To the editor:
While primary central nervous system lymphoma (PCNSL) may be classified among the 3 established molecular subclasses of large B-cell lymphoma: germinal center, activated B-cell, and type 3 based upon gene expression profiles,1 2 recent microarray-based expression profiling studies, one performed by our group1 and another by Tun et al,2 identified molecular characteristics that distinguish CNS lymphomas from nodal and/or extranodal large B-cell lymphoma. Using a cDNA-microarray–based platform, we identified more than 400 clones that distinguished 23 cases of PCNSL from 9 cases of nodal large B-cell lymphoma. Only a subset (29) of these clones were identified in our report. Using an oligonucleotide-based platform, Tun et al identified more than 60 genes that distinguished 13 cases of PCNSL from 11 nodal and 19 extranodal large B-cell lymphomas.2
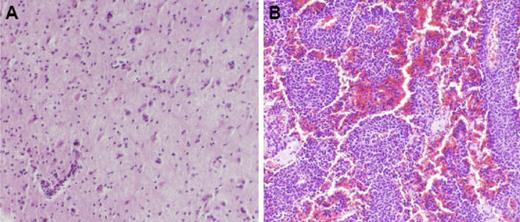
Determination of the molecular features of CNS lymphoma is significantly more challenging than for systemic non-Hodgkin lymphoma. The architecture of CNS lymphoma is often heterogeneous, with variable cell density and neovascularization, and infiltration by immune effector cells.1,3,4 The molecular profiling of individual small brain biopsy specimens may, therefore, provide an incomplete analysis of the tumor and its microenvironment. We proposed that there are at least 2 major growth patterns of PCNSL as evidenced by the histopathology of diagnostic tumor specimens: approximately one-half of cases of PCNSL are of low tumor cellularity in which normal brain elements are evident within the neoplasm; the remainder of tumors are of high cellular density without intervening normal brain elements between the neoplastic cells (Figure 1). High-density tumors that express activated STAT-6 are associated with a worse prognosis after treatment with standard methotrexate-based regimens than tumors of low cell density, independent of STAT-6 activation status.1
Low- and high-density PCNSL tumors. Low-density (A) and high-density (B) PCNSL tumors, each of large B-cell histology, are distinguished by the presence (A) or absence (B) of intervening normal brain elements between the tumor cells in pathologic specimens. Magnification ×100 (hematoxylin and eosin). Image were photographed using an Olympus Vanox AHBT3 microscope with Olympus DP70 digital camera (Olympus Optical, Tokyo, Japan). Images were captured onto a personal computer using Olympus DP controller software, version 1.2.1.108.
Low- and high-density PCNSL tumors. Low-density (A) and high-density (B) PCNSL tumors, each of large B-cell histology, are distinguished by the presence (A) or absence (B) of intervening normal brain elements between the tumor cells in pathologic specimens. Magnification ×100 (hematoxylin and eosin). Image were photographed using an Olympus Vanox AHBT3 microscope with Olympus DP70 digital camera (Olympus Optical, Tokyo, Japan). Images were captured onto a personal computer using Olympus DP controller software, version 1.2.1.108.
Neurons and glia are a rich source of molecules involved in signal transduction, including proto-oncogenes such as bcl-2.5-7 Therefore, molecular profiling of PCNSL specimens of low tumor density would clearly be prone to sampling error, given the presence of these interspersed normal brain elements. Given that both our study and that of Tun et al profiled specimens of dense tumor cellularity, it follows that these studies may be biased toward the most aggressive cases of PCNSL. We believe this issue has not previously been discussed in the literature.
Finally, the significance of these studies is limited because each lacks an independent validation set and used distinct microarray platforms. Therefore, we believe it to be essential to identify the subset of overlapping genes that are concordant in distinguishing CNS from non-CNS large B-cell lymphoma in each study, especially since only a subset of our data were presented in our publication (Table 1).1 The evidence for a unique CNS lymphoma molecular signature is underscored by the fact that, while all of the non-CNS DLBCL cases profiled in our study were obtained from lymph node biopsies, 19 of 30 (63%) of the non-CNS lymphoma cases profiled by Tun et al were extranodal in origin and from a variety of organ sites (eg, tonsil, bone, testes). This heterogeneity likely contributes to the discordance in differentially expressed genes reported in these studies.
Differentially expressed genes in the 2 studies
| CNS to non-CNS . | Symbol . | Rubenstein study . | Tun study . | ||
|---|---|---|---|---|---|
| Fold change . | P . | Fold change . | P . | ||
| Up-regulated | |||||
| Osteopontin | SPP1 | 11.4 | 8.00 × 10−6 | 9.73 | 3.03 × 10−8 |
| Complement component 1, q, subcomponent | C1QB | 2.8 | 8.29 × 10−6 | 2 | 2.40 × 10−2 |
| Hemoglobin, alpha2 | HBA2 | 2 | 6.26 × 10−4 | 2.5 | 1.90 × 10−2 |
| Regulator of G-protein signaling 13 | RGS13 | 2.3 | 3.34 × 10−2 | 2.4 | 2.70 × 10−2 |
| Chitinase 3-like 1 | CH13L1 | 2.8 | 5.02 × 10−2 | 2.72 | 5.10 × 10−5 |
| VT-cell leukemia/lymphoma 1A | TCL1A | 2.8 | 1.07 × 10−1 | 2.96 | 5.55 × 10−5 |
| Down-regulated | |||||
| Nicotinamide N-methyltransferase | NNMT | 0.56 | 5.26 × 10−3 | 0.43 | 8.99 × 10−4 |
| Vascular endothelial growth factor C | VEGFS | 0.51 | 7.16 × 10−3 | 0.4 | 1.00 × 10−3 |
| Collagen, type VI. alpha 1 | COL6A1 | 0.45 | 1.32 × 10−2 | 0.45 | 3.25 × 10−4 |
| Latexin | LXN | 0.76 | 1.99 × 10−2 | 0.48 | 2.67 × 10−2 |
| Lumican | LUM | 0.5 | 2.27 × 10−2 | 0.28 | 8.45 × 10−3 |
| Laminin, alpha 4 | LAMA4 | 0.74 | 4.54 × 10−2 | 0.37 | 8.00 × 10−3 |
| CNS to non-CNS . | Symbol . | Rubenstein study . | Tun study . | ||
|---|---|---|---|---|---|
| Fold change . | P . | Fold change . | P . | ||
| Up-regulated | |||||
| Osteopontin | SPP1 | 11.4 | 8.00 × 10−6 | 9.73 | 3.03 × 10−8 |
| Complement component 1, q, subcomponent | C1QB | 2.8 | 8.29 × 10−6 | 2 | 2.40 × 10−2 |
| Hemoglobin, alpha2 | HBA2 | 2 | 6.26 × 10−4 | 2.5 | 1.90 × 10−2 |
| Regulator of G-protein signaling 13 | RGS13 | 2.3 | 3.34 × 10−2 | 2.4 | 2.70 × 10−2 |
| Chitinase 3-like 1 | CH13L1 | 2.8 | 5.02 × 10−2 | 2.72 | 5.10 × 10−5 |
| VT-cell leukemia/lymphoma 1A | TCL1A | 2.8 | 1.07 × 10−1 | 2.96 | 5.55 × 10−5 |
| Down-regulated | |||||
| Nicotinamide N-methyltransferase | NNMT | 0.56 | 5.26 × 10−3 | 0.43 | 8.99 × 10−4 |
| Vascular endothelial growth factor C | VEGFS | 0.51 | 7.16 × 10−3 | 0.4 | 1.00 × 10−3 |
| Collagen, type VI. alpha 1 | COL6A1 | 0.45 | 1.32 × 10−2 | 0.45 | 3.25 × 10−4 |
| Latexin | LXN | 0.76 | 1.99 × 10−2 | 0.48 | 2.67 × 10−2 |
| Lumican | LUM | 0.5 | 2.27 × 10−2 | 0.28 | 8.45 × 10−3 |
| Laminin, alpha 4 | LAMA4 | 0.74 | 4.54 × 10−2 | 0.37 | 8.00 × 10−3 |
Authorship
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Contribution: J.L.R., P.T., and M.A.S. designed research. J.L.R., C.K., A.S., and P.T. performed research. T.T.B. provided specimens for the research. J.L.R., C.K., A.S., P.T., and M.A.S. analyzed and interpreted data. J.L.R., C.K., P.T., T.T.B., and M.A.S. drafted the paper.
This work was supported by a National Cancer Institute Research Career Award, University of California San Francisco Brain Tumor Specialized Program of Research Excellence (SPORE), and by grants from Gabrielle's Angel Foundation for Cancer Research (New York, NY) and the American Cancer Society (J.L.R.).
Correspondence: James L. Rubenstein, MD, PhD, University of California, San Francisco, Division of Hematology/Oncology, M1282 Box 1270, San Francisco, CA 94143; e-mail: jamesr@medicine.ucsf.edu.
References
National Institutes of Health

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal