Abstract
Chromosomal translocations involving 11q23 are frequent in infant acute leukemia and give rise to the formation of MLL fusion genes. The mechanism of leukemic transformation by these fusions has been the subject of numerous investigations. However, the dependence of acute leukemia on MLL fusion activity in vivo and the efficacy of targeting this activity to eliminate disease have not been established. We have developed a model for conditional expression of MLL-ENL in hematopoietic progenitor cells, in which expression of the fusion oncogene is turned off by doxycycline. Conditionally immortalized myeloblast cells derived from these progenitors were found to induce leukemia in vivo. Leukemic cells isolated from primary recipient mice were shown to have acquired additional genetic abnormalities and, when transplanted into secondary recipients, induced leukemia with shortened latencies. However, the leukemic cells remained dependent on MLL-ENL expression in vitro and in vivo, and its ablation resulted in regression of established leukemias. This study demonstrates that even genetically complex leukemias can be reversed on inactivation of the initiating MLL fusion and has important implications for the design of novel leukemia therapies.
Introduction
A central goal of cancer research is the development of specific therapies for malignancies, which will avoid or minimize the toxicities associated with current treatment protocols. Development of such therapies relies on understanding the underlying biology of the cancer in question. An example of this approach is the study of MLL fusion oncogenes in leukemia.1–4 These fusions are generated by chromosomal translocations of the MLL gene on chromosome 11q235 and are prevalent in infant acute lymphoid (ALL) and myeloid (AML) leukemia, and in treatment-related AML.6,7
Several studies have addressed the importance of MLL fusion activity for the initiation and maintenance of hematopoietic transformation. We and others have used conditional expression models to demonstrate that immortalization of cells transformed by MLL-ENL is abrogated in vitro on loss of fusion protein activity.8–10 Elegant studies have also shown that cofactors, such as Menin, and downstream transcriptional targets, such as Meis-1, Hoxa genes, and Mef2c, are required for in vitro transformation by certain MLL fusions8,11–14 and, in some cases, leukemia engraftment.15 However, it remains to be demonstrated whether interfering with MLL fusion activity will translate into elimination of established leukemia in vivo. In this regard, it is important to exclude that the bone marrow microenvironment may support the survival of leukemic stem cells in the absence of MLL fusion activity and that acquisition of secondary mutations by leukemic cells in vivo may render them resistant to MLL fusion inhibition.
The importance of secondary mutations and genetic aberrations for the initiation and progression of acute leukemias associated with MLL translocations is not fully understood. The brief latencies of infant acute leukemias involving MLL/11q23 rearrangements and their high concordance in monozygotic twins16 suggest that MLL fusions may be sufficient to cause overt leukemia. This is consistent with the rapid onset of malignancy on de novo formation of the MLL-ENL fusion in mice17 and leukemia induction by human cells expressing MLL fusion genes.18
However, not all models of MLL fusion induced leukemia have short latencies; and in some cases, a preleukemic phase in disease progression has been documented.19,20 It has been suggested that the discrepancy between these models and leukemia in infants may be the result of the MLL fusions themselves rendering cells more susceptible to further DNA damage by genotoxic agents originally causing the MLL translocation.21 This would result in the rapid acquisition of secondary mutations in cells expressing MLL fusions. Furthermore, although a recent study detected very few additional genetic abnormalities in MLL/11q23 rearranged ALL,22 analysis of MLL-ENL-positive ALL and AML found that half contained additional cytogenetic abnormalities, in most cases, single trisomies.23
This suggests that, at least for some MLL/11q23 rearranged leukemias, secondary mutations may be required for overt leukemia and gives rise to the question of whether targeting MLL fusion activity would be effective in these leukemias. The possibility that a small number of genes, for example, the initiating oncogene, are required for maintaining malignancy despite the presence of numerous additional genetic abnormalities is suggested by the “oncogene addiction” theory.24 Indeed, conditional Myc25 and BCR-ABL26 expression has been used to demonstrate regression of hematopoietic malignancies on loss of oncogene expression. In this study, we have used conditional MLL-ENL expression to demonstrate that mice with established leukemias are cured by ablation of MLL-ENL expression in vivo, despite acquisition of secondary genetic abnormalities by the leukemic cells. These experiments suggest that targeting the transcriptional/signaling networks established by MLL fusion oncogenes will lead to effective therapies for MLL/11q23-rearranged leukemias.
Methods
Mice
All mice were maintained in the animal facilities of the Institute of Child Health (London, United Kingdom), and experiments were performed according to United Kingdom Home Office regulations and Institute of Child Health institutional guidelines.
Retroviral constructs and virus production
The pMSCV-MLL-ENL-neo and the pMSCV-tTA-IRES-EGFP were both constructed as described previously.10 The pMSCV-neo-TRE-MLL-ENL vector, containing the full-length flag-tagged MLL-ENL cDNA, was constructed by replacing the Myc-tagged TRE-mMLL fragment from the pMSCV-neo-TRE-mMLL-ENL vector10 with a flag-tagged TRE-MLL fragment. Retroviral supernatants were produced as described previously.27
Isolation and infection of hematopoietic progenitor cells
Bone marrow hematopoietic progenitor cells (HPCs) form C57BL/6 mice were purified by magnetic-activated cell sorting using the Lineage Cell Depletion Kit (Miltenyi Biotec, Surrey, United Kingdom). Lineage-negative HPCs were cultured for 24 hours in Dulbecco modified Eagle medium (Invitrogen, Paisley, United Kingdom) supplemented with 10% heat-inactivated fetal calf serum (Sigma-Aldrich, Poole, United Kingdom), 100 U/mL penicillin (Invitrogen), 100 μg/mL streptomycin (Invitrogen), and 2 mM l-glutamine (Invitrogen; complete medium), 50 μM 2-mercaptoethanol (Sigma-Aldrich), 100 ng/mL stem cell factor (SCF), 10 ng/mL interleukin-3 (IL-3), and 10 ng/mL IL-6 (PeproTech, Rocky Hill, NJ). HPCs were then infected with retrovirus supplemented with the same growth factors on 2 consecutive days by spinoculation (centrifugation at 700g, 25°C, 45 minutes) in the presence of 5 μg/mL polybrene (Sigma-Aldrich), as described previously.10
Methylcellulose culture and generation of cell lines
Colony-forming assays were performed in Methocult M3434 (StemCell Technologies, Vancouver, BC) supplemented with 10 ng/mL granulocyte-macrophage colony-stimulating factor to generate immortalized cell lines, as described previously.10 Cell lines were maintained in RPMI 1640 (Invitrogen) with 10% fetal calf serum, 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM l-glutamine, and 50 μM 2-mercaptoethanol supplemented with 100 ng/mL SCF, 10 ng/mL IL-3, and 10 ng/mL IL-6. In some cases, 2 μg/mL doxycycline (Clontech–Takara Bio Europe, Saint-Germain-en-Laye, France) was added to the cultures in vitro. Colonies were stained with 1 mg/mL p-iodonitrotetrazolium (Sigma-Aldrich). The viability of cells after culture with different cytokine combinations (SCF, IL-3, IL-6, or granulocyte-macrophage colony-stimulating factor) was measured using an MTS assay kit according to the manufacturer's instructions (Promega, Southampton, United Kingdom). Cytospins were made using a Shandon cytospin 3 (Thermo Electron, Ulm, Germany) and fixed and stained with Wright-Giemsa.
Leukemogenesis assays
Sublethally γ-irradiated (6.5 Gy) C57BL/6-CD45.1 mice were injected intravenously with 106 immortalized cells. Mice were killed when they developed clinical signs of disease. For secondary transplantation, sublethally irradiated C57BL/6-CD45.1 or C57BL/6 mice were injected intravenously with 106 primary leukemic splenocytes. Peripheral blood was analyzed for presence of leukemic cells at various time points after transplantation. In some cases, after detection of leukemic cells in the peripheral blood, recipient mice were administered 200 μg/mL doxycycline and 5% sucrose in the drinking water for a specified period of time. Mice were killed when they developed clinical signs of disease or at specified time points after transplantation, and the liver, splenocytes, and bone marrow cells harvested for analysis. Liver tissue was fixed in 10% buffered formalin (Sigma-Aldrich) and set in paraffin. Hematoxylin-and-eosin stain was performed on 4-μm sections.
Flow cytometry
Cells were stained using phycoerythrin- or allophycocyanin-conjugated antibodies to c-Kit, Mac-1, Gr-1, or CD45.2 (eBioscience, Wembley, United Kingdom). Cells were resuspended in PBS, 0.5% bovine serum albumin, and 0.05% sodium azide, and preincubated with unlabeled anti–Fcγ III/II receptor mAb (2.4G2), before staining with primary antibody. Flow cytometry was performed using a Cyan ADP analyser and Summit 4.3 software (Beckman Coulter, High Wycombe, United Kingdom).
Southern blot analysis
Genomic DNA was digested with BamHI. Southern blotting was performed according to standard protocols, using the 0.8-kb neomycin resistance gene cDNA labeled with [32P] α-deoxycytidine triphosphate by random primer labeling.
Cytogenetic analysis and comparative genomic hybridization
A total of 10 μg/mL Colecemid (KaryoMAX; Invitrogen) was added to the cultures for at least 1 hour. Cells were then treated with 75 mM KCl, and cell pellets were fixed using a 3:1 methanol/acetic acid mixture. Chromosomes were visualized by Giemsa banding. Array-based comparative genomic hybridization, on genomic DNA isolated from immortalized and leukemic cells, was performed by Miltenyi Biotec using 4× 44K Agilent Mouse Genome CGH Microarrays (Agilent Technologies, South Queensferry, United Kingdom). The original microarray data can be found in the ArrayExpress database under accession number E-MEXP-1911.
Real-time quantitative PCR
Total RNA was isolated from cells using the TRIzol reagent (Invitrogen). RNA quality was determined using the Agilent 2100 Bioanalyzer (Agilent Technologies). A total of 2 μg RNA was converted into cDNA using Moloney murine leukemia virus reverse transcriptase, amplification grade DNAse I, random primers, RNAseOUT, and dNTPs (all from Invitrogen), according to the manufacturer's instructions. Real-time quantitative polymerase chain reaction (PCR) was performed using TaqMan probe based chemistry and an ABI Prism 7900HT fast Sequence Detection System (Applied Biosystems, Warrington, United Kingdom). A specific primer and probe set spanning the MLL-ENL breakpoint region was designed with PrimerExpress software version 2.0 (Applied Biosystems) and used at 900 nM forward primer (5′-CAGGGTGGTTTGCTTTCTCTGT-3′), 300 nM reverse primer (5′-GCGATGCCCCAGCTCTAA-3′), and 150 nM probe (5′-FAM-TGGACGGTGCACTCTACATGCCCACTA-TAMRA-3′). RNA expression levels of MLL-ENL, Mpo (Mm00447877_g1), Ltf (Mm00434787_m1), and Mmp9 (Mm00600164_g1) were normalized to 18S RNA expression (Hs99999901_s1), and relative copy number of MLL-ENL DNA was normalized to mouse β-Actin DNA (4352933E). The 2−ΔΔCT relative quantitation method was used to determine the relative expression level or copy number of MLL-ENL.28 All primer/probe sets were from Applied Biosystems.
Statistical analysis
Numbers of mice used in each experiment are provided in the figures or figure legends. Statistical analysis of survival curves was performed using the Mantel-Haenszel log-rank test, where P values less than .05 were considered statistically significant.
Results
Conditional MLL-ENL–immortalized cells induce leukemia
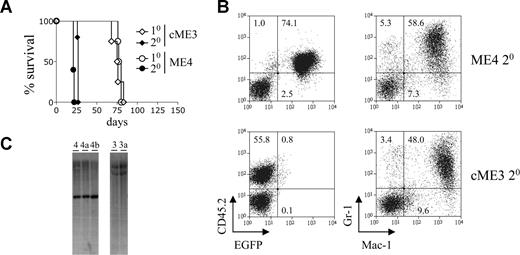
The retroviral expression constructs used in this study are depicted in Figure S1A (available on the Blood website; see the Supplemental Materials link at the top of the online article). These constructs were used to generate immortalized myeloid cells with either conditional or constitutive expression of MLL-ENL (Figure S1B), as previously described.10 The ability of these cells to induce leukemia in vivo was assessed by transplanting them into sublethally irradiated congenic mice. The conditional and constitutive MLL-ENL immortalized cells induced AML in primary recipients with approximately the same latency (Figure 1A; Table 1). To assess whether the disease could be transplanted, primary leukemic splenocytes were transplanted into sublethally irradiated secondary recipients. All secondary recipients developed AML with a much shorter latency than the primary recipients (Figure 1A; Table 1). They exhibited extensive leukemic cell infiltration in their livers (Figure S1C). Secondary leukemias derived from both conditional and constitutive immortalized cells expressed high levels of the myeloid marker Mac-1 and intermediate to high levels of the granulocyte marker Gr-1 (Figure 1B). This phenotype is very similar to that of the immortalized cells from which they were derived (data not shown).
Leukemic cells with conditional expression of MLL-ENL induce disease with shortened latency in secondary recipients. (A) Survival curves of primary and secondary recipients transplanted with immortalized or leukemic myeloid cells. Immortalized myeloid cells with conditional (ME4, ○) or constitutive (cME3, ◇) MLL-ENL expression were transplanted into primary recipient mice (n = 4 for each group). The resulting leukemic cells were transplanted into secondary recipients (● and ♦, n = 5 for each group). (B) Myeloid marker expression by leukemic cells. Dot plots show expression of CD45.2 and EGFP (left panel), and Gr1 and Mac1 (right panel), by leukemic ME4 and cME3 cells within the spleens of primary recipient mice. Numbers in the plots refer to the percentages of cells within each quadrant. (C) Immortalized cells and their leukemic derivatives show an identical pattern of retroviral integration. Southern blot analysis of genomic DNA isolated from immortalized cells ME4 and cME3 and their leukemic progeny ME4a, ME4b, and cME3a. Blots show 5′ (ME4, 4a and 4b) and 3′ (cME3, 3a) end-fragments produced by digestion of the integrated provirus and genomic DNA with BamHI.
Leukemic cells with conditional expression of MLL-ENL induce disease with shortened latency in secondary recipients. (A) Survival curves of primary and secondary recipients transplanted with immortalized or leukemic myeloid cells. Immortalized myeloid cells with conditional (ME4, ○) or constitutive (cME3, ◇) MLL-ENL expression were transplanted into primary recipient mice (n = 4 for each group). The resulting leukemic cells were transplanted into secondary recipients (● and ♦, n = 5 for each group). (B) Myeloid marker expression by leukemic cells. Dot plots show expression of CD45.2 and EGFP (left panel), and Gr1 and Mac1 (right panel), by leukemic ME4 and cME3 cells within the spleens of primary recipient mice. Numbers in the plots refer to the percentages of cells within each quadrant. (C) Immortalized cells and their leukemic derivatives show an identical pattern of retroviral integration. Southern blot analysis of genomic DNA isolated from immortalized cells ME4 and cME3 and their leukemic progeny ME4a, ME4b, and cME3a. Blots show 5′ (ME4, 4a and 4b) and 3′ (cME3, 3a) end-fragments produced by digestion of the integrated provirus and genomic DNA with BamHI.
Latencies of primary and secondary leukemias
| Leukemic cells . | Latency of primary AML, d . | Latency of secondary AML . | Latency of secondary AML after culture . |
|---|---|---|---|
| cME3a | 80 | 21.4 ± 0.6 (n = 5) | ND |
| ME4a | 76 | 26.2 ± 1.1 (n = 5) | 17.8 ± 9.2 (n = 5)* |
| ME4b | 83 | 26.8 ± 0.5 (n = 5) | ND |
| ME4c | 75 | 22.4 ± 2.5 (n = 5) | ND |
| ME5a | 83 | 16.6 ± 2.2 (n = 10) | 22.0 ± 9.5 (n = 10)* |
| ME7a | 81 | 34.2 ± 3.3 (n = 5) | 14.2 ± 3.3 (n = 5)* |
| ME7b | 89 | 30.5 ± 2.4 (n = 4) | ND |
| ME7c | 83 | 37.0 ± 7.4 (n = 5) | ND |
| Leukemic cells . | Latency of primary AML, d . | Latency of secondary AML . | Latency of secondary AML after culture . |
|---|---|---|---|
| cME3a | 80 | 21.4 ± 0.6 (n = 5) | ND |
| ME4a | 76 | 26.2 ± 1.1 (n = 5) | 17.8 ± 9.2 (n = 5)* |
| ME4b | 83 | 26.8 ± 0.5 (n = 5) | ND |
| ME4c | 75 | 22.4 ± 2.5 (n = 5) | ND |
| ME5a | 83 | 16.6 ± 2.2 (n = 10) | 22.0 ± 9.5 (n = 10)* |
| ME7a | 81 | 34.2 ± 3.3 (n = 5) | 14.2 ± 3.3 (n = 5)* |
| ME7b | 89 | 30.5 ± 2.4 (n = 4) | ND |
| ME7c | 83 | 37.0 ± 7.4 (n = 5) | ND |
Values represent the latency of the primary leukemias and the mean latency and SD (numbers of mice per group are indicated) of secondary leukemias resulting from transplantation of the primary leukemias ex vivo or after culture for 1 month in vitro.
ND indicates not determined.
In vitro culture of leukemic cells caused a small but significant increase in ME5a latency (P = .038, Mantel-Haenszel log-rank test), had no effect on ME4a latency (P = .269), and reduced the latency of ME7a (P = .002).
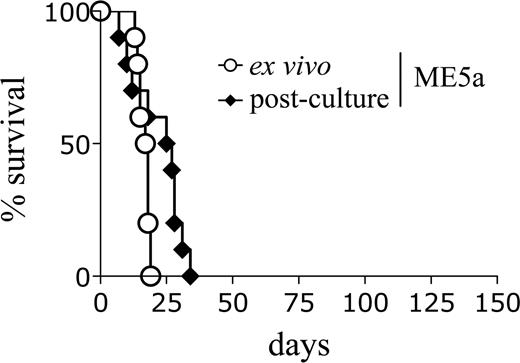
An explanation for the shortened latency of the secondary recipients may be the outgrowth of a minor clone in vivo which was present in the original line but not detected by Southern blot analysis. However, this does not appear to be the case because the leukemic cells had an identical retroviral integration pattern to the original immortalized cells (Figures 1C, S1D). Alternatively, it is possible that the leukemic cells became “conditioned” to the microenvironment in vivo such that, on secondary transplantation, they were better able to respond to environmental signals and engrafted more efficiently. To test this hypothesis, primary leukemic splenocytes were either transplanted directly into secondary recipients or maintained in liquid culture for 1 month and then transplanted. Interestingly, the cultured leukemic cells did not lose the ability to induce AML with shortened latencies (Figure 2; Table 1). This result suggests that the decreased secondary latency is not simply the result of an enhanced ability of the leukemic cells to respond to environmental signals in vivo.
Leukemic cells retain the ability to induce accelerated leukemia after in vitro culture. Survival curves of mice transplanted with ME5a leukemic cells, either freshly isolated (○, n = 10) or after 1-month culture in vitro (♦, n = 10).
Leukemic cells retain the ability to induce accelerated leukemia after in vitro culture. Survival curves of mice transplanted with ME5a leukemic cells, either freshly isolated (○, n = 10) or after 1-month culture in vitro (♦, n = 10).
Leukemic cells differentiate on loss of MLL-ENL expression in vitro
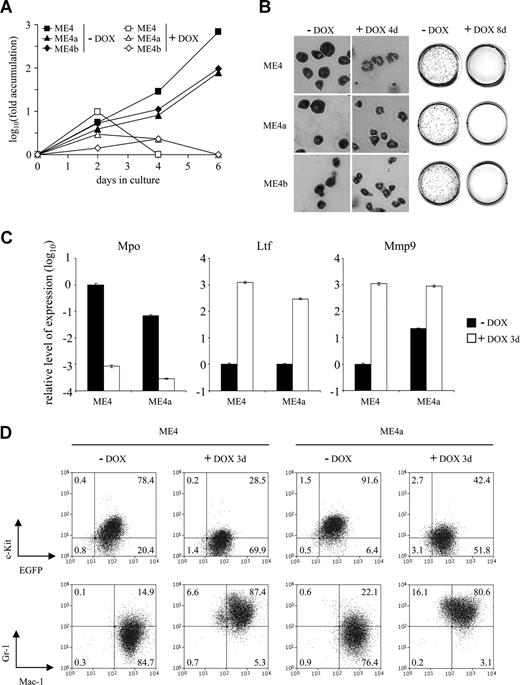
Leukemic splenocytes were isolated from individual primary recipients and cultured in vitro. In all cases, the leukemic cells proliferated more slowly than the immortalized cells from which they were derived (Figure S2A and data not shown). The cells remained growth factor dependent and shared the same growth factor requirements as the immortalized cells (Figure S2B). Both the conditional immortalized cells and their leukemic progeny ceased proliferating in vitro and failed to form colonies in methylcellulose on inhibition of MLL-ENL expression by doxycycline treatment (Figure 3A,B). Furthermore, they exhibited changes in morphology and neutrophil granule gene expression consistent with terminal differentiation (Figure 3B,C). Leukemic cells differentiated at a similar rate and exhibited similar changes in cell surface marker expression as the immortalized cells (Figure 3D). As expected, the proliferation and phenotype of the constitutive immortalized cells and their leukemic progeny were not altered in response to doxycycline (data not shown).
Differentiation of leukemic cells in response to doxycycline. (A) Fold accumulation (log10) in cell number of ME4 (□, ■), ME4a (▵, ▴), and ME4b (◇, ♦) with (□, ▵, ◇) or without (■, ▴, ♦) 2 μg/mL doxycycline. (B) Morphology of the cell lines (left panel, original magnification ×400) and p-iodonitrotetrazolium stains of methylcellulose cultures (right panel) after culture with or without doxycycline (DOX) for 4 and 8 days, respectively. (C) Relative level of neutrophil granule mRNA expression, measured by quantitative reverse-transcribed PCR, in ME4 and ME4a cells after culture with (□) or without (■) doxycycline for 3 days. Plots show a decrease in primary granule myeloperoxidase (Mpo) and an increase in secondary granule lactotransferrin (Ltf) and tertiary granule matrix metallopeptidase 9 (Mmp9) gene expression after doxycycline treatment. Values for each gene were normalized to untreated ME4 cells. Columns represent the mean of triplicate measurements; error bars represent the SD. (D) Changes in surface antigen expression in ME4 (left panel) and ME4a (right panel) cells after culture with or without doxycycline for 3 days. (Top panels) c-Kit and EGFP expression. (Bottom panels) Gr-1 and Mac-1 expression. Numbers in the plots refer to the percentages of cells within each quadrant.
Differentiation of leukemic cells in response to doxycycline. (A) Fold accumulation (log10) in cell number of ME4 (□, ■), ME4a (▵, ▴), and ME4b (◇, ♦) with (□, ▵, ◇) or without (■, ▴, ♦) 2 μg/mL doxycycline. (B) Morphology of the cell lines (left panel, original magnification ×400) and p-iodonitrotetrazolium stains of methylcellulose cultures (right panel) after culture with or without doxycycline (DOX) for 4 and 8 days, respectively. (C) Relative level of neutrophil granule mRNA expression, measured by quantitative reverse-transcribed PCR, in ME4 and ME4a cells after culture with (□) or without (■) doxycycline for 3 days. Plots show a decrease in primary granule myeloperoxidase (Mpo) and an increase in secondary granule lactotransferrin (Ltf) and tertiary granule matrix metallopeptidase 9 (Mmp9) gene expression after doxycycline treatment. Values for each gene were normalized to untreated ME4 cells. Columns represent the mean of triplicate measurements; error bars represent the SD. (D) Changes in surface antigen expression in ME4 (left panel) and ME4a (right panel) cells after culture with or without doxycycline for 3 days. (Top panels) c-Kit and EGFP expression. (Bottom panels) Gr-1 and Mac-1 expression. Numbers in the plots refer to the percentages of cells within each quadrant.
Leukemic cells acquire genetic abnormalities in vivo
Cytogenetic analysis revealed that some leukemic cells had acquired gross chromosomal abnormalities, which were not detected in the immortalized cells from which they were derived. For example, the ME7-immortalized cells possessed a normal karyotype yet their leukemic progeny (ME7a) displayed clonal trisomy 6 (Figure 4A). In most cases, the leukemic cells displayed more single-cell abnormalities than the immortalized cells from which they were derived (Table S1).
Leukemic cells have acquired genetic abnormalities. (A) Mouse GTG-banded karyotype of ME7a leukemic cells. Chromosomes are arranged alongside their corresponding ideograms (original magnification ×1000). (B) Comparative genomic hybridization of ME4 and ME4b DNA samples. The increase in copy number of a region of chromosome 4 (left panel) and decrease in copy number of a region of chromosome 15 (right panel) detected in ME4b leukemic cells compared with ME4 immortalized cells. Each point on the graph represents the log2 ratio value of a single oligonucleotide probe. The shaded area and vertical line represent an aberrant region identified by the Z-score algorithm (CGH Analytics version 3.4; Agilent Technologies).
Leukemic cells have acquired genetic abnormalities. (A) Mouse GTG-banded karyotype of ME7a leukemic cells. Chromosomes are arranged alongside their corresponding ideograms (original magnification ×1000). (B) Comparative genomic hybridization of ME4 and ME4b DNA samples. The increase in copy number of a region of chromosome 4 (left panel) and decrease in copy number of a region of chromosome 15 (right panel) detected in ME4b leukemic cells compared with ME4 immortalized cells. Each point on the graph represents the log2 ratio value of a single oligonucleotide probe. The shaded area and vertical line represent an aberrant region identified by the Z-score algorithm (CGH Analytics version 3.4; Agilent Technologies).
More sensitive comparative genomic hybridization analysis was used to examine chromosomal copy number in the ME4b and ME7b leukemic cells, both of which appeared cytogenetically normal. This analysis revealed that ME4b leukemic cells had acquired an increase in copy number of a chromosome 4 region and a decrease in copy number of a chromosome 15 region, compared with the parental ME4-immortalized cells (Figures 4B, S3A). In addition, the ME7b leukemic cells possessed an increase in copy number of a small region of chromosome 6 (Figure S3B). These changes in copy number were not associated with changes in the copy number of the MLL-ENL provirus (Figure S4A) or increased MLL-ENL transcripts (Figure S4B).
Continued MLL-ENL expression is required to maintain leukemia in vivo
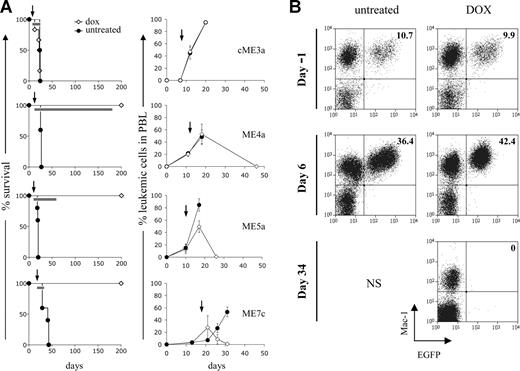
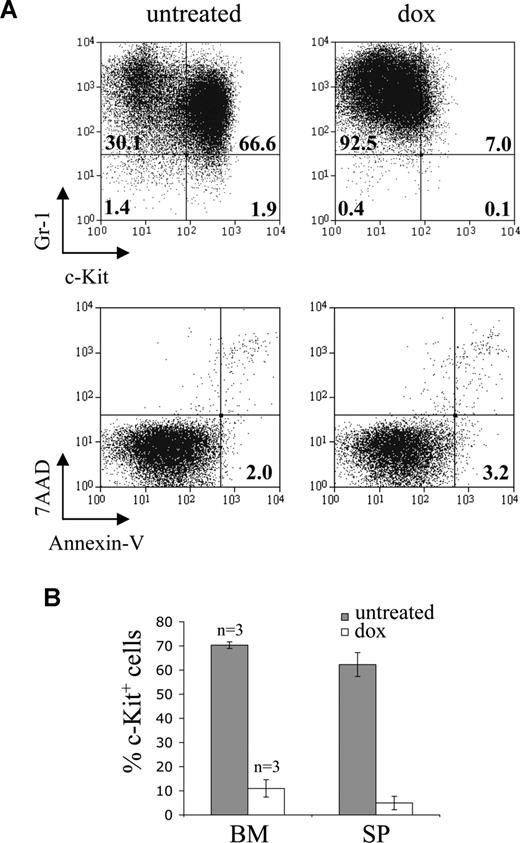
It is possible that the acquired genetic abnormalities could confer resistance to loss of MLL-ENL expression by the leukemic cells in vivo. To examine this, secondary recipient mice with established leukemia were treated with doxycycline. The peripheral blood of secondary recipients was analyzed at regular intervals after transplantation, and doxycycline was administered after detection of leukemic cells. As expected, doxycycline had no effect on the accumulation of leukemic cells in the peripheral blood of mice transplanted with constitutive cells and did not alter their survival (Figure 5A). In contrast, doxycycline treatment of mice transplanted with conditional cells caused the complete elimination of leukemic cells from the peripheral blood in all cases examined (Figures 5A,B, S5). Most of these mice remained healthy over the observation period studied (Figures 5A, S5). Mice transplanted with conditional cells and left untreated rapidly died of fatal AML. The loss of conditional leukemic cells from peripheral blood after doxycycline treatment was probably the result of their terminal differentiation in vivo. Thus, leukemic cells isolated from the spleen and bone marrow of doxycycline-treated mice showed loss of c-Kit expression but no detectable apoptosis (Figure 6).
Abrogating MLL-ENL expression leads to elimination of established leukemias in vivo. (A) Effect of doxycycline on survival of recipient mice (left panel) and elimination of leukemic cells from peripheral blood (right panel). Recipient mice were transplanted with the indicated leukemic cells and after their detection in the peripheral blood, one group was given doxycycline in their water (◇: cME3a, n = 6; ME4a, n = 5; ME5a, n = 5; ME7c, n = 4) and the other left untreated (●: cME3a, n = 4; ME4a, n = 5; ME5a, n = 5; ME7c, n = 5). Arrows represent the point at which doxycycline treatment started; bars on the survival curves represent the length of treatment in each experiment. Doxycycline-treated mice are still alive 350, 270, and 360 days after transplantation of ME4a, ME5a, and ME7c cells, respectively. The graphs depict the presence of leukemic cells (Mac1+CD45.2+ cells in CD45.1 recipient mice for cME3a, and Mac1+EGFP+ cells for ME4a, ME5a, and ME7a) as a percentage of total Mac1+ cells in the peripheral blood of recipient mice. Points on the graphs represent mean values; bars represent SD. (B) The presence of Mac1+EGFP+ leukemic ME4a cells in the peripheral blood of recipient mice before (day −1) and after (day 6 and 34) their treatment (right panel), or not (left panel), with doxycycline. Numbers in the top right quadrant represent Mac1+EGFP+ cells as a percentage of total Mac1+ cells. No mice in the untreated group survived until the last time point shown, indicated by NS.
Abrogating MLL-ENL expression leads to elimination of established leukemias in vivo. (A) Effect of doxycycline on survival of recipient mice (left panel) and elimination of leukemic cells from peripheral blood (right panel). Recipient mice were transplanted with the indicated leukemic cells and after their detection in the peripheral blood, one group was given doxycycline in their water (◇: cME3a, n = 6; ME4a, n = 5; ME5a, n = 5; ME7c, n = 4) and the other left untreated (●: cME3a, n = 4; ME4a, n = 5; ME5a, n = 5; ME7c, n = 5). Arrows represent the point at which doxycycline treatment started; bars on the survival curves represent the length of treatment in each experiment. Doxycycline-treated mice are still alive 350, 270, and 360 days after transplantation of ME4a, ME5a, and ME7c cells, respectively. The graphs depict the presence of leukemic cells (Mac1+CD45.2+ cells in CD45.1 recipient mice for cME3a, and Mac1+EGFP+ cells for ME4a, ME5a, and ME7a) as a percentage of total Mac1+ cells in the peripheral blood of recipient mice. Points on the graphs represent mean values; bars represent SD. (B) The presence of Mac1+EGFP+ leukemic ME4a cells in the peripheral blood of recipient mice before (day −1) and after (day 6 and 34) their treatment (right panel), or not (left panel), with doxycycline. Numbers in the top right quadrant represent Mac1+EGFP+ cells as a percentage of total Mac1+ cells. No mice in the untreated group survived until the last time point shown, indicated by NS.
Leukemic cells differentiate in vivo in response to doxycycline. (A) Response of leukemic ME4b cells to doxycycline in vivo. Dot plots show Gr-1 and c-Kit expression (top panel), and staining with 7-amino-actinomycin D (7-AAD) and annexin V (bottom panel), on gated EGFP+ ME4b cells in the bone marrow of mice 18 days after transplantation with ME4b cells and 3 days after their treatment (right plot), or not (left plot), with doxycycline. Numbers in the plots represent the percentage of cells in each quadrant (top panel) and of apoptotic (7-amino-actinomycin D−annexin-V+) EGFP+ cells (bottom panel). The mean fluorescence intensity of Gr-1 expression by ME4b cells (top panel) increased from 684 to 1259 after doxycycline treatment. (B) Percentage of EGFP+ ME4b cells expressing c-Kit in the bone marrow and spleen of the recipient mice from panel A, treated (□) or not (■) with doxycycline. Bars represent mean values (n = 3); error bars represent SD.
Leukemic cells differentiate in vivo in response to doxycycline. (A) Response of leukemic ME4b cells to doxycycline in vivo. Dot plots show Gr-1 and c-Kit expression (top panel), and staining with 7-amino-actinomycin D (7-AAD) and annexin V (bottom panel), on gated EGFP+ ME4b cells in the bone marrow of mice 18 days after transplantation with ME4b cells and 3 days after their treatment (right plot), or not (left plot), with doxycycline. Numbers in the plots represent the percentage of cells in each quadrant (top panel) and of apoptotic (7-amino-actinomycin D−annexin-V+) EGFP+ cells (bottom panel). The mean fluorescence intensity of Gr-1 expression by ME4b cells (top panel) increased from 684 to 1259 after doxycycline treatment. (B) Percentage of EGFP+ ME4b cells expressing c-Kit in the bone marrow and spleen of the recipient mice from panel A, treated (□) or not (■) with doxycycline. Bars represent mean values (n = 3); error bars represent SD.
In some recipients, complete remission was not sustained and leukemic cells reappeared in the peripheral blood. All of the mice transplanted with ME4c leukemic cells relapsed while on doxycycline treatment (Figure S5). In addition, 2 of the ME4b recipients relapsed: one while on doxycycline treatment and one 41 days after doxycycline withdrawal (Figure S5). Because leukemias that relapsed on doxycycline demonstrated significant expression of MLL-ENL mRNA, it is probable that doxycycline treatment selected for outgrowth of leukemic cells in which MLL-ENL expression had become tTA-independent (Figure S6A). This is also the most probable explanation for the ME4b relapse after doxycycline withdrawal (Figure S5). Leukemic cells from this mouse were able to form colonies in methylcellulose but were slower growing and expressed lower levels of MLL-ENL mRNA than cells from the ME4b recipient that relapsed while on doxycycline. Interestingly, MLL-ENL expression in these leukemic cells was not abrogated by doxycycline treatment (Figure S6B).
Discussion
This study shows that immortalized myeloid cells with conditional expression of MLL-ENL gave rise to highly aggressive and invasive AML in vivo and that this disease regressed completely on loss of MLL-ENL expression. Despite acquisition of additional genetic abnormalities, the leukemic cells remained dependent on continued MLL-ENL expression and in its absence underwent terminal myeloid differentiation in vitro and in vivo. Strikingly, treatment for as little as 10 days with doxycycline was sufficient to achieve complete remission in some cases, suggesting that the putative bone marrow leukemic stem cells were effectively targeted.
Previous studies have reported that leukemias initiated by MLL fusions exhibit a reduced latency on secondary transplantation.15,19,29 It is unclear whether this is the result of acquired genetic abnormalities and epigenetic modification or to enhanced engraftment potential resulting from in vivo conditioning to microenvironmental signals. We reasoned that the latter would be lost on in vitro culture of leukemic cells. However, freshly isolated leukemic splenocytes were observed to give rise to AML in secondary recipients with similar latencies to those of cells transplanted after culture for one month in vitro. These data suggest that conditioning of leukemic cells does not account for the reduced secondary latencies in our model but is compatible with acquisition of heritable mutations during leukemia progression in vivo.
In several cases, we demonstrated that the leukemic cells had acquired additional genetic abnormalities, not present in the parental immortalized cells from which they were derived. These mutations included a trisomy of chromosome 6 in one case and gains and losses of chromosomal regions in a further 2 cases. This is consistent with cytogenetic analysis of human leukemias associated with the MLL-ENL fusion.23 Whether these abnormalities are required for or merely a consequence of leukemic progression remains unclear. However, it is probable that the acquisition and selection of these abnormalities in vivo, or of mutations which our analyses were not sensitive enough to detect, account for the reduced latency of AML in secondary recipients. The possible impact of secondary mutations on MLL fusion-induced leukemias is illustrated by a study, which found that coexpression of mutant FLT3, with an internal tandem duplication, with MLL-ENL in hematopoietic cells resulted in leukemias with considerably shortened latencies.30
Induction of reversible hematopoietic malignancies has been demonstrated previously using the “Tet-Off” system in which expression of the Myc25 and BCR-ABL26,31,32 oncogenes was controlled by a variety of different tTA transgenes. Experiments with Myc expressing lymphoid tumors demonstrated that, although many tumors regressed on prolonged Myc inactivation, others relapsed. Interestingly, all the relapsed tumors had additional chromosomal rearrangements and had become Myc-independent.33 Using a transgenic model in which tTA was placed under the control of the murine stem cell leukemia gene regulatory elements, conditional expression of BCR-ABL resulted in reversible chronic myeloid leukemia (CML) and B lymphoid blast crisis.32 In these experiments, one of the leukemias was found to progress even during tetracycline treatment and on analysis of tumor cells was found to express BCR-ABL despite exposure to tetracycline.32 We found a similar loss of conditional oncogene expression in a few leukemias that relapsed, suggesting in vivo selection for leukemic cells in which MLL-ENL expression had become doxycycline-insensitive.
In conclusion, in all of the leukemias induced by MLL-ENL, leukemic progression was entirely dependent on continued MLL-ENL expression. It is important to note that the use of retroviral vectors in our study may have resulted in overexpression of the MLL-ENL oncogene. A recent study has demonstrated that the relatively low expression levels of the MLL-AF9 fusion achieved under the control of mouse MLL promoter elements resulted in a reduced capacity of the fusion to transform committed progenitor cells.34 This suggests that cooperating secondary mutations may have a more important role in leukemic progression in AML patients, where the MLL fusions are expressed under the control of endogenous promoters, than in our model. For these reasons, it will be important to examine the dependence of primary AML cells on expression of the appropriate MLL fusion.
However, if extended to AML patient cells and to MLL fusions in general, our data suggest that targeting MLL fusion oncogenes, or downstream pathways regulated by them, will be effective at eradicating established leukemia. The paradigm for targeting the initiating oncogene in hematopoietic malignancy is treatment of BCR-ABL-associated CML with the pharmacologic ABL kinase inhibitor imatinib mesylate.35 This treatment is relatively nontoxic and has few side effects. Interestingly, the addiction of these leukemias to BCR-ABL is underlined by the frequent presence of point mutations within BCR-ABL kinase domain, rendering it refractory to imatinib, in relapsed leukemia after imatinib therapy.35
Unlike BCR-ABL, MLL fusions regulate gene expression, and it is doubtful that they can be targeted directly because of the inherent difficulty in inhibiting transcription factor activity. A more promising approach would be to block downstream pathways regulated by MLL fusions. The demonstration that inhibiting or ablating the expression of specific MLL fusion target genes can reverse immortalization13,15 suggests that this may be feasible. We and others have recently demonstrated that native MLL is required for normal hematopoiesis and hematopoietic stem cell activity.36,37 This suggests that specific therapies will have to be carefully calibrated against inhibition of normal MLL function to avoid hematopoietic toxicity. However, the “oncogene addiction” theory suggests that, because of the novel or “bizarre” circuitry set up by the initiating oncogene, leukemic cells may be more sensitive to inhibition of MLL activity than normal cells.38,39 This “addiction” to high levels of oncogene activity probably explains why normal hematopoietic cells are not affected by ABL inhibition, although this results in elimination of CML cells.38
In conclusion, this study demonstrates that leukemic cells are addicted to MLL-ENL expression and suggests that targeting oncogenic pathways maintained by MLL fusions in patients with 11q23 rearrangements would be a major therapeutic advance.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the staff of the Institute of Child Health (ICH) Western Laboratories for excellent animal husbandry, Gemma Williams for secretarial assistance, Eleanor Ashton for technical support, and A. Biondi (Milan, Italy) and D. C. Tkachuk (Toronto, ON) for MLL-fusion cDNAs.
This work was supported by grants from the Children with Leukaemia, the Leukaemia Research Fund, and the Great Ormond Street Hospital Special Purpose Fund, United Kingdom.
Authorship
Contribution: S.J.H. and V.W.-V. designed and performed experiments, analyzed and interpreted results, and wrote the paper; S.J.C. performed cytogenetic analysis; N.J.S. performed the histopathology; J.d.B. analyzed and interpreted results; and O.W. supervised the project, designed and performed experiments, analyzed and interpreted results, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Owen Williams, Molecular Haematology and Cancer Biology Unit, UCL Institute of Child Health and Great Ormond Street Hospital, 30 Guilford St, London WC1N 1EH, United Kingdom; e-mail: owen.williams@ich.ucl.ac.uk.
References
Author notes
*S.J.H. and V.W.-V. contributed equally to this paper.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal