Abstract
Cytokine signaling via various transcription factors regulates receptor activator of nuclear factor (NF)–κB ligand (RANKL)–mediated osteoclast differentiation from monocyte/macrophage lineage cells involved in propagation and resolution of inflammatory bone destruction. Protein inhibitor of activated STAT3 (PIAS3) was initially identified as a molecule that inhibits DNA binding of STAT3 and regulates many transcription factors through distinct mechanisms. To analyze PIAS3 function in osteoclasts in vivo, we have generated transgenic mice in which PIAS3 is specifically expressed in the osteoclast lineage using the tartrate-resistant acid phosphatase (TRAP) gene promoter. PIAS3 transgenic mice showed an osteopetrotic phenotype due to impairment of osteoclast differentiation. Overexpression of PIAS3 in RAW264.7 cells suppressed RANKL-induced osteoclastogenesis by inhibiting the expression of c-Fos and NFATc1. Interestingly, PIAS3 inhibits the transcriptional activity of microphthalmiaassociated transcription factor (MITF) independent of sumoylation. Down-regulation of PIAS3 markedly enhances RANKL-mediated osteoclastogenesis in RAW264.7 cells. Furthermore, overexpression of PIAS3 in mouse primary osteoblast (POB), down-regulates RANKL expression induced by interleukin-6 (IL-6) cytokine family, and inhibits osteoclast formation from bone marrow macrophages (BMMs) in vitro coculture system. Down-regulation of PIAS3 leads to the accelerated expression of RANKL in POB stimulated with IL-6 and soluble IL-6 receptor (sIL-6R). Taken together, our results clearly indicate that PIAS3 negatively regulates RANKL-mediated osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblasts.
Introduction
Hematopoietic precursors of the monocyte/macrophage lineage differentiate into osteoclasts in response to receptor activator of nuclear factor (NF)–κB ligand (RANKL).1,2 Several cytokine factors are known to influence RANKL-induced osteoclast formation and function. RANKL expressed on osteoblasts binds to its receptor, RANK, on osteoclast precursors, leading to the activation of osteoclast-specific transcription factors, such as microphthalmia-associated transcription factor (MITF), PU.1, c-Fos, and nuclear factor of activated T cells c1 (NFATc1).3,4 However, the RANKL-mediated transcriptional program for osteoclastogenesis has remained elusive
Mutation of the mouse mi locus is associated with a loss of pigmentation, small eyes, a reduced number of mast cells, and a failure of secondary bone resorption, which also results in osteopetrosis. The mi locus encodes a basic helix-loop-helix transcription factor, MITF5,6 that is important for osteoclastogenesis in vitro and in vivo.7,8 MITF induces the expression of osteoclast-specific genes, including tartrate-resistant acid phosphatase (TRAP), cathepsin K, and osteoclast-associated receptor (OSCAR), by binding to the canonical E-box sequence (CANNTG) in their promoter region.9-12
STAT3 is a transcription factor involved in signal transduction pathways that are activated by several extracellular stimuli, including the interleukin-6 (IL-6) cytokine family. IL-6 cytokine family members (IL-6, IL-11, leukemia inhibitory factor [LIF], oncostatin M [OSM], and cardiotropin-1 [CT-1]) promote osteoclastogenesis and bone resorption in bone marrow cultures.13,14 STAT3 is tyrosine phosphorylated by Jak kinase (JAK) and translocates as a dimer into the nucleus, where it activates specific genes.15 Activated STAT3 is involved in the regulation of cell growth, differentiation, and survival and is essential for gp130-mediated osteoclast formation, during which it induces the expression of RANKL.16,17
Protein inhibitor of activated STAT3 (PIAS3) was originally isolated as a molecule that binds to signal transducer and activator of transcription 3 (STAT3) and blocks its DNA binding ability, thereby inhibiting STAT3-mediated gene activation.18 In addition, PIAS3 binds directly to MITF and blocks MITF's binding and transcription of target genes in mast cells and melanocytes.19 PIAS proteins mediate their functions in either a sumoylation-dependent (as an E3 ligase) or -independent manner. Sumoylation has emerged as an important regulatory mechanism, especially in transcription and signal transduction.20,21 However, the effect of PIAS3-induced MITF sumoylation on MITF transcriptional activity is unclear.22
In this study, we further examined the role of PIAS3 in RANKL-mediated osteoclastogenesis. It was recently reported that PIAS3 acts as a negative regulator of RANKL-mediated osteoclastogenesis by down-regulating NFTAc1 and OSCAR, in part by recruiting the histone deacetylase 1 (HDAC1) corepressor.23 Here we performed in vivo and in vitro analyses to elucidate the mechanisms underlying PIAS3's negative regulation of the RANKL-mediated transcriptional network. Our results revealed that (1) TRAP–PIAS3-transgenic (Tg) mice exhibit an osteopetrotic phenotype due to impaired osteoclast differentiation; (2) in osteoclast precursors, PIAS3 suppresses the transcriptional activity of MITF independent of sumoylation, thereby inhibiting the expression of c-Fos and NFATc1; and (3) in osteoblasts, PIAS3 attenuates the RANKL expression induced by IL-6–type cytokine-gp130-STAT3 signaling.
Methods
Reagent, plasmids, and antibodies
1,25-Dihydroxyvitamin D3 (1,25(OH)2D3) was purchased from Wako (Osaka, Japan). Recombinant mouse macrophage colony-stimulating factor (M-CSF), IL-11, and OSM were from R&D Systems (Minneapolis, MN). Recombinant mouse soluble RANKL, IL-6, and soluble IL-6 receptor (sIL-6R) were from PeproTech (London, United Kingdom). Prostaglandin E2 (PGE2) was from Sigma-Aldrich (St Louis, MO). FuGENE HD was from Roche (Indianapolis, IN). Lipofectamine RNAiMAX was from Invitrogen (Tokyo, Japan). The following plasmids were kindly provided: pcDNA-MITF-myc,24 c-Fos promoter reporter plasmid,25 pEGFP-PIAS3,26 and pCI-PIAS3(C334S).27 Anti-PIAS3, STAT3, pSTAT3, MITF, c-Fos, and NFATc1 Ab were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti–β-actin Ab was from Sigma-Aldrich. Anti-myc Ab was from MBL (Nagoya, Japan). Anti-GFP Ab was from Nacalai Tesque (Kyoto, Japan).
Generation of transgenic mice
Mouse PIAS3 cDNA was fused to 1.8 kb of the mouse TRAP gene promoter.28-30 The transgenic mice were generated by pronuclear injection methods into C57BL/6 mice. Genomic DNA from the tail was analyzed by polymerase chain reaction (PCR) and Southern blot analysis. For PCR analysis, specific primers (P1: 5′-CACTTCGCTAGATGAACAGGAC-3′, P2: 5′-TGAGTTTGGACAAACCACAAC-3′) were used. For Southern blot analysis, tail DNA was digested NheI and HpaI, fractionated on 0.8% agarose gel, transferred to Hybond-N+ membrane (GE Healthcare, Tokyo, Japan), and probed with a 403-bp alkaline phosphatase-labeled PCR fragment of mouse PIAS3 cDNA. Southern blot probe was generated by PCR with specific primers (P3: 5′-ACTGCCCTTCTATGAAGTCTATGG-3′, P4: 5′-CTCAGATGACCAATTAACTACGATG-3′). All experiments were performed with sex- and age-matched mice according to the protocol approved by the Laboratory Animal Care and Use Committee of Keio University School of Medicine.
Radiologic analysis and microcomputed tomographic analysis
Femurs of 8-week-old mice were fixed with 70% ethanol, and radiologic analysis of long bones was performed with a soft x-ray instrument (CMB-II; SOFTEX, Tokyo, Japan). Bone mineral density (BMD) was measured by DXA using a bone mineral analyzer (DCS-600EX-III; ALOKA, Tokyo, Japan). Quantitative microcomputed tomography (microCT) was performed with Scan Xmate-A090S (Comuscantecno, Yokohama, Japan). Data from scanned slices was used for 3-dimensional analysis to calculate femoral morphometric parameters by TRY/3D BON (Ratoc System Engineering, Tokyo, Japan). The nomenclature and units used were according to the recommendation of the Nomenclature Committee of the American Society for Bone and Mineral Research.31
Histologic and histomorphometric analysis
The harvested lower extremities were fixed in 10% neutral-buffered formalin, decalcified in 10% ethylenediaminetetraacetic acid (EDTA), pH 7.4, embedded in paraffin, and then cut into 4-μm sections. Van Gieson stain was performed according to the standard procedure. The undecalcified bone was embedded in glycolmethacrylate. A 3-μm-thick section was cut longitudinally in the proximal region of the tibia and stained for toluidine blue O and TRAP. We analyzed the total 28 to 33 fields to generate the bone histomorphometric data by the semiautomatic image analyzing system (Osteoplan II; Carl Zeiss, Thornwood, NY). The nomenclature and units used were according to the recommendation of the Nomenclature Committee of the American Society for Bone and Mineral Research.31
Retroviral gene transfer
To generate retroviral stocks, pMX-PIAS3 was transfected into the packaging cell line Plat-E. Viral supernatant was collected from culture media 48 hours after transfection. Bone marrow macrophages (BMMs) and primary osteoblasts (POBs) were incubated with viral supernatant for 48 hours, selected by 1 μg/mL blasticidine for additional 48 hours, and used for following experiments. The transducing efficiency was approximately 10%. RAW264.7 cells were also incubated with viral supernatant for 48 hours and were selected by culturing for 2 weeks at the presence of 10 μg/mL blasticidine.
Osteoclast formation
Osteoclasts were induced from RAW264.7 cells by plating at 5 × 103 cells/48-well plate in α-minimum essential medium (MEM) containing 10% fetal bovine serum (FBS) and 50 ng/mL RANKL. After 3 days of culture, the cells were stained for TRAP. TRAP-positive multinuclear cells that possessed more than 3 or 10 nuclei were counted as osteoclasts. Bone marrow cells were isolated from femur and tibia of 6- to approximately 8-week-old ddY mice as described.32 Nonadherent bone marrow cells (2 × 106 cells/48-well plate) were cultured with 30 ng/mL M-CSF for 2 days. The adherent cells were used as BMMs. These osteoclast precursors were further cultured in the presence of 50 ng/mL RANKL and 30 ng/mL M-CSF to generate osteoclasts. To generate osteoclasts from coculture with bone marrow cells and POBs, these cells were seeded at 5 × 105 cells and 2 × 104 cells/48-well plate, respectively, were cocultured for 7 days in the presence of OSM, as indicated concentrations.
Immunoprecipitation and immunoblotting
HEK 293T cells were transiently transfected with the indicated plasmids using FuGENE HD, and at 48 hours after transfection, the cells were treated with 10 μM MG-132 for 3 hours. After the cells were washed with ice-cold phosphate-buffered saline (PBS), they were lysed with IP buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40 [NP-40], 40 μM MG-132, 1 mM N-ethylmaleimide and protease inhibitor mixture from Nacalai Tesque). Extracts were clarified by centrifugation at 15 000 rpmfor 30 minutes. For each IP, cell extract proteins were mixed with primary antibody for 1 hour and then incubated with 25 μL Immobilized Protein G (Pierce, Rockford, IL) overnight with gentle rotation at 4°C. The immunoprecipitations were washed 5 times with IP buffer, boiled in sodium dodecyl sulfate (SDS) sample buffer, and then used for immunoblotting. For Western blot analysis, the whole-cell lysate and immunoprecipitates were resolved by SDS–polyacrylamide gel electrophoresis (SDS-PAGE), and proteins were detected by immunoblotting.
DNA binding enzyme-linked immunosorbent assay
The TransFactor kits (Clontech, Mountain View, CA) are highly sensitive enzyme-linked immunosorbent assay (ELISA)–based kits that facilitate the study of the DNA binding interactions of several different transcription factors. Whole-cell lysates were extracted from mock- and PIAS3-transduced RAW264.7 cells stimulated with 50 ng/mL RANKL for 24 hours; we analyzed the DNA binding activity of p65, p50, c-Fos, CREB-1, ATF2, c-Rel, and NFATc1. We examined the DNA binding activity of STAT3 for mock- and PIAS3-transduced POBs stimulated with 100 ng/mL OSM.
Luciferase reporter assay
For the transfection of reporter plasmids, HEK 293T cells were seeded at 5 × 104/48-well plate 1 day before transfection. Plasmid DNA was mixed with FuGENE HD and transfected into the cells following the manufacturer's protocol. After 48 hours of transfection, luciferase activity was measured with a luciferase assay system (Promega, Madison, WI) according to the manufacturer's instructions.
Chromatin immunoprecipitation (ChIP)
Mock- and PIAS3-transduced RAW264.7 cells were cultured with 50 ng/mL RANKL for 24 hours. These cells were fixed with 1% formaldehyde and harvested in SDS lysis buffer (10 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM ethyleneglycoltetraacetic acid [EGTA], 1% Triton X-100, 0.1% sodium-deoxycholate, and 0.1% SDS). Samples were sonicated, and divided into 2 aliquots. Each sample was incubated at 4°C overnight with 50 μL protein A-Sepharose beads (Invitrogen) that had been incubated overnight with 10 μg antibodies against immunoglobulin G (IgG) or MITF. After being washed, the immunoprecipitated protein-DNA was eluted from the beads by heating for 20 minutes at 65°C in elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, and 1% SDS). The cross-links were then reversed by incubating overnight at 65°C. The DNA was extracted using a MinElute kit (QIAGEN, Valencia, CA) and quantified by real-time PCR specific for the promoter region containing the MITF binding site. Primers used are shown in Table S1C (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Statistical analysis
Statistical analysis was performed using Student t test or analysis of variance (ANOVA) followed by Bonferroni correction when applicable. P values less than .05 were considered significant.
Results
Generation of transgenic mice expressing PIAS3 in osteoclasts
To clarify the role of PIAS3 in vivo, we used the promoter of the gene for TRAP, an enzyme that is highly expressed in osteoclasts.28-30 Mouse PIAS3 cDNA was fused with a 1.8-kb sequence located upstream of the TRAP gene (Figure 1A). Ten lines of offspring were generated by pronuclear microinjection, and the presence of the transgene was detected by PCR of the tail DNA using transgene-specific primers (Figure 1B). Twenty-seven founder lines were established by mating founders with C57/BL6 wild-type (WT) mice. We selected 4 Tg lines in which PIAS3 was highly expressed as determined by real-time PCR of cDNA derived from the tail (data not shown). Two of these lines (nos. 55 and 56) expressed P1AS3 at higher levels in BMMs than the other lines (nos. 44 and 51) and had integrated the transgene, as determined by Southern blot analysis (Figure 1B,C).
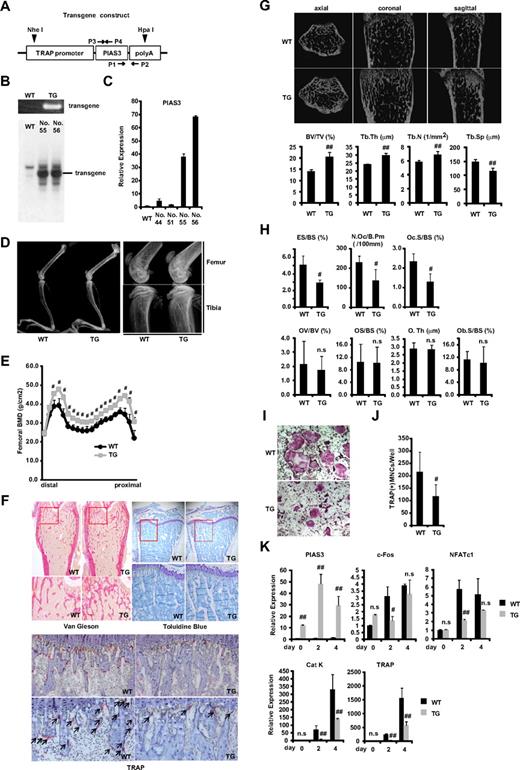
PIAS3-Tg mice show an osteopetrotic phenotype and impair osteoclast differentiation. (A) Schematic of the PIAS3 transgene construct containing the TRAP promoter followed by mouse full-length PIAS3 cDNA and poly(A) sequence. ▼ indicates a digestion site by HpaI and NheI for Southern blot analysis. →, ← indicate specific primers for genomic PCR (P1,2) and for generation of Southern blot probe (P3,4). (B) Genomic PCR with tail DNAs from WT and Tg mice (top). Southern blot of tail DNA from WT and PIAS3-Tg mice (bottom). (C) Expression of PIAS3 mRNA by real-time PCR. Total RNA was extracted from BMMs of WT and PIAS3-Tg mice for real-time PCR (right). (D) Radiologic analysis of long bones of TRAP–PIAS3-Tg mice and WT littermates. Right panel shows a maginified view of left panel. (E) Bone mineral density of 20 equal longitudinal divisions of femurs from 8-week-old WT and PIAS3-Tg mice. (F) Histologic analysis of long bone of PIAS3-Tg and WT (top left, Van Gieson; top right, toluidine blue; bottom, TRAP/hematoxylin staining). Arrows indicate the TRAP-positive mature osteoclasts. (G) MicroCT of axial, coronal, and sagittal sections of distal femoral metaphysis from representative 8-week-old PIAS3-Tg and WT. BV/TV (bone volume/tissue volume), Tb.Th (trabecular thickness), Tb.N (trabecular number), and Tb.Sp (trabecular separation) were analyzed by TRY/3D BON. (H) The parameters for osteoclastic bone resorption (ES/BS, N.Oc/B.Pm, and Oc.S/BS) and parameters for osteoblastic bone formation (OV/BV, OS/BS, O.Th, and Ob.S/BS) in the bone morphometrical analysis. ES/BS, eroded surface/bone surface; N.Oc/B.Pm, osteoclast number/bone perimeter; Oc.S/BS, osteoclast surface/bone surface; OV/BV, osteoid volume/bone volume; OS/BS, osteoid surface/bone surface; O.Th, osteoid thickness; Ob.S/BS, osteoblast surface/bone surface. (I) BMMs isolated from PIAS3-Tg and WT mice were cultured for 5 days with 30 ng/mL M-CSF and 50 ng/mL RANKL. Cultured cells were fixed and stained for TRAP. (J) TRAP-positive MNCs having more than 3 nuclei were counted as osteoclast. (K) Expression of PIAS3 and osteoclastogenic markers during RANKL-induced osteoclastogenesis. BMMs isolated from PIAS3-Tg and WT mice were cultured in the presence of 30 ng/mL M-CSF and 50 ng/mL RANKL for 0, 2, and 4 days. Total RNA was extracted from cultured cells, and real-time PCR was performed with primers specific for PIAS3, c-Fos, NFATc1, cathepsin K, and TRAP. Error bars in all panels represent standard deviation (SD). #P < .05; ##P < .01; n.s, not significant.
PIAS3-Tg mice show an osteopetrotic phenotype and impair osteoclast differentiation. (A) Schematic of the PIAS3 transgene construct containing the TRAP promoter followed by mouse full-length PIAS3 cDNA and poly(A) sequence. ▼ indicates a digestion site by HpaI and NheI for Southern blot analysis. →, ← indicate specific primers for genomic PCR (P1,2) and for generation of Southern blot probe (P3,4). (B) Genomic PCR with tail DNAs from WT and Tg mice (top). Southern blot of tail DNA from WT and PIAS3-Tg mice (bottom). (C) Expression of PIAS3 mRNA by real-time PCR. Total RNA was extracted from BMMs of WT and PIAS3-Tg mice for real-time PCR (right). (D) Radiologic analysis of long bones of TRAP–PIAS3-Tg mice and WT littermates. Right panel shows a maginified view of left panel. (E) Bone mineral density of 20 equal longitudinal divisions of femurs from 8-week-old WT and PIAS3-Tg mice. (F) Histologic analysis of long bone of PIAS3-Tg and WT (top left, Van Gieson; top right, toluidine blue; bottom, TRAP/hematoxylin staining). Arrows indicate the TRAP-positive mature osteoclasts. (G) MicroCT of axial, coronal, and sagittal sections of distal femoral metaphysis from representative 8-week-old PIAS3-Tg and WT. BV/TV (bone volume/tissue volume), Tb.Th (trabecular thickness), Tb.N (trabecular number), and Tb.Sp (trabecular separation) were analyzed by TRY/3D BON. (H) The parameters for osteoclastic bone resorption (ES/BS, N.Oc/B.Pm, and Oc.S/BS) and parameters for osteoblastic bone formation (OV/BV, OS/BS, O.Th, and Ob.S/BS) in the bone morphometrical analysis. ES/BS, eroded surface/bone surface; N.Oc/B.Pm, osteoclast number/bone perimeter; Oc.S/BS, osteoclast surface/bone surface; OV/BV, osteoid volume/bone volume; OS/BS, osteoid surface/bone surface; O.Th, osteoid thickness; Ob.S/BS, osteoblast surface/bone surface. (I) BMMs isolated from PIAS3-Tg and WT mice were cultured for 5 days with 30 ng/mL M-CSF and 50 ng/mL RANKL. Cultured cells were fixed and stained for TRAP. (J) TRAP-positive MNCs having more than 3 nuclei were counted as osteoclast. (K) Expression of PIAS3 and osteoclastogenic markers during RANKL-induced osteoclastogenesis. BMMs isolated from PIAS3-Tg and WT mice were cultured in the presence of 30 ng/mL M-CSF and 50 ng/mL RANKL for 0, 2, and 4 days. Total RNA was extracted from cultured cells, and real-time PCR was performed with primers specific for PIAS3, c-Fos, NFATc1, cathepsin K, and TRAP. Error bars in all panels represent standard deviation (SD). #P < .05; ##P < .01; n.s, not significant.
PIAS3-transgenic mice showed increased bone mass and impaired osteoclast differentiation
To examine the phenotype of the PIAS3-Tg mice, we first compared their long bones with those of WT littermates by radiologic analysis. As shown in Figure 1D and E, the long bones of the PIAS3-Tg mice showed greater radiodensity, especially at the metaphyses of the femur and tibia, and a clear increase in bone mineral density, compared with their WT littermates. Histologic analysis showed increased trabecular bone formation in the metaphysis and a distinct reduction in the number of osteoclasts in the Tg mice (Figure 1F). Furthermore, microCT analysis revealed a significant increase in bone volume (BV/TV) due to increased trabecular number (Tb.N) and thickness (Tb.Th) and reduced trabecular separation (Tb.Sp) in the Tg mice (Figure 1G). These results indicate that the PIAS3-Tg mice have an osteopetrotic phenotype. Histomorphometric analysis indicated an increase in bone volume associated with a decrease in osteoclast number (N.Oc/B.Pm) and a decrease in indicators of osteoclastic bone resorption (ES/BS and Oc.S/BS) in the PIAS3-Tg mice (Figure 1H). On the other hand, the osteoblastic parameters (OV/BV, OS/BS, O.Th, and Ob.S/BS) were the same as in WT mice. These results indicate that the PIAS3-Tg mice exhibit an increase in bone mass that is caused by impaired osteoclastic bone resorption owing to a defect in osteoclast differentiation.
To confirm whether the osteopetrotic phenotype observed in the Tg mice was due to the impairment of osteoclast differentiation, we examined osteoclast formation in primary cultures of BMMs. BMMs were cultured in the presence of 30 ng/mL M-CSF and 50 ng/mL RANKL, and the osteoclastogenic activity of the cells was examined. The number of TRAP-positive multinucleated cells (MNCs) that differentiated from the Tg BMMs was consistently and markedly lower than the number of those that differentiated from WT BMMs (Figure 1I,J). Furthermore, real-time PCR analysis revealed that the expression of c-Fos and NFATc1 had attenuated 2 days after RANKL stimulation of the BMMs from PIAS3-Tg mice. However, there was no significant difference in the expression of these genes between PIAS3-Tg mice and their WT littermates 4 days after RANKL stimulation. The expression of cathepsin K and TRAP was inhibited during the RANKL-mediated osteoclastogenesis from the BMMs of PIAS3-Tg mice (Figure 1K). Together, these results indicate that PIAS3-Tg mice exhibit an osteopetrotic phenotype due to the impairment of osteoclast differentiation and that PIAS3 down-regulates the expression of c-Fos and NFATc1 during RANKL-mediated osteoclastogenesis, resulting in the inhibition of osteoclastogenesis in vivo.
PIAS3 inhibited RANKL-mediated osteoclastogenesis in vitro
We next examined the expression of PIAS3 during osteoclast differentiation from BMMs. Reverse transcription PCR (RT-PCR) revealed that markers of osteoclasts, MMP-9 and NFATc1, increased during the 3 days after the addition of RANKL. However, the expression of PIAS3 was constant from day 0 to day 3 (Figure 2A). Next, we analyzed the expression of PIAS3 in RAW264.7 cells. As in BMMs, the expression of PIAS3 in the RAW264.7 cells treated with 50 ng/mL RANKL was the same as in untreated cells (data not shown).
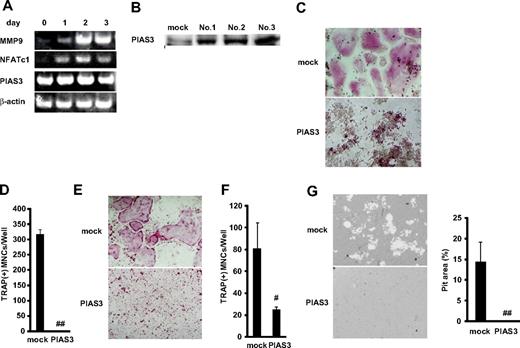
Overexpression of PIAS3 inhibits RANKL-mediated osteoclastogenesis in vitro. (A) Expression of MMP9, NFATc1, PIAS3, and β-actin mRNA. Total RNA from BMMs treated with 30 ng/mL M-CSF and 50 ng/mL RANKL for 0, 1, 2, and 3 days were measured by RT-PCR. (B) Western blot analysis of PIAS3 in RAW264.7 cells transduced with mock or PIAS3. (C) Analysis of osteoclast differentiation by TRAP staining. Mock- and PIAS3-transduced RAW264.7 cells were cultured for 3 days in the presence of 50 ng/mL RANKL. Cultured cells were fixed and stained for TRAP. (D) TRAP-positive MNCs having more than 3 nuclei were counted as osteoclast. (E) Analysis of osteoclast differentiation by TRAP staining. Mock- and PIAS3-transduced BMMs were cultured for 5 days in the presence of 30 ng/mL M-CSF and 50 ng/mL RANKL. Cultured cells were fixed and stained for TRAP. (F) TRAP-positive MNCs having more than 3 nuclei were counted as osteoclast. (G) Bone resorption assay using osteologic discs. Mock- or PIAS3-transduced RAW264.7 cells were cultured with 50 ng/mL RANKL on osteologic discs for 5 days. Pit area was quantified using ImageJ software. Error bars in all panels represent SD. #P < .05; ##P < .001.
Overexpression of PIAS3 inhibits RANKL-mediated osteoclastogenesis in vitro. (A) Expression of MMP9, NFATc1, PIAS3, and β-actin mRNA. Total RNA from BMMs treated with 30 ng/mL M-CSF and 50 ng/mL RANKL for 0, 1, 2, and 3 days were measured by RT-PCR. (B) Western blot analysis of PIAS3 in RAW264.7 cells transduced with mock or PIAS3. (C) Analysis of osteoclast differentiation by TRAP staining. Mock- and PIAS3-transduced RAW264.7 cells were cultured for 3 days in the presence of 50 ng/mL RANKL. Cultured cells were fixed and stained for TRAP. (D) TRAP-positive MNCs having more than 3 nuclei were counted as osteoclast. (E) Analysis of osteoclast differentiation by TRAP staining. Mock- and PIAS3-transduced BMMs were cultured for 5 days in the presence of 30 ng/mL M-CSF and 50 ng/mL RANKL. Cultured cells were fixed and stained for TRAP. (F) TRAP-positive MNCs having more than 3 nuclei were counted as osteoclast. (G) Bone resorption assay using osteologic discs. Mock- or PIAS3-transduced RAW264.7 cells were cultured with 50 ng/mL RANKL on osteologic discs for 5 days. Pit area was quantified using ImageJ software. Error bars in all panels represent SD. #P < .05; ##P < .001.
To elucidate the role of PIAS3 in RANKL-mediated osteoclastogenesis, we overexpressed PIAS3 in RAW264.7 cells and BMMs using retroviral techniques. We infected RAW264.7 cells with retrovirus that harbored PIAS3 and established 3 immortalized cell lines (Figure 2B). All 3 cell lines transduced with PIAS3 gave similar results in the RANKL-mediated osteoclastogenesis assay, and therefore we present here the results obtained from 1 representative cell line. The addition of RANKL induced the formation of TRAP-positive MNCs from mock-transduced cells, whereas the PIAS3-overexpressing RAW264.7 cells failed to form any TRAP-positive MNCs in response to RANKL (Figure 2C,D). We also confirmed that PIAS3 played a negative role in the in vitro osteoclastogenesis from BMMs (Figure 2E,F). Furthermore, the bone resorption activity was inhibited in the PIAS3-transduced RAW264.7 cells (Figure 2G). However, the overexpression of PIAS3 did not affect the proliferation or survival of RAW264.7 cells in a cell proliferation assay or DNA fragmentation assay (see Document S1 and Figure S1A,B). These results together indicate that PIAS3 negatively regulates RANKL-mediated osteoclastogenesis in vitro and that the PIAS3-induced inhibition of osteoclastogenesis is due to its effects on differentiation itself, rather than on cell proliferation or survival.
Overexpression of PIAS3 suppresses the expression of c-Fos during RANKL-mediated osteoclastogenesis
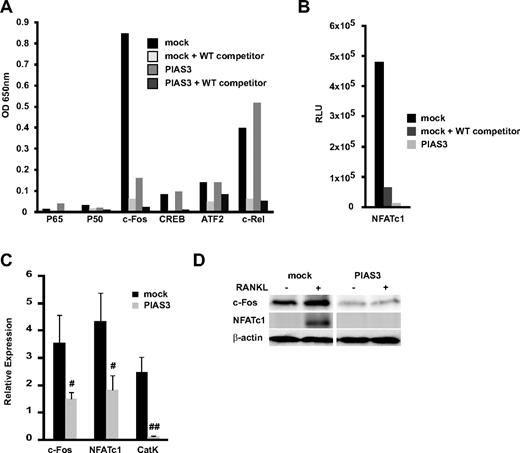
We examined the transcriptional activity of the osteoclast-related transcription factors p65, p50, c-Fos, CREB, ATF2, c-Rel, and NFATc1 using a DNA binding ELISA kit. This kit is designed to quantify the interaction between a specific transcription factor and DNA.33 Mock- and PIAS3-transduced RAW264.7 cells were stimulated with RANKL for 24 hours. In the mock-transduced cells, c-Fos was transactivated by the RANKL stimulation, and this activation was inhibited by WT oligonucleotide competitors. However, in the PIAS3-transduced cells, the c-Fos transcriptional activity was significantly attenuated compared with the mock-transduced cells (Figure 3A). The NFATc1 transcriptional activity stimulated by RANKL also decreased in the PIAS3-transduced cells (Figure 3B). The activity of the other transcription factors (p65, p50, CREB, ATF2, and c-Rel) did not differ between the mock- and PIAS3-transduced cells. It is reported that PIAS3 suppresses NF-κb–mediated transcription by interacting with the p65 subunit,34 and that p65 promotes osteoclast differentiation by blocking RANKL-induced apoptosis.35 However, we demonstrated that PIAS3 did not inhibit the p65 transactivation and had no effect on cell survival in RAW264.7 cells (Figure S1). Therefore, it is likely that PIAS3 does not interact with the NF-κb–mediated signal transduction in osteoclast precursors.
Overexpression of PIAS3 suppresses the expression and transcriptioan activity of c-Fos during RANKL-mediated osteoclastogenesis. (A,B) The whole-cell extracts from both mock- or PIAS3-transduced cells were stimulated with 50 ng/mL RANKL for 24 hours. Thirty micrograms of each cell lysate was used for the DNA binding ELISA assay. RLU, relative luciferase unit. (C) Mock- or PIAS3-transduced RAW264.7 cells were cultured with 50 ng/mL RANKL for 2 days. Total RNA was collected from cultured cells and real-time PCR was performed with primers specific for the indicated genes. (D) Mock- or PIAS3-transduced RAW264.7 cells were cultured with 50 ng/mL RANKL for 24 hours. Cells were harvested and lysates were analyzed by Western blot analysis for the indicated genes. Error bars in all panels represent SD. #P < .05; ##P < .01.
Overexpression of PIAS3 suppresses the expression and transcriptioan activity of c-Fos during RANKL-mediated osteoclastogenesis. (A,B) The whole-cell extracts from both mock- or PIAS3-transduced cells were stimulated with 50 ng/mL RANKL for 24 hours. Thirty micrograms of each cell lysate was used for the DNA binding ELISA assay. RLU, relative luciferase unit. (C) Mock- or PIAS3-transduced RAW264.7 cells were cultured with 50 ng/mL RANKL for 2 days. Total RNA was collected from cultured cells and real-time PCR was performed with primers specific for the indicated genes. (D) Mock- or PIAS3-transduced RAW264.7 cells were cultured with 50 ng/mL RANKL for 24 hours. Cells were harvested and lysates were analyzed by Western blot analysis for the indicated genes. Error bars in all panels represent SD. #P < .05; ##P < .01.
The transcription factor NFATc1 plays a central role in the osteoclastogenesis induced by RANKL.32 NFATc1 expression is regulated by c-Fos, a family of proteins that form the transcription factor AP-1 when dimerized.32,36 These results are consistent with the notion that PIAS3 lowers the c-Fos transcriptional activity, resulting in a decrease in NFATc1 induction, which leads to the inhibition of osteoclastogenesis. We therefore examined the expression of c-Fos and NFATc1 induced by RANKL stimulation. Real-time PCR revealed that the overexpression of PIAS3 attenuated the RANKL-induced expression of c-Fos, NFATc1, and cathepsin K in RAW264.7 cells (Figure 3C). Western blot analysis revealed that the levels of c-Fos and NFATc1 protein induced by RANKL were markedly lower in the PIAS3-transduced RAW264.7 cells than in the mock-transduced cells (Figure 3D). In contrast, the overexpression of PIAS3 did not alter the expression of MITF or STAT3, which are PIAS3-interacting partner molecules in mast cells and melanocytes (data not shown).37,38 These results suggest that the inhibitory effect of PIAS3 on RANKL-mediated osteoclastogenesis is probably owing to the inhibition of c-Fos expression in the early stage of osteoclast differentiation.
Down-regulation of PIAS3 enhances RANKL-mediated osteoclastogenesis
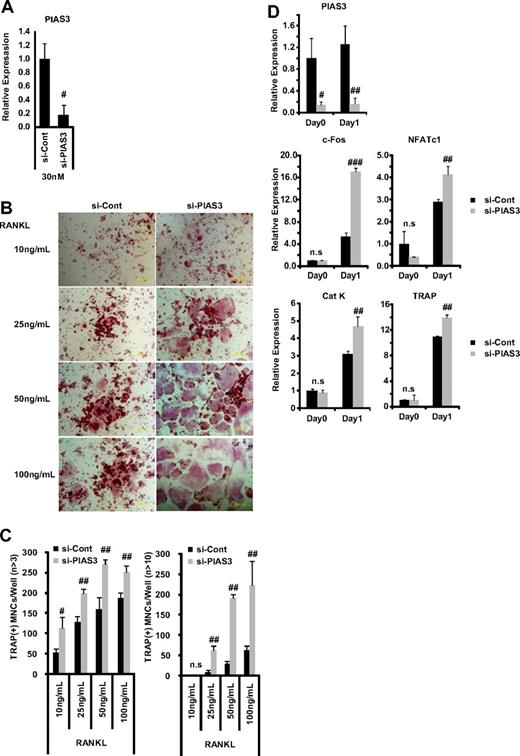
Since the overexpression of PIAS3 inhibited RANKL-mediated osteoclastogenesis, we investigated PIAS3's physiologic role in osteoclastogenesis using the small interfering RNA (siRNA) technique. The mRNA expression of PIAS3 was reduced to 20% of the control level by 30 nM PIAS3-specific siRNA in RAW264.7 cells (Figure 4A). The down-regulation of PIAS3 in the RAW264.7 cells enhanced the RANKL-mediated osteoclastogenesis (Figure 4B,C) and markedly increased the expression of c-Fos, NFATc1, cathepsin K, and TRAP (Figure 4D). Furthermore, the down-regulation of PIAS3 using lower concentrations of the specific siRNA (5 and 10 nM) also resulted in a significant increase in the number of TRAP-positive MNCs (Figure S2A,B). Similarly, BMMs transduced with PIAS3 siRNA also exhibited enhanced RANKL-mediated osteoclastogenesis (Figure S2C,D). These results clearly indicate that PIAS3 has a negative role in RANKL-mediated osteoclastogenesis.
Down-regulation of PIAS3 enhances RANKL-mediated osteoclastogenesis. (A) RAW264.7 cells transfected with 30 nM negative control or PIAS3 siRNA were cultured with 50 ng/mL RANKL for 2 days. Real-time PCR showed that PIAS3 siRNA decreased PIAS3 mRNA by 80%. (B) Analysis of osteoclast differentiation by TRAP staining. RAW264.7 cells were transfected with 30 nM negative control or PIAS3 siRNA and were stimulated by various concentrations of RANKL as indicated. (C) TRAP-positive MNCs having more than 3 and 10 nuclei were separately counted. (D) RAW264.7 cells transfected with negative control or PIAS3 siRNA were cultured with 50 ng/mL RANKL for the indicated times. Total RNA was recovered from cultured cells, and real-time PCR was performed with primers specific for PIAS3, c-Fos, NFATc1, cathepsin K, and TRAP. Error bars in all panels represent SD. #P < .05; ##P < .01; ###P < .001; n.s, not significant.
Down-regulation of PIAS3 enhances RANKL-mediated osteoclastogenesis. (A) RAW264.7 cells transfected with 30 nM negative control or PIAS3 siRNA were cultured with 50 ng/mL RANKL for 2 days. Real-time PCR showed that PIAS3 siRNA decreased PIAS3 mRNA by 80%. (B) Analysis of osteoclast differentiation by TRAP staining. RAW264.7 cells were transfected with 30 nM negative control or PIAS3 siRNA and were stimulated by various concentrations of RANKL as indicated. (C) TRAP-positive MNCs having more than 3 and 10 nuclei were separately counted. (D) RAW264.7 cells transfected with negative control or PIAS3 siRNA were cultured with 50 ng/mL RANKL for the indicated times. Total RNA was recovered from cultured cells, and real-time PCR was performed with primers specific for PIAS3, c-Fos, NFATc1, cathepsin K, and TRAP. Error bars in all panels represent SD. #P < .05; ##P < .01; ###P < .001; n.s, not significant.
PIAS3 inhibits the transcriptional activity of MITF independent of sumoylation
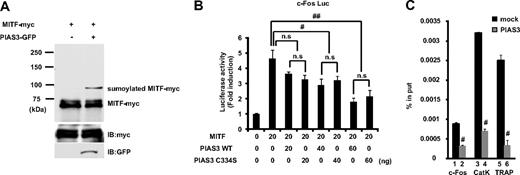
PIAS proteins can act as small ubiquitin-like modifier (SUMO)–E3-ligases, and SUMO modification has emerged as an important regulatory mechanism of transcription and signal transduction.20,21 PIAS3 has been reported to stimulate MITF sumoylation,22 and we confirmed that the overexpression of PIAS3 could sumoylate MITF in HEK 293T cells (Figure 5A). MITF is known to transactivate the c-Fos promoter and up-regulate c-Fos gene expression.25 Since PIAS3 overexpression decreased the c-Fos expression in RAW264.7 cells, we examined whether PIAS3 inhibited the MITF transcriptional activity in a reporter assay. We used the 0.8-kb c-Fos promoter reporter gene, which contains several potential MITF binding sites.25 MITF produced a 5-fold increase in the basal c-Fos promoter activity of HEK 293T cells. WT PIAS3 suppressed this transcriptional activity of MITF in a dose-dependent manner, and a PIAS3 mutant (C334S), which is a SUMO-E3-ligase defective mutant,27 exhibited a suppressive activity similar to that of WT PIAS3 (Figure 5B). These results indicate that PIAS3 inhibits the transcriptional activity of MITF independent of sumoylation, after the down-regulation of c-Fos gene expression.
PIAS3 inhibits the transcriptional activity of MITF through blocking the DNA binding, independent of sumoylation. (A) HEK 293T cells were transfected expression vectors encoding MITF-myc and PIAS3-GFP. Whole-cell extracts were immunoprecipitated with anti-myc antibody and immunoblotted using anti-myc antibody (top). Whole-cell extracts were immunoblotted by anti-myc and anti-green fluorescent protein (GFP) antibody (middle and bottom). (B) HEK 293T cells were cotransfected with 0.8 kb c-Fos promoter luciferase reporter, MITF, with the indicated amounts of PIAS3 (WT or C334S). Each well was also cotransfected with Renilla luciferase reporter to normalize the transfection efficiency (#P < .05; ##P < .01). (C) Chromatin immunoprecipitation for mock- or PIAS3-transduced RAW264.7 cells. Immunoprecipitated chromatin was analyzed by real-time PCR for the presence of c-Fos, cathepsin K, and TRAP promoter that were normalized to the respective input levels. Error bars in all panels represent SD. #P < .01.
PIAS3 inhibits the transcriptional activity of MITF through blocking the DNA binding, independent of sumoylation. (A) HEK 293T cells were transfected expression vectors encoding MITF-myc and PIAS3-GFP. Whole-cell extracts were immunoprecipitated with anti-myc antibody and immunoblotted using anti-myc antibody (top). Whole-cell extracts were immunoblotted by anti-myc and anti-green fluorescent protein (GFP) antibody (middle and bottom). (B) HEK 293T cells were cotransfected with 0.8 kb c-Fos promoter luciferase reporter, MITF, with the indicated amounts of PIAS3 (WT or C334S). Each well was also cotransfected with Renilla luciferase reporter to normalize the transfection efficiency (#P < .05; ##P < .01). (C) Chromatin immunoprecipitation for mock- or PIAS3-transduced RAW264.7 cells. Immunoprecipitated chromatin was analyzed by real-time PCR for the presence of c-Fos, cathepsin K, and TRAP promoter that were normalized to the respective input levels. Error bars in all panels represent SD. #P < .01.
PIAS3 directly inhibits MITF binding to the c-Fos, cathepsin K, and TRAP promoters
PIAS3 directly binds to MITF, thereby blocking its binding to the promoters of downstream genes.19 In this study, we showed that PIAS3 inhibited the c-Fos promoter activity produced by MITF and attenuated the c-Fos gene expression. To examine whether PIAS3 could inhibit the c-Fos promoter activity through an interaction with MITF in osteoclast precursors, we performed a ChIP assay using mock- and PIAS3-transduced RAW264.7 cells that were treated with 50 ng/mL RANKL for 24 hours. The c-Fos promoter DNA fragment containing the serum-responsive element (SRE) region, which is essential for the promoter activation by MITF,25 was precipitated with an anti-MITF antibody from mock- and PIAS3-infected RAW264.7 cells. The amount of precipitated DNA fragments relative to total input DNA was then quantified by real-time PCR for the c-Fos promoter region. In PIAS3-transduced RAW264.7 cells, the amount of DNA fragments precipitated was 30% of that in the mock-transduced cells (Figure 5C bars 1 and 2). These results demonstrate that PIAS3 inhibits MITF binding to the c-Fos promoter region.
Moreover, we investigated whether PIAS3 inhibited MITF's binding to the promoter region of the cathepsin K and TRAP genes. Previous reports demonstrated that PIAS3 inhibits MITF binding to the E-box of mMCP6 genes.19 The cathepsin K and TRAP genes have an E-box sequence in their promoter regions.9,10,39 In PIAS3-transduced RAW264.7 cells, the amount of precipitated DNA fragments relative to total input DNA was 20% of that found in the mock-transduced cells (Figure 5C bars 3-6). These observations suggest that PIAS3 suppresses RANKL-mediated osteoclastogenesis by inhibiting MITF's binding to osteoclast-specific gene promoters, such as the promoters for c-Fos, cathepsin K, and TRAP.
PIAS3 negatively regulates RANKL expression in POBs and inhibits osteoclast formation in an in vitro coculture system
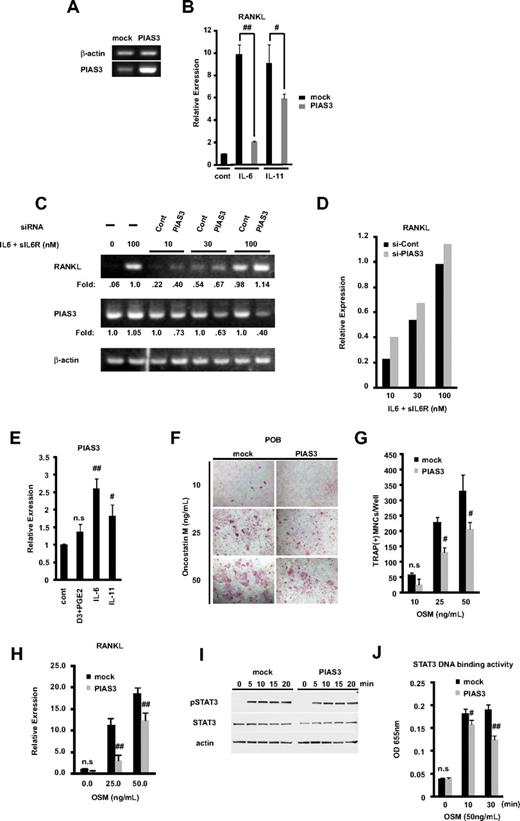
RANKL expressed on the membrane of osteoblasts binds to its receptor RANK, on the osteoclast precursors, resulting in osteoclastogenesis, which constitutes the cellular basis of bone remodeling.1,2,40 Members of the IL-6 cytokine family transmit their signals through STAT3 activation and induce RANKL expression.16,17 Since PIAS3 is a specific inhibitor of STAT3-mediated gene activation,18 we examined whether PIAS3 inhibits the RANKL expression induced by STAT3. We prepared POBs transduced with mock or PIAS3 using a retroviral technique. We confirmed the presence of endogenous and transduced PIAS3 in mouse osteoblasts by RT-PCR (Figure 6A). These mock- or PIAS3-transduced POBs were stimulated with 100 ng/mL IL-6 and 100 ng/mL sIL-6R or 20 ng/mL IL-11 for 24 hours. As shown in Figure 6B, real-time PCR analysis revealed that the overexpression of PIAS3 significantly attenuated the IL-6– and IL-11–induced RANKL expression in POBs, to 21% and 65% of the level in mock-transduced POBs, respectively.
PIAS3 negatively regulates RANKL expression in primary osteoblasts and inhibits osteoclast formation in vitro coculture system. (A) Expression of PIAS3 and β-actin mRNA in mock- or PIAS3-transduced mouse POBs was measured by RT-PCR. (B) Expression of RANKL mRNA in POBs that were stimulated with 100 ng/mL IL-6 and 100 ng/mL sIL-6R, or 20 ng/mL IL-11, was determined by real-time PCR. (C) POBs were transfected with negative control and PIAS3 siRNA. Expression of RANKL and PIAS3 mRNA in POBs that were stimulated by adding graded concentrations of IL-6 and sIL-6R. (D) Densitometric values for each band of panel C were quantified and normalized using ImageJ software. (E) Expression of PIAS3 mRNA in POBs that were stimulated with 1,25(OH)2D3 + PGE2, IL-6 + sIL6R, and IL-11, was determined by real-time PCR. (F) Analysis of osteoclast differentiation by TRAP staining. POBs transduced with mock or PIAS3 were cocultured with WT BMMs for 7 days in the presence of indicated concentration of OSM. Cultured cells were fixed and stained for TRAP. (G) TRAP-positive MNCs having more than 3 nuclei were counted as osteoclast. (H) Expression of RANKL mRNA in mock- or PIAS3-transduced POBs was measured by real-time PCR. (I) Mock- and PIAS3-transduced POBs were incubated with 50 ng/mL OSM for the indicated times. Cells lysates were analyzed by Western blot analysis for the pSTAT3, STAT3, and β-actin. (J) Mock- and PIAS3-transduced POBs were stimulated with 50 ng/mL OSM for 0, 10, and 30 minutes. A total of 60 μg of each cell lysate was used for the STAT3 DNA binding ELISA kit assay. Error bars in all panels represent SD. #P < .05; ##P < .01; n.s, not significant.
PIAS3 negatively regulates RANKL expression in primary osteoblasts and inhibits osteoclast formation in vitro coculture system. (A) Expression of PIAS3 and β-actin mRNA in mock- or PIAS3-transduced mouse POBs was measured by RT-PCR. (B) Expression of RANKL mRNA in POBs that were stimulated with 100 ng/mL IL-6 and 100 ng/mL sIL-6R, or 20 ng/mL IL-11, was determined by real-time PCR. (C) POBs were transfected with negative control and PIAS3 siRNA. Expression of RANKL and PIAS3 mRNA in POBs that were stimulated by adding graded concentrations of IL-6 and sIL-6R. (D) Densitometric values for each band of panel C were quantified and normalized using ImageJ software. (E) Expression of PIAS3 mRNA in POBs that were stimulated with 1,25(OH)2D3 + PGE2, IL-6 + sIL6R, and IL-11, was determined by real-time PCR. (F) Analysis of osteoclast differentiation by TRAP staining. POBs transduced with mock or PIAS3 were cocultured with WT BMMs for 7 days in the presence of indicated concentration of OSM. Cultured cells were fixed and stained for TRAP. (G) TRAP-positive MNCs having more than 3 nuclei were counted as osteoclast. (H) Expression of RANKL mRNA in mock- or PIAS3-transduced POBs was measured by real-time PCR. (I) Mock- and PIAS3-transduced POBs were incubated with 50 ng/mL OSM for the indicated times. Cells lysates were analyzed by Western blot analysis for the pSTAT3, STAT3, and β-actin. (J) Mock- and PIAS3-transduced POBs were stimulated with 50 ng/mL OSM for 0, 10, and 30 minutes. A total of 60 μg of each cell lysate was used for the STAT3 DNA binding ELISA kit assay. Error bars in all panels represent SD. #P < .05; ##P < .01; n.s, not significant.
We then examined whether the knockdown of PIAS3 could up-regulate the expression of RANKL. POBs were transfected with PIAS3 siRNA or negative control siRNA (see Document S1) and cultured for 2 days. RANKL expression was stimulated by adding graded concentrations of IL-6 and sIL-6R. The expression of RANKL and PIAS3 was determined by RT-PCR. PIAS3 siRNA decreased the endogenous PIAS3 expression, and it increased the RANKL expression at all concentrations of stimulators tested. Densitometric analysis revealed the consistent up-regulation of RANKL expression by PIAS3 siRNA knockdown (Figure 6C,D). These results demonstrate that PIAS3 acts as a negative regulator of the RANKL expression induced by IL-6–type cytokines in POBs.
Next, we examined whether IL-6–type cytokines regulated the expression of PIAS3 in POBs. The expression of PIAS3 was significantly up-regulated by IL-6 and IL-11. On the other hand, 1,25(OH)2D3 + PGE2 did not induce the expression of PIAS3 (Figure 6E). These results suggest a possible negative feedback mechanism of RANKL expression involving PIAS3 induced by cytokine signaling.
To examine whether overexpressed PIAS3 in POBs inhibited osteoclast formation from BMMs, we used an in vitro coculture system with POBs and BMMs. First, we prepared mock- or PIAS3-transduced POBs and incubated each osteoblast population with BMMs for 7 days in the presence of OSM, which promotes osteoclast differentiation in this coculture system. OSM stimulates osteoclast formation by activating the gp130 receptor subunit on stromal/osteoblastic cells, which in turn leads to the STAT3-mediated expression of RANKL.16,17,41 The number of TRAP-positive MNCs in the coculture with PIAS3-transduced POBs was significantly reduced in the presence of 25 and 50 ng/mL OSM compared with cultures of mock-transduced POBs (Figure 6F,G). As shown in Figure 6H, PIAS3 attenuated the expression of RANKL induced by each concentration of OSM. However, PIAS3 did not affect the expression of STAT3 or the phosphorylation of STAT3 induced by OSM-gp130 signaling (Figure 6I). Next, we examined whether PIAS3 inhibited the STAT3 DNA binding activity induced by gp130 signaling using a DNA binding ELISA kit. As expected, the STAT3 DNA binding activity was inhibited in the PIAS3-transduced POBs compared with mock-transduced POBs (Figure 6J). These observations clearly indicate that PIAS3 inhibits the OSM-gp130-STAT3 signaling by suppressing the DNA binding activity of STAT3 in POBs, resulting in the down-regulation of RANKL expression and the inhibition of osteoclast formation.
Discussion
The differentiation of osteoclasts is regulated by a group of transcription factors that includes PU.1, MITF, c-Fos, NF-κb, and NFATc1.3,4 PIAS proteins (PIAS1, PIAS3, PIASxβ, and PIASy) are expressed during osteoclast differentiation (data not shown), which suggests that the PIAS family is involved in the control of osteoclast differentiation or function. Here, we demonstrated that PIAS3 negatively regulates RANKL-mediated osteoclastogenesis, using in vivo analysis and in vitro gain-of-function and loss-of-function methods. The skeleton is a dynamic organ in which the mineralized bone is continuously resorbed by osteoclasts and new bone is formed by osteoblasts. Bone remodeling depends on a precise balance between bone formation and resorption.3,42 Radiologic analysis demonstrated an osteopetrotic bone phenotype in the TRAP–PIAS3-Tg mice, and bone histomorphometric analysis clearly revealed that this phenotype was due to an inhibition of osteoclastic activity. The formation of TRAP-positive MNCs induced by RANKL was markedly inhibited in the Tg mice. c-Fos and NFATc1 expression stimulated by RANKL in PIAS3-Tg BMMs was attenuated in the early stage of osteoclast differentiation (2 days after RANKL stimulation), compared with WT BMMs. However, the expression of these genes was no different from the WT levels in the late stage (4 days after RANKL stimulation). It is likely that the protein level of PIAS3 in vivo was not high enough to block the RANKL signaling cascade.
PIAS3 also down-regulated the expression of c-Fos and NFATc1 in vitro, in RAW264.7 cells and BMMs. The signal pathway of c-Fos, a family of dimeric AP-1 transcription factors, is required for RANKL-induced NFATc1 expression.32,36 Grigoriadis et al reported that c-Fos–deficient mice show an osteopetrotic phenotype due to a complete block of osteoclast differentiation.43 We therefore propose that the effect of PIAS3 on osteoclast differentiation is upstream of c-Fos and that PIAS3 negatively regulates RANKL-mediated osteoclastogenesis at the early stage. If this is the case, by what kind of mechanism does PIAS3 control the expression of c-Fos? PIAS proteins are reported to have various functions and to regulate transcription through several mechanisms, including blocking the DNA binding activity of transcription factors, recruiting transcriptional corepressors or coactivators, promoting protein sumoylation, and sequestering transcription factors in distinct subnuclear structures.44 It was recently suggested that the SUMO ligase activity of the PIAS3 protein may be involved in the regulation of transcription factors.22,27 However, we found that a mutant PIAS3 that is defective in SUMO ligase activity was still able to repress MITF transcription in a reporter assay. We also demonstrated that the overexpression of PIAS3 in RAW264.7 cells inhibited the DNA binding activity of MITF to osteoclast-specific gene promoters, such as those for c-Fos, cathepsin K, and TRAP, in a ChIP assay. Taken together, our findings indicate that PIAS3 exerts its negative effect in osteoclastogenesis by inhibiting the DNA binding activity of MITF, independent of PIAS3's sumoylation activity. Consistent with this mechanism, LEF1,45 C/EBPα,46 PR,47 AR,48 and STAT149 are all inhibited by PIAS protein overexpression, independent of their ability to be directly modified by SUMO. However, the addition of PIAS3 to reporter assays led to inhibitory effects on transcription that were not specific to MITF and therefore are difficult to interpret.
In stromal/osteoblastic cells, RANKL expression is strictly regulated by osteotropic factors, including 1,25-dihydroxyvitamin D3, parathyroid hormone, and cytokines such as IL-1, IL-6, and IL-11.3,4,13,41 Cytokines of the IL-6 type, such as IL-6, IL-11, and OSM, cause dimerization of gp130 at the plasma membrane. Dimerized gp130 activates Jak, a tyrosine kinase, and the activated Jak phosphorylates STAT3. Phosphorylated STAT3 is translocated to the nucleus and promotes the transcription of RANKL in stromal/osteoblastic cells.16,17 Thus, we examined the possible effect of PIAS3 on RANKL expression mediated by the IL-6 cytokine family in POBs. In POBs, the overexpression of PIAS3 attenuated the RANKL expression induced by IL6 and IL11. Moreover, the PIAS3 overexpression in POBs inhibited the osteoclast formation from BMMs stimulated by OSM in a coculture system (Figure 6B,F,G). On the other hand, the knockdown of PIAS3 increased the RANKL expression induced by IL-6 (Figure 6D,E). These results demonstrate that endogenous PIAS3 suppresses the IL-6 cytokine family–gp130-STAT3 signal transduction and inhibits the expression of RANKL in stromal/osteoblastic cells, leading to the inhibition of RANKL-mediated osteoclastogenesis. The question remains as to how PIAS3 inhibits STAT3 signal transduction, since PIAS3 does not affect the expression or phosphorylation of STAT3. Our DNA binding ELISA showed that PIAS3 inhibited the STAT3 DNA binding activity in primary osteoblasts, which is consistent with a previous report demonstrating that PIAS3 blocks STAT3 DNA binding activity and inhibits STAT3-mediated gene activation.18 The IL-6 cytokine family is involved in osteoarticular pathologies, mainly in osteolytic diseases (bone metastasis, Paget disease, giant cell tumors) and in rheumatoid arthritis.41 Therefore, PIAS3 has a protective role against bone destruction under inflammatory conditions.
Moreover, we demonstrated that IL-6 and IL-11 up-regulated the expression of PIAS3, which is a negative regulator of RANKL expression in POBs. Previous reports showed that the suppressor of cytokine signaling (SOCS) family of proteins is up-regulated in response to cytokine stimulation, and the corresponding SOCS proteins inhibit the cytokine-induced signaling pathway. Thus, SOCS proteins form part of a classical negative feedback circuit.50 Similarly, the negative feedback mechanism for RANKL expression involving PIAS3 would prevent excessive osteoclastogenesis, especially in pathologic states in which inflammatory cytokines such as IL-6 or IL-11 are produced.
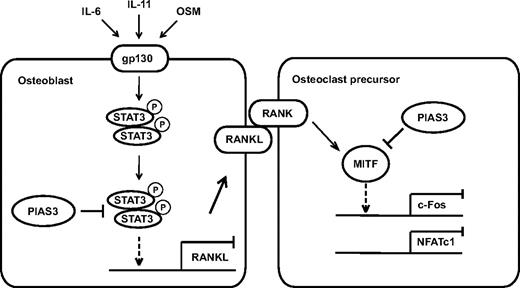
In conclusion, we present evidence that PIAS3 plays a critical role in the regulation of osteoclast differentiation and bone resorption, directly in osteoclast precursors and indirectly via osteoblasts (Figure 7). Further elucidation of the detailed mechanisms of PIAS3 gene regulation and function should contribute to the discovery of novel molecular therapeutic approaches for treating various types of inflammatory bone destruction.
A schematic model of PIAS3 in osteoclastogenesis. In osteoclast precursors, PIAS3 inhibits the transcriptional activity of MITF, resulting in attenuation of the expression of c-Fos and NFATc1. In osteoblasts, PIAS3 inhibits the IL-6 cytokine family-gp130-STAT3 signaling by suppressing the DNA binding activity of STAT3, resulting in attenuation of the RANKL expression and inhibition of osteoclastogenesis.
A schematic model of PIAS3 in osteoclastogenesis. In osteoclast precursors, PIAS3 inhibits the transcriptional activity of MITF, resulting in attenuation of the expression of c-Fos and NFATc1. In osteoblasts, PIAS3 inhibits the IL-6 cytokine family-gp130-STAT3 signaling by suppressing the DNA binding activity of STAT3, resulting in attenuation of the RANKL expression and inhibition of osteoclastogenesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr D. Fisher (Harvard Medical School, Boston, MA) for C5 MITF antibody; Dr T. Kitamura (Tokyo University, Tokyo, Japan) for Plat-E cells and pMX retroviral vector; Dr H. Yokosawa (Hokkaido University, Sapporo, Japan) for PIAS3 C334S expression plasmid; Dr H. Yasuda (M. D. Anderson Cancer Center, Houston, TX) for EGFP-PIAS3 expression plasmid; Dr M. Yoda, Dr T. Tomonda, and Ms S. Tomita for technical support; Ms Y. Hashimoto for technical assistance in histology; and Dr H. Murayama (Kureha Chemical Industry, Tokyo, Japan) for bone histomorphometrical analysis. We thank Teijin Pharma (Tokyo, Japan) for financial support.
Authorship
Contribution: T.H., J.T., A.H., M.F., S.U., T.K., and Y.O. performed research and analyzed data; H.T. and Y.T. designed the research and analyzed data; M.M., A.Y., R.N., S.V.R., and H.A. contributed vital new reagents; and T.H. and H.T. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hironari Takaishi, 35 Shinanomachi Shinjuku-ku, Tokyo, Japan; e-mail: hironari@sc.itc.keio.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal