cells. Modeling this heterogeneity in a leukemia model of MN1 and HOX gene coexpression allowed us to investigate pathways that mediate an increased LIC frequency and accelerated disease progression.
Methods
Retroviral vectors and vector production
Mice and retroviral infection of primary bone marrow cells
C57BL/6J mice were bred and maintained in the animal research center of the British Columbia Cancer Agency as approved by the University of British Columbia Animal Care Committee. Homozygous C.129-Stat5btm1Hwd/J mice crossed to BALB/c mice and wild-type BALB/c littermates were obtained from The Jackson Laboratory.32 Homozygous 129S6/SvEv-Stat1tm1Rds mice and wild-type 129S6/SvEv littermates were obtained from Taconic.33 Primary mouse bone marrow cells were transduced as previously described.34 Briefly, bone marrow cells were harvested from mice treated with 150 mg 5-fluorouracil/kg 4 days before harvest (Faulding) and stimulated for 48 hours in Dulbecco modified Eagle medium (DMEM) supplemented with 15% fetal bovine serum (FBS), 10 ng/mL human interleukin-6 (hIL-6), 6 ng/mL murine interleukin-3 (mIL-3), and 20 ng/mL murine stem cell factor (mSCF; all from StemCell Technologies Inc). The cells were infected by cocultivation with irradiated (4000 cGy) GP+ E86 viral producer cells in the presence of 5 μg/mL protamine sulfate (Sigma-Aldrich). Cells were then sorted for green fluorescent protein (GFP) expression and/or selected in puromycin (Sigma-Aldrich) or geneticin (Invitrogen), where applicable, and maintained in 6 ng/mL mIL-3, 10 ng/mL hIL-6, and 20 ng/mL mSCF unless otherwise specified.
Bone marrow transplantation and monitoring of mice
Transduced bone marrow cells from C57BL/6J and 129SvEv mice were injected into the tail vein of lethally irradiated syngeneic recipient mice that were exposed to a single dose of 750 cGy total-body irradiation accompanied by a life-sparing dose of 2.5 × 105 freshly isolated bone marrow cells from syngeneic mice. Transduced cells from Stat5b-null or wild-type BALB/c mice were injected into the tail vein of nonobese diabetic/severe combined immunodeficient/IL2Rγ-null mice irradiated with a single dose of 250 cGy total-body irradiation. Engraftment of donor cells was monitored by tail vein bleeds and fluorescence-activated cell sorting (FACS) analysis of GFP-expressing cells every 4 weeks. Blood counts with differential white blood cell (WBC) analysis were performed using an ABC Vet Automated Blood Counter (VetNovations Canada). Lineage distribution was determined by FACS analysis (FACSCalibur; Becton Dickinson) as previously described.18 Monoclonal antibodies used were phycoerythrin (PE)–labeled Gr-1, Mac-1-PE, B220-PE, CD4-PE/CD8-PE, Ter119-PE, Sca-1-PE, and allophycocyanin–labeled c-kit (Pharmingen). Morphologic and histologic analysis of peripheral blood and bone marrow was performed as previously described.18 Briefly, mouse tissues were dissected and fixed overnight in 10% neutral buffered formalin solution, embedded in paraffin, and sectioned at 4 to 7 μm. Cytospin preparations and tissue sections were stained with hematoxylin and eosin. Images were visualized using a Nikon Eclipse 80i microscope (Nikon) and a 40 ×/0.65 numeric aperture objective, or a 100 ×/1.4 numeric aperture objective with Zeiss Immersol medium. A Nikon Coolpix 4500 camera (Nikon) and Canon ZoomBrowser EX 2.0 software (Canon) were used to capture images.
Expression analysis by RT-PCR
RNA was extracted and reverse transcribed as previously described.35 Quantitative reverse-transcriptase polymerase chain reaction (RT-PCR) was done as previously described using the 7900HT Fast Real-Time PCR system (Applied Biosystems).34 Relative expression was determined with the 2−ΔΔCT method,36 and the housekeeping gene transcript Abl1 was used to normalize the results. Primers were manufactured by Invitrogen. Primer sequences (5′ to 3′) are provided in supplemental Table 6 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
In vitro proliferation assays
Proliferation assays were performed in vitro in 15% FBS containing DMEM supplemented with either a combination of 3 cytokines (6 ng/mL mIL-3, 10 ng/mL hIL-6, 20 ng/mL mSCF) or only one of the following cytokines: 10 ng/mL murine granulocyte-macrophage colony-stimulating factor (mGM-CSF), 5 ng/mL murine granulocyte colony-stimulating factor (mG-CSF), 20 ng/mL murine macrophage–colony-stimulating factor (mM-CSF) or 3 U/mL human erythropoietin. All culture media and growth factors were obtained from StemCell Technologies Inc. Cells were seeded at a cell density 1 × 104/mL and were counted every 3 to 4 days with the Vi-Cell XR Cell Viability Analyzer (Beckman Coulter Inc). Fresh media and cytokines were added upon replating. In vitro cytotoxicity assays were performed as described.37 Tyrphostin AG 490 (Sigma-Aldrich) was dissolved in ethanol, and added to the culture medium at the specified concentrations as 1/1000th of the final volume.
Stimulation experiments and Western blot analysis
MN1+CTL–, MN1+ND13–, ND13+CTL–, MN1+HOXA9–, or HOXA9+CTL–transduced (immortalized) bone marrow cells were washed 3 times with PBS and cytokine-starved for 16 hours in DMEM containing 0.5% FBS. The cells were then stimulated with growth factors (50 ng/mL GM-CSF or 50 ng/mL mIL-3, 50 ng/mL hIL-6, 50 ng/mL mSCF) for 5 minutes at 37°C. For the stimulation experiments, 1 × 106 stimulated and nonstimulated cells were lysed with 80 μL 1 × sodium dodecyl sulfate (SDS) sample buffer, and passaged 5 times through a 26-gauge needle to shear DNA. The samples were boiled for 1 minute, separated on a 10% SDS–polyacrylamide gel electrophoresis, transferred to polyvinylidene fluoride, and probed with antibodies for pSTAT1 Tyr701, pSTAT3 Tyr705, pSTAT5 Tyr694, p-AKT Ser473 (all Cell Signaling Technology), p-ERK1/2 (phosphor-p44/42MAPK Thr202/Tyr204 E10 monoclonal antibody; New England Biolabs), SHIP P1C1 and BCL-2 (both Santa Cruz Biotechnology), or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a loading control (Research Diagnostics Inc). Blots were incubated with primary antibodies at dilutions recommended by the manufacturer for 2 hours at room temperature on an orbital shaker, washed 3 times for 10 minutes with Tris-buffered saline Tween-20 at room temperature, and incubated for 45 minutes with secondary antibody. Horseradish peroxidase–conjugated donkey anti–mouse or goat anti–rabbit antibodies (Sigma-Aldrich) in 0.1% Tween-20, 5% bovine serum albumin Tris-buffered saline were used for protein detection. Proteins were visualized using enhanced chemiluminescence reagent (Renaissance). Blots were exposed to Kodak X-OMAT Blue film for 5 seconds to 1 minute depending on the intensity of the signal. Densitometry was performed using the Versadoc 3000 Imaging System (Bio-Rad) and the Quantity One software (Bio-Rad) using GAPDH signals for normalization.
Gene expression profiling and statistical analysis
Gene expression profiling, cytogenetic, and molecular analyses of 201 diagnostic AML patient samples and 5 healthy controls from peripheral blood or bone marrow were performed as described by Valk et al.38 Patient samples were grouped according to recurrent cytogenetic or molecular aberrations. The gene set enrichment analysis software39 (http://www.broad.mit.edu/gsea/index.jsp)40 was used to compare enrichment of 13 Stat signaling-related and 5 interleukin/Jak signaling-related gene sets (available from the Molecular Signature database V2.5,39 (http://www.broad.mit.edu/gsea/msigdb/index.jsp, supplemental Tables 4-5) in cytogenetic or molecular patient subgroups to healthy controls, and to calculate normalized enrichment scores (NESs) for each gene set (1000 phenotype permutations). Significant enrichment of gene sets was set at a less than 25% false discovery rate (the estimated probability that a gene set represents a false-positive finding39 ). Pairwise comparisons were performed by Student t test for continuous variables and by chi-squared test for categoric variables. The 2-sided level of significance was set at P values less than .05. Comparison of survival curves were performed using the log-rank test. Statistical analyses were performed with Excel (Microsoft Canada) or GraphPad Prism 5 (GraphPad Software). LIC frequencies were calculated with the L-Calc software (StemCell Technologies).
Results
The leukemia-initiating cell frequency and expansion potential are greatly increased in a 2-oncogene compared with a 1-oncogene model of AML
Combined retroviral overexpression of 2 oncogenes in murine bone marrow cells has been used for leukemic complementation studies to identify synergy between genes that, alone, are nonleukemic, but may induce leukemia when expressed together. We exploited the MN1 leukemia model to study the impact on leukemia stem cell properties when an additional oncogene is overexpressed in MN1-transformed cells. For coexpression with MN1, we chose 2 genes of the HOX gene family, the fusion protein NUP98HOXD13 found in several AML patients and HOXA9 that is frequently overexpressed in AML. Cells retrovirally transduced with NUP98HOXD13 or HOXA9 grow as cytokine-dependent myeloid cell lines in vitro but do not induce leukemia in mice. We retrovirally transduced fluorouracil–treated murine bone marrow cells with MN1 and a control YFP vector (hereafter labeled CTL) or MN1+ND13 and selected/sorted for double-transduced cells in vitro. The leukemia stem cell frequency was determined by limiting dilution transplant assay for cells with leukemia-initiating cell (LIC) activity (Figure 1A). Limiting dilution analysis from 2 independent experiments demonstrated that the LIC frequency in MN1+ND13 cells was 33-fold higher than in MN1+CTL cells (1 LIC in 165 cells [0.6%] and 1 LIC in 5465 cells [0.018%], respectively, P < .01, Figure 1B). Mice that received a transplant of the lowest cell dose that engrafted mice (limiting dilution) had a significantly shorter survival when they received a transplant of MN1+ND13 cells compared with MN1+CTL cells (P = .009, Figure 1C), indicating that MN1+ND13 LICs had a greater proliferative and/or self-renewal potential than MN1+CTL LICs. To directly compare the self-renewal potential between the 2 leukemia models, we determined the fold increase of LICs in culture during a 6-day culture period. Limiting dilution transplant assays were performed at the beginning and at the end of the 6-day culture period (IL-3, IL-6, SCF), and the in vitro cell number increase was monitored. Strikingly, the absolute number of LICs increased 9001-fold in MN1+ND13 cells compared with 68-fold in MN1+CTL cells, corresponding to a 132-fold higher self-renewal expansion potential of MN1+ND13 cells (P < .01 for day-6 LIC frequency MN1+CTL vs MN1+ND13; Figure 1D and supplemental Table 1).
The leukemia-initiating cell frequency and expansion potential are greatly increased in a 2-oncogene compared with a 1-oncogene model of acute myeloid leukemia. (A) Experimental design of limiting dilution assay for LIC and assessment of LIC expansion potential during 6 days of in vitro culture. (B) LIC assay from bone marrow cells expressing 1 (MN1+CTL) or 2 (MN1+ND13) oncogenes. Cells were cultured ex vivo for infection and selection for 21 days. Results of 2 independent experiments were analyzed together after observation of mice for 16 weeks (n = 4-7 per cell dose, 95% CI). LIC frequency and 95% CI were calculated by Poisson statistics. (C) Survival curves for mice that received a transplant of a cell dose at limiting dilution either transduced with MN1+CTL (n = 4) or MN1+ND13 (n = 4). (D) Consecutive CRU assays (5 cell doses) before and after a 6-day culture period from MN1+CTL- compared with MN1+ND13-expressing cells. Cells were cultured ex vivo for infection and selection for 13 days (corresponds to day 0 of expansion experiment), and 105 cells were plated on day 0. LIC frequency was calculated by Poisson statistics. *P < .01 for day-6 LIC frequency MN1+CTL versus MN1+ND13. AB indicates antibiotics for selection of transgene-expressing cells; CTL, control; and ND13, NUP98HOXD13 fusion gene
The leukemia-initiating cell frequency and expansion potential are greatly increased in a 2-oncogene compared with a 1-oncogene model of acute myeloid leukemia. (A) Experimental design of limiting dilution assay for LIC and assessment of LIC expansion potential during 6 days of in vitro culture. (B) LIC assay from bone marrow cells expressing 1 (MN1+CTL) or 2 (MN1+ND13) oncogenes. Cells were cultured ex vivo for infection and selection for 21 days. Results of 2 independent experiments were analyzed together after observation of mice for 16 weeks (n = 4-7 per cell dose, 95% CI). LIC frequency and 95% CI were calculated by Poisson statistics. (C) Survival curves for mice that received a transplant of a cell dose at limiting dilution either transduced with MN1+CTL (n = 4) or MN1+ND13 (n = 4). (D) Consecutive CRU assays (5 cell doses) before and after a 6-day culture period from MN1+CTL- compared with MN1+ND13-expressing cells. Cells were cultured ex vivo for infection and selection for 13 days (corresponds to day 0 of expansion experiment), and 105 cells were plated on day 0. LIC frequency was calculated by Poisson statistics. *P < .01 for day-6 LIC frequency MN1+CTL versus MN1+ND13. AB indicates antibiotics for selection of transgene-expressing cells; CTL, control; and ND13, NUP98HOXD13 fusion gene
These dramatic differences in the measured LIC self-renewal characteristics between MN1+ND13 and MN1 mice were not evident by standard evaluation of leukemic mice (Figure 2). Mice that received a transplant of a high dose of MN1+ND13 or MN1 transgene–positive cells (106 cells per mouse, transplanted directly after selection/sorting) had similar disease latencies (Figure 2A), similarly enlarged spleens, increased WBC counts, and reduced red blood cell (RBC) counts in peripheral blood when they became moribund (Figure 2B-D). From both leukemia models, 10% to 20% of bone marrow cells expressed myeloid markers (Gr-1 and Mac-1) but lacked lymphoid markers. C-kit expression was significantly higher in MN1 cells compared with MN1+ND13 cells (81% vs 54%, respectively, P < .001, Figure 2E), consistent with previous reports of MN1 and ND13+MEIS1 leukemias.18,27 Morphologic analysis of bone marrow cells confirmed the diagnosis of AML, and spleen and liver sections showed infiltration of these organs by both leukemia models (Figure 2F). Disease latency of secondary transplants was not significantly different for MN1 mice (as previously reported18 ). However, secondary transplants of MN1+ND13 leukemias had a significantly shorter survival compared with the primary transplants (P < .001, Figure 2G), indicating either the increased self-renewal expansion potential of these cells or acquisition of additional somatic mutations.
Phenotypically the 2-oncogene (MN1+ND13) and the 1-oncogene (MN1) leukemia model produce similar leukemias. (A) Survival analysis of mice that received a transplant of MN1-transduced (n = 18), MN1+ND13-transduced (n = 13), or control vector-transduced (n = 7) bone marrow cells. (B) Spleen weight, (C) WBC, and (D) RBC counts of moribund leukemic MN1 (n = 10) and MN1+ND13 (n = 5) mice, and of normal control mice (n = 3; mean ± SD). (E) Immunophenotype of leukemic bone marrow cells from moribund MN1 (n = 9) and MN1+ND13 (n = 7) mice gated on transgene-expressing cells (mean ± SD). (F) Representative Wright-Giemsa–stained cytospin preparation of bone marrow cells from leukemic mice, and hematoxylin and eosin–stained tissue sections from spleen and liver of leukemic mice (scale bars represent 10 μm). (G) Survival analysis of mice that received a transplant of MN1+ND13-expressing cells (primary, n = 13) and of leukemic MN1+ND13-expressing leukemic bone marrow (secondary, n = 9).
Phenotypically the 2-oncogene (MN1+ND13) and the 1-oncogene (MN1) leukemia model produce similar leukemias. (A) Survival analysis of mice that received a transplant of MN1-transduced (n = 18), MN1+ND13-transduced (n = 13), or control vector-transduced (n = 7) bone marrow cells. (B) Spleen weight, (C) WBC, and (D) RBC counts of moribund leukemic MN1 (n = 10) and MN1+ND13 (n = 5) mice, and of normal control mice (n = 3; mean ± SD). (E) Immunophenotype of leukemic bone marrow cells from moribund MN1 (n = 9) and MN1+ND13 (n = 7) mice gated on transgene-expressing cells (mean ± SD). (F) Representative Wright-Giemsa–stained cytospin preparation of bone marrow cells from leukemic mice, and hematoxylin and eosin–stained tissue sections from spleen and liver of leukemic mice (scale bars represent 10 μm). (G) Survival analysis of mice that received a transplant of MN1+ND13-expressing cells (primary, n = 13) and of leukemic MN1+ND13-expressing leukemic bone marrow (secondary, n = 9).
Enhanced STAT and Erk signaling in the 2-oncogene compared with the 1-oncogene model
Bone marrow cells transduced with MN1 alone, ND13 alone, or MN1+ND13 grow as cytokine-dependent myeloid cell lines in vitro. We hypothesized that differences in the cytokine responsiveness of these cells may reflect differences in the self-renewal behavior of these cells and underlying active signaling pathways. Thus we investigated the growth properties of these cells under various cytokine conditions. MN1+CTL and MN1+ND13 cells proliferated with similar growth rates when cultured in either IL-3 (6 ng/mL) or SCF (20 ng/mL; Figure 3A-B), but failed to grow when cultured in either IL-6 (10 ng/mL), G-CSF (5 ng/mL), M-CSF (20 ng/mL), or erythropoietin (3U/mL; data not shown). Interestingly, MN1+ND13 cells proliferated in the presence of GM-CSF (10 ng/mL), whereas MN1+CTL cells did not (P < .05, Figure 3C). Increased proliferation in response to GM-CSF stimulation was also observed in cells expressing MN1+HOXA9 compared with MN1+CTL (supplemental Figure 1A-B).
GM-CSF hypersensitivity in the 2-oncogene compared with the 1-oncogene model. (A-C) Cytokine stimulation assay of murine bone marrow cells stably transduced with MN1+ND13 or MN1+CTL. Cells were cultured in the presence of the indicated cytokines (6 ng/mL IL-3, 10 ng/mL IL-6, 20 ng/mL SCF, or 10 ng/mL GM-CSF), and viable cells were counted and replated every 3 to 4 days (mean ± SD, n = 4). (D) Quantitative gene expression analysis of Gm-csfrα, Il-3rα, and G-csfrα in bone marrow cells of moribund mice that received a transplant of MN1+ND13- and MN1+CTL-expressing cells (mean ± SD). (E-F) Quantitative gene expression analysis of agonistic (E) and antagonistic (F) signaling molecules downstream of the GM-CSFR in leukemic cell lines MN1+CTL and MN1+ND13 in relation to gene expression in the nonleukemic cell line ND13 (mean ± SD, n = 4). $ND13 was used as a nonleukemic calibrator.
GM-CSF hypersensitivity in the 2-oncogene compared with the 1-oncogene model. (A-C) Cytokine stimulation assay of murine bone marrow cells stably transduced with MN1+ND13 or MN1+CTL. Cells were cultured in the presence of the indicated cytokines (6 ng/mL IL-3, 10 ng/mL IL-6, 20 ng/mL SCF, or 10 ng/mL GM-CSF), and viable cells were counted and replated every 3 to 4 days (mean ± SD, n = 4). (D) Quantitative gene expression analysis of Gm-csfrα, Il-3rα, and G-csfrα in bone marrow cells of moribund mice that received a transplant of MN1+ND13- and MN1+CTL-expressing cells (mean ± SD). (E-F) Quantitative gene expression analysis of agonistic (E) and antagonistic (F) signaling molecules downstream of the GM-CSFR in leukemic cell lines MN1+CTL and MN1+ND13 in relation to gene expression in the nonleukemic cell line ND13 (mean ± SD, n = 4). $ND13 was used as a nonleukemic calibrator.
GM-CSF binding to its receptor alpha chain (CSF2RA, GM-CSFRα) recruits the beta common chain that is shared by IL-3, IL-5, and GM-CSF, and activates the intracellular signaling JAK/STAT, ERK, and PI3K/AKT pathways.41 Gm-csfra and Mpl gene expression was not significantly different in bone marrow cells or cell lines of leukemic MN1+ND13 or MN1+CTL mice, respectively (Figure 3D-E). Expression analysis of kinases downstream of the GM-CSFR showed decreased expression of Jak1, Jak2, Lck, and Lyn in leukemic MN1+ND13 and MN1+CTL cell lines compared with nonleukemic ND13+CTL cells. However, Lyn expression was higher in MN1+ND13 cells compared with MN1+CTL cells (P = .02, Figure 3E). The 50% inhibitory concentration for the Jak2 inhibitor tyrphostin was not significantly different in MN1+CTL compared with MN1+ND13 cells (supplemental Figure 2). Suppressor of cytokine signaling (SOCS) family proteins can terminate GM-CSF–induced agonistic signals. Interestingly, Socs1 and Socs2 expression was significantly reduced in MN1+ND13 compared with MN1+CTL cells (Figure 3F), indicating a potential mechanism for the increased GM-CSF response of MN1+ND13 cells involving a lack of signal termination. Expression levels of potential autocrine or paracrine factors were not significantly different between MN1+CTL and MN1+ND13 cells (supplemental Figure 3). Gene expression analysis of HoxA and HoxB cluster genes showed that genes of the HoxB cluster were expressed at higher levels in MN1+ND13 cells compared with MN1+CTL cells (supplemental Figure 4). To directly assess downstream signaling events in our cell lines, Western blot analysis of phosphorylated STAT1, STAT3, STAT5, ERK1/2, and AKT was performed in cytokine-stimulated cells (Figure 4A-B). Densitometry analysis of Western blot results normalized to GAPDH demonstrated that phosphorylation of STAT3, STAT1, and ERK1/2 was increased in MN1+ND13 cells compared with ND13+CTL or MN1+CTL cells when stimulated with IL-3/IL-6/SCF (Figure 4C). Stimulation with GM-CSF resulted in increased phosphorylation of STAT5, STAT3, STAT1, and ERK1/2 in MN1+ND13 compared with MN1+CTL and ND13+CTL cells (Figure 4D). No difference in phosphorylation of AKT and in protein expression of the negative regulator of GM-CSF signaling SHIP (also known as SHIP1) and the antiapoptotic protein BCL-2 were observed (Figure 4A). This experiment was repeated in a similar leukemia model where MN1 was co-overexpressed with HOXA9. Stimulation with GM-CSF resulted in increased phosphorylation of STAT3, STAT1, and ERK1/2 almost exclusively in MN1+HOXA9 cells, and in increased levels of phosphorylated STAT3, STAT1, and ERK1/2 in MN1+HOXA9 cells when stimulated with IL-3/IL-6/SCF (Figure 4B). Collectively these data demonstrate that STAT1, STAT3, STAT5, and ERK1/2 signaling is enhanced in MN1+ND13 and MN1+HOXA9 compared with MN1+CTL and ND13+CTL leukemias.
Enhanced STAT and ERK signaling in the 2-oncogene compared with the 1-oncogene model. (A) Western blot analysis of pSTAT1, pSTAT3, pSTAT5, pERK1/2, pAKT, SHIP, and BCL-2 proteins in the indicated cell lines. Cells were starved for 16 hours in 0.5% FBS medium and then stimulated with starve media or with saturating concentrations (*) of mIL-3 (50 ng/mL), hIL-6 (50 ng/mL), mSCF (50 ng/mL), or GM-CSF (50 ng/mL) for 5 minutes at 37°C. Total-cell lysates were separated by SDS-polyacrylamide gel electrophoresis, transferred to polyvinylidene fluoride, and probed with indicated antibodies or GAPDH as a loading control. (B) Western blot analysis of pSTAT and pERK1/2 proteins in MN1+CTL, MN1+HOXA9, and HOXA9+CTL cells pretreated as described for panel A. (C-D) Densitometry analysis of Western blot analysis of phosphorylated STAT and ERK1/2 proteins in the indicated cell lines stimulated with IL-3/IL-6/SCF (C) or GM-CSF (D; normalized to GAPDH, mean ± SD, n = 2 [see panel A and data not shown]).
Enhanced STAT and ERK signaling in the 2-oncogene compared with the 1-oncogene model. (A) Western blot analysis of pSTAT1, pSTAT3, pSTAT5, pERK1/2, pAKT, SHIP, and BCL-2 proteins in the indicated cell lines. Cells were starved for 16 hours in 0.5% FBS medium and then stimulated with starve media or with saturating concentrations (*) of mIL-3 (50 ng/mL), hIL-6 (50 ng/mL), mSCF (50 ng/mL), or GM-CSF (50 ng/mL) for 5 minutes at 37°C. Total-cell lysates were separated by SDS-polyacrylamide gel electrophoresis, transferred to polyvinylidene fluoride, and probed with indicated antibodies or GAPDH as a loading control. (B) Western blot analysis of pSTAT and pERK1/2 proteins in MN1+CTL, MN1+HOXA9, and HOXA9+CTL cells pretreated as described for panel A. (C-D) Densitometry analysis of Western blot analysis of phosphorylated STAT and ERK1/2 proteins in the indicated cell lines stimulated with IL-3/IL-6/SCF (C) or GM-CSF (D; normalized to GAPDH, mean ± SD, n = 2 [see panel A and data not shown]).
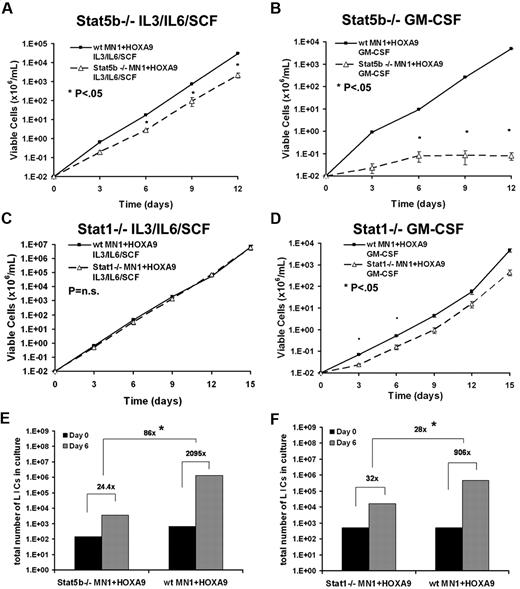
STAT5b, and to a lesser extent STAT1, mediate massive leukemia stem cell expansion
Two differentially activated signaling molecules, STAT1 and STAT5b, were chosen for further study due to the availability of knockout mice. The requirement of STAT5b and STAT1 for GM-CSF hypersensitivity and self-renewal expansion of LICs in MN1+HOXA9 leukemias was tested using bone marrow from Stat5b- or Stat1-null mice and compared with their respective wild-type littermates. Freshly isolated bone marrow cells were retrovirally transduced with MN1+HOXA9 and sorted/selected for transgene double-positive cells. With these cells, we performed cytokine stimulation assays and limiting dilution assays for LIC. In contrast to wild-type cells, Stat5b-null MN1+HOXA9 cells demonstrated a reduced proliferation rate compared with wild-type cells in IL-3/IL-6/SCF–supplemented media and were unresponsive to GM-CSF stimulation (P < .05, Figure 5A-B). Stat1-null MN1+HOXA9 cells proliferated at the same rate as wild-type cells in response to IL-3/IL-6/SCF and showed only mild growth retardation in response to GM-CSF (Figure 5C-D). Next, LIC frequencies were measured at the beginning and end of a 6-day culture period (IL-3, IL-6, SCF), and the cell number increase in vitro was monitored to compare the self-renewal expansion potential of wild-type and knockout leukemia models. Strikingly, the number of LICs increased 2095-fold in wild-type cells transduced with MN1+HOXA9 cells compared with 24.4-fold in Stat5b−/− cells transduced with MN1+HOXA9 cells, corresponding to an 86-fold lower self-renewal expansion potential when STAT5b was absent (Figure 5E and supplemental Table 2). Proliferation of bulk STAT5b-null cells was only 7.1-fold lower than proliferation of wild-type cells, suggesting that STAT5b had a major direct effect on self-renewal. The self-renewal expansion potential of MN1+HOXA9 cells in Stat1−/− cells was 28-fold reduced during a 6-day in vitro culture period compared with wild-type MN1+HOXA9 cells (Figure 5F and supplemental Table 3). Proliferation of bulk STAT1-null cells was only 2-fold lower than proliferation of wild-type cells. Differences in the absolute level of self-renewal expansion may be accounted for by different mouse strains used for the knockout studies. Equal LIC frequencies in STAT1-null and wild-type cells on day 0 may be due to the overall smaller effect of STAT1 on self-renewal and the limited sensitivity of the assay. These experiments demonstrate a major contribution of STAT5b, and to a lesser extent STAT1, to the self-renewal of leukemia stem cells in the MN1+HOXA9 combination leukemia model.
STAT5b and to a lesser extent STAT1 mediate massive leukemia stem cell expansion. (A-B) Cytokine stimulation assay of wild-type or Stat5b−/− murine bone marrow cells stably transduced with MN1+HOXA9. Cells were cultured in the presence of the indicated cytokines (6 ng/mL IL-3, 10 ng/mL IL-6, 20 ng/mL SCF, or 10 ng/mL GM-CSF), and viable cells were counted and replated every 3 days (mean ± SD, n = 3). (C-D) Cytokine stimulation assay of wild-type or Stat1−/− murine bone marrow cells stably transduced with MN1+HOXA9. Cells were cultured in the presence of the indicated cytokines (6 ng/mL IL-3, 10 n/mL IL-6, 20 ng/mL SCF, or 10 ng/mL GM-CSF), and viable cells were counted and replated every 3 days (mean ± SD, n = 3). (E) Consecutive limiting dilution assays for LIC (4 cell doses) before and after a 6-day culture period from wild-type compared with Stat5b−/− bone marrow cells expressing MN1+HOXA9. Cells were cultured ex vivo for infection and selection for 16 days (corresponds to day 0 of expansion experiment), and 105 cells were plated on day 0. LIC frequency was calculated by Poisson statistics. (F) Consecutive limiting dilution assays for LIC (4 cell doses) before and after a 6-day culture period from wild-type compared with Stat1−/− bone marrow cells expressing MN1+HOXA9. Cells were cultured ex vivo for infection and selection for 16 days (corresponds to day 0 of expansion experiment), and 105 cells were plated on day 0. LIC frequency was calculated by Poisson statistics. *P < .01 for day-6 LIC frequency MN1+HOXA9 in null versus MN1+HOXA9 in wild-type cells.
STAT5b and to a lesser extent STAT1 mediate massive leukemia stem cell expansion. (A-B) Cytokine stimulation assay of wild-type or Stat5b−/− murine bone marrow cells stably transduced with MN1+HOXA9. Cells were cultured in the presence of the indicated cytokines (6 ng/mL IL-3, 10 ng/mL IL-6, 20 ng/mL SCF, or 10 ng/mL GM-CSF), and viable cells were counted and replated every 3 days (mean ± SD, n = 3). (C-D) Cytokine stimulation assay of wild-type or Stat1−/− murine bone marrow cells stably transduced with MN1+HOXA9. Cells were cultured in the presence of the indicated cytokines (6 ng/mL IL-3, 10 n/mL IL-6, 20 ng/mL SCF, or 10 ng/mL GM-CSF), and viable cells were counted and replated every 3 days (mean ± SD, n = 3). (E) Consecutive limiting dilution assays for LIC (4 cell doses) before and after a 6-day culture period from wild-type compared with Stat5b−/− bone marrow cells expressing MN1+HOXA9. Cells were cultured ex vivo for infection and selection for 16 days (corresponds to day 0 of expansion experiment), and 105 cells were plated on day 0. LIC frequency was calculated by Poisson statistics. (F) Consecutive limiting dilution assays for LIC (4 cell doses) before and after a 6-day culture period from wild-type compared with Stat1−/− bone marrow cells expressing MN1+HOXA9. Cells were cultured ex vivo for infection and selection for 16 days (corresponds to day 0 of expansion experiment), and 105 cells were plated on day 0. LIC frequency was calculated by Poisson statistics. *P < .01 for day-6 LIC frequency MN1+HOXA9 in null versus MN1+HOXA9 in wild-type cells.
MN1 and HOXA9 coexpression in AML patients is associated with highly active STAT signaling and poor risk AML
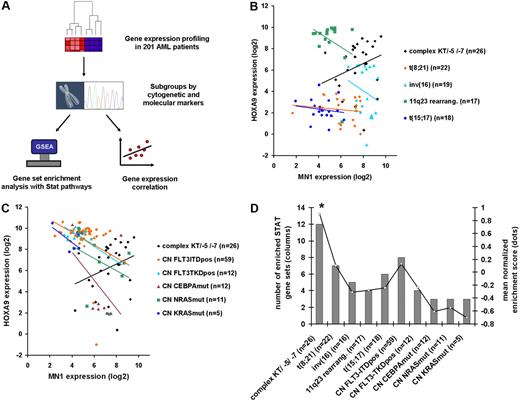
In AML patients with normal karyotypes, it is well documented that HOX gene expression (HOXA4, A5, A6, A7, A9, A10, B2, B3, B5, B6) is inversely correlated with MN1 gene expression.13,14 Patients with mutated NPM1 genes have high HOX gene expression, whereas patients with nonmutated NPM1 genes have high MN1 expression. Consistent with this, we found that gene expression of MN1 and HOXA9 was inversely correlated in the majority of the 201 AML patients with various cytogenetic and molecular subgroups analyzed (Figure 6B-C). Strikingly, however, we found that MN1 and HOXA9 gene expression was only positively correlated in patients with complex karyotype/loss of chromosome 5 or 7 (R = 0.38; Figure 6B-C). We next evaluated the gene expression levels of target genes of STAT family members as a measure of the activation status of STAT proteins using the gene set enrichment analysis (GSEA) algorithm in previously published gene expression profiles of 201 AML patients characterized by defined cytogenetic or molecular markers38 (Figure 6A). Thirteen curated gene sets were available from the Molecular Signatures Database (http://www.broad.mit.edu/gsea/msigdb), consisting of genes regulated by various STAT family members (supplemental Table 4). Differential expression of the genes contained in one gene set was calculated for each subgroup of patients and represented by the normalized enrichment score (NES). Twelve of the 13 STAT gene sets were significantly enriched in patients with complex karyotype or other high-risk karyotypes (loss of chromosome 5 or 7), but at a much lower proportion in all other subgroups (Figure 6D). The mean NES of the 13 STAT gene sets was significantly higher in patients with complex karyotype/loss of chromosome 5 or 7 (NES 0.9) compared with all other cytogenetic or molecular subgroups (NES 0.13 in patients with normal karyotype and internal tandem duplication of FLT3; NES 0.1 in patients with t(8;21)(q22;q22); negative NES in all other cytogenetic or molecular subgroups [P < .02, Figure 6D]). Interleukin/JAK signaling-related gene sets (supplemental Table 5) were depleted in all patient groups compared with healthy controls (supplemental Figure 5). Taken together, MN1 and HOXA9 co-overexpression was found in the most aggressive subgroup of AML and was associated with strongly activated STAT signaling, corresponding to the results of the MN1+HOX mouse models.
STAT signaling is most highly activated in AML patients with poor prognosis and is associated with MN1 and HOXA9 co-overexpression. (A) Experimental design and analysis algorithm of gene expression and pathway analysis in 201 AML patients of defined cytogenetic or molecular subgroups. (B-C) Correlation of gene expression of MN1 and HOXA9 genes in 201 AML patients according to cytogenetic (B) and molecular (C) subgroups. Complex karyotype/loss of chromosome 5/loss of chromosome 7 AML is the only subgroup with positively correlated MN1 and HOXA9 expression, and with high expression of both genes. (D) Results of gene set enrichment analysis with 13 STAT signaling gene sets in AML patient subgroups using previously published gene expression profiles. Bars represent number of significantly enriched gene sets in subgroup (scale on left axis); line represents mean normalized enrichment score of the 13 gene sets tested (scale on right axis). *P < .02 for comparison of mean NES score in patients with complex karyotype/−5/−7 with any other cytogenetic or molecular subgroup.
STAT signaling is most highly activated in AML patients with poor prognosis and is associated with MN1 and HOXA9 co-overexpression. (A) Experimental design and analysis algorithm of gene expression and pathway analysis in 201 AML patients of defined cytogenetic or molecular subgroups. (B-C) Correlation of gene expression of MN1 and HOXA9 genes in 201 AML patients according to cytogenetic (B) and molecular (C) subgroups. Complex karyotype/loss of chromosome 5/loss of chromosome 7 AML is the only subgroup with positively correlated MN1 and HOXA9 expression, and with high expression of both genes. (D) Results of gene set enrichment analysis with 13 STAT signaling gene sets in AML patient subgroups using previously published gene expression profiles. Bars represent number of significantly enriched gene sets in subgroup (scale on left axis); line represents mean normalized enrichment score of the 13 gene sets tested (scale on right axis). *P < .02 for comparison of mean NES score in patients with complex karyotype/−5/−7 with any other cytogenetic or molecular subgroup.
Discussion
We quantified the frequency and expansion potential of LICs in a progression model of AML driven by MN1 and HOX oncogenes. Both LIC frequency and expansion potential were greatly increased in the 2-oncogene compared with the 1-oncogene model, documenting the functional heterogeneity of LICs. Coexpression of MN1 and HOXA9 enhanced Jak/STAT and Erk signaling, and loss-of-function studies demonstrated that STAT5b and to a lesser extent STAT1 are critical for the excess expansion potential in MN1/HOXA9 compared with MN1 leukemias. Corresponding to our murine studies, MN1 and HOXA9 coexpression was associated with strong activation of STAT target genes in AML patients, and with the cytogenetic subgroup representing the most aggressive leukemia (complex karyotype/loss of chromosome 5 or 7).
By genetic modeling, we demonstrated the enormous functional heterogeneity of LICs in murine models of constitutive expression of MN1 with or without a HOX gene. Taking advantage of the leukemogenic nature of the MN1 oncogene allowed the generation of a more aggressive leukemia through the coexpression of only 2 genes. Surprisingly, MN1 and MN1+HOX leukemias had very similar phenotypes and disease latencies when transplanted at a high cell dose. Therefore, rather than relying on phenotypic enumeration of LICs, we assessed LIC frequency and expansion potential functionally. The LIC frequency was higher in the MN1+ND13 than in the MN1 model but was similar in each model before and after a 6-day in vitro culture period (supplemental Table 1). However, MN1+ND13 cells proliferated at a much higher rate during the expansion period in vitro than the MN1 cells, thus accounting for the increased expansion potential. During cell division, stem cells either divide into 2 more differentiated (non–stem cell) daughter cells, 1 differentiated and 1 stem cell (self-renewal maintenance), or 2 stem cells (self-renewal expansion). Based on the observed increase in LICs during the 6-day in vitro culture, we concluded that LICs preferentially underwent self-renewal expansion divisions. Interestingly, the fold increase of LICs was even higher than the total fold increase of all cells. Although we cannot assess the rate of proliferation, senescence, or apoptosis for individual cells, this finding suggests that LICs proliferated at a higher rate (or apoptosed less) than their progeny. This is in contrast to a recently reported model of CML in which quiescence of phenotypically defined LICs was associated with an increased self-renewal potential.42 However, our tests of the LIC expansion potential in vitro may not reflect the complex interactions of leukemic cells with their in vivo environment.
From cytokine stimulation studies, hypersensitivity to GM-CSF stimulation was identified as the major difference between the 1- and 2-oncogene models in vitro. This appeared to be a true synergistic effect of MN1 and ND13 or HOXA9, as none of the single-oncogene models were responsive to GM-CSF. Hypersensitivity to GM-CSF is found in patients with juvenile myelomonocytic leukemia, a rare form of chronic infant or childhood leukemia,43 and is used as a diagnostic criterion. Genetically, mutations in PTPN11,44,45 the RAS family of oncogenes,46 and NF147 are frequently found in these patients as well as STAT5 activation.48,49 Analysis of JAK/STAT and ERK pathways showed preferential activation of these pathways in the 2-oncogene model, whereas the PI3K/AKT pathway that is also activated by GM-CSFR signaling was not differentially activated. In addition, Gm-csfr was not differentially expressed between the different leukemia models, although Socs1 and Socs2, which are induced by STAT5 and function as negative regulators of STAT5 and STAT3,50,51 were significantly down-regulated in the MN1+ND13 model. Inhibition of JAK2 did not yield differential cytotoxicity in MN1+ND13 compared with MN1+CTL cells. In fact, Jak1 and Jak2 were down-regulated in the 2 cell lines, and interleukin pathways that signal through JAK2 were down-regulated in AML patients. Our data suggest that the observed GM-CSF hypersensitivity in MN1+ND13 cells is more a result of a lack of termination signal of the JAK/STAT pathway and less a result of JAK2 hyperactivation.
The family of STAT signaling proteins comprises 7 members that transduce extracellular signals to the nucleus to regulate transcription of target genes.41 There is considerable structural and functional overlap between the family members, and STAT5A and STAT5B are 96% conserved at the protein level.52 To characterize the significance of STAT signaling for LIC self-renewal, MN1 and HOXA9 were coexpressed in Stat1- or Stat5b-deficient bone marrow cells and their LIC expansion potential was compared with MN1+HOXA9-transduced wild-type cells. The LIC expansion potential was reduced in Stat5b- and, to a lesser extent, Stat1-deficient cells, suggesting a critical role for STAT signaling in the MN1+HOXA9 model. Constitutive STAT5 activation has been found in a large proportion of AML patients,53,54 and in some patients STAT5B was selectively phosphorylated.55 Although LIC expansion was reduced in Stat5b-deficient cells these cells were still leukemogenic. Other STAT members, namely STAT5a, may have been recruited to replace STAT5b function. Other studies have shown in a compound knockout of Stat5a and Stat5b that STAT5 is indispensable for the leukemogenicity of BCR-ABL56 and TEL-PDGFβR.57 Future studies will be needed to clarify whether STAT signaling mediates only the excess expansion found in MN1+HOX leukemias compared with MN1 leukemias or whether it is required also for MN1-induced LIC expansion. This will have important implications for STAT proteins as therapeutic targets.
The heterogeneity of AML is evident at the phenotypic, genetic, and molecular level, as well as in the different survival times of AML patients. Whereas patients with fusion genes AML1/ETO, PML/RARA, or CBFβ/MYH11 have a good prognosis, other patients with loss of chromosomes 5 or 7 or a complex karyotype have a very poor prognosis.58,59 In 50% of patients without cytogenetic aberrations, NPM1 mutations have been identified.1 It has been shown that NPM1-mutated patients overexpress HOXA9 and other HOX genes, whereas nonmutated patients overexpress MN1 and other genes.13-15 MN1 and HOXA9 expression was not positively correlated in any of the cytogenetic or molecular subgroups in the 201 analyzed AML patients except in the very poor risk AML group of complex karyotype or loss of chromosome 5 or 7. In this subgroup, STAT signaling was most active compared with all other groups, and the mean normalized enrichment score of STAT-regulated gene sets was much lower in FLT3-ITD–positive patients (a mutation known to activate STAT signaling60,61 ), or even negative in other subgroups compared with patients with complex karyotype or loss of chromosome 5 or 7. Interestingly, Schepers et al investigated the effect of shRNA-mediated knockdown of STAT5 in primary CD34+ AML cells and found that STAT5 knockdown impaired growth and expansion potential of AML progenitor cells.62 We suggest that the MN1 and HOXA9 coexpression leukemia may serve as a model of complex karyotype/poor risk AML. Our findings suggest that STAT signaling may be most effectively targeted in poor risk AML, and that treatment of aggressive leukemias will require inhibition of multiple pathways.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by grants from the Terry Fox Foundation, the Canadian Stem Cell Network, the Cancer Research Society, and the Deutsche Forschungsgemeinschaft (M.H., grant number He 5240/1-1). B.A. is a recipient of a Leukemia Research Fund of Canada fellowship. F.K. is supported by the Deutsche Forschungsgemeinschaft Germany (grant number Ku 2288/1-1).
Authorship
Contribution: M.H. designed and performed the research, analyzed the data, and wrote the paper; L.M.S. designed and performed the research and analyzed the data; B.A., F.K., C.L., A.W., M.L., G.L., S.F., P.J.V., and R.D. performed the research and analyzed the data; C.B. performed the research; B.L. and G.K. analyzed the data; and R.K.H designed the research, analyzed the data, and wrote the paper. All authors read and agreed to the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Corresponding author: R. Keith Humphries, Terry Fox Laboratory, British Columbia Cancer Agency, 675 West 10th Ave, Vancouver, BC, V5Z 1L3 Canada; e-mail: khumphri@bccrc.ca.




![Figure 4. Enhanced STAT and ERK signaling in the 2-oncogene compared with the 1-oncogene model. (A) Western blot analysis of pSTAT1, pSTAT3, pSTAT5, pERK1/2, pAKT, SHIP, and BCL-2 proteins in the indicated cell lines. Cells were starved for 16 hours in 0.5% FBS medium and then stimulated with starve media or with saturating concentrations (*) of mIL-3 (50 ng/mL), hIL-6 (50 ng/mL), mSCF (50 ng/mL), or GM-CSF (50 ng/mL) for 5 minutes at 37°C. Total-cell lysates were separated by SDS-polyacrylamide gel electrophoresis, transferred to polyvinylidene fluoride, and probed with indicated antibodies or GAPDH as a loading control. (B) Western blot analysis of pSTAT and pERK1/2 proteins in MN1+CTL, MN1+HOXA9, and HOXA9+CTL cells pretreated as described for panel A. (C-D) Densitometry analysis of Western blot analysis of phosphorylated STAT and ERK1/2 proteins in the indicated cell lines stimulated with IL-3/IL-6/SCF (C) or GM-CSF (D; normalized to GAPDH, mean ± SD, n = 2 [see panel A and data not shown]).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/19/10.1182_blood-2009-06-227603/4/m_zh89990942920004.jpeg?Expires=1767719319&Signature=pCw2WjAYWHW0t2mbk0tqPFDOUJafouVfD~fO6nGcU4aXu9x8El2KF~UPhPrxeezxTHjrxHo2kMHO1~WvC3273bAHdMNODh8H~2mXmCXHG4eVe63-JHp3LQOlsdB~ny3nL3R1lDvW5yp1SkSI-XPP2qyivBx9nhdYloW2cmPLXSq11eTb-cojZUNJJ-tL8-wndUP-pZsqHvp846zIdg7awzsAnNRdyFNWV2w83tleBW8hgsBVc2DDknIBLl81Nd3rESnEbLdpLnKVjuuzlOJy6ifQpiRqjnYTcrQdaF3bQ1k1cRcRXAc8uThX21psx1JKAbn1wf652XZtdWlcL74z9A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)