Abstract
Primary central nervous system (CNS) lymphoma (PCNSL) is a diffuse large B-cell lymphoma (DLBCL) confined to the CNS. A genome-wide gene expression comparison between PCNSL and non-CNS DLBCL was performed, the latter consisting of both nodal and extranodal DLBCL (nDLBCL and enDLBCL), to identify a “CNS signature.” Pathway analysis with the program SigPathway revealed that PCNSL is characterized notably by significant differential expression of multiple extracellular matrix (ECM) and adhesion-related pathways. The most significantly up-regulated gene is the ECM-related osteopontin (SPP1). Expression at the protein level of ECM-related SPP1 and CHI3L1 in PCNSL cells was demonstrated by immunohistochemistry. The alterations in gene expression can be interpreted within several biologic contexts with implications for PCNSL, including CNS tropism (ECM and adhesion-related pathways, SPP1, DDR1), B-cell migration (CXCL13, SPP1), activated B-cell subtype (MUM1), lymphoproliferation (SPP1, TCL1A, CHI3L1), aggressive clinical behavior (SPP1, CHI3L1, MUM1), and aggressive metastatic cancer phenotype (SPP1, CHI3L1). The gene expression signature discovered in our study may represent a true “CNS signature” because we contrasted PCNSL with wide-spectrum non-CNS DLBCL on a genomic scale and performed an in-depth bioinformatic analysis.
Introduction
Primary central nervous system (CNS) lymphoma (PCNSL) is a diffuse large B-cell lymphoma (DLBCL) with a tropism for the CNS microenvironment and is confined to the CNS. Biologically, PCNSL is interesting in that it is a B-cell lymphoma in the CNS where very few B lymphocytes, if any, are found under normal circumstances.1 Some studies have indicated that PCNSL is of germinal center B-cell origin.2,3 According to a gene expression study, non-CNS DLBCL has been classified into 3 groups: germinal center B cell type, activated B cell type (ABC), and type 3.4 PCNSL has been shown to have immunophenotypic features of ABC.5 These findings taken together indicate that PCNSL develops from a B cell that has been exposed to a germinal center influence outside the CNS. Therefore, understanding the mechanisms that mediate B-cell migration and adaptation to the CNS microenvironment are important goals in research into the biology of PCNSL.
PCNSL remains incurable in most patients.6 Obviously, a better understanding of its biology is crucial to improve the prognosis. To this end, many studies, including DNA microarray studies, have been performed comparing PCNSL to non-CNS DLBCL, usually of nodal type. Of these, the largest microarray study to date compared PCNSL to nodal DLBCL and revealed several important molecular properties, including features linked to angiotropism.7 In our opinion, it is important to contrast PCNSL with all types of non-CNS DLBCL (both nDLBCL and enDLBCL) on a genomic scale and to use in-depth bioinformatic analysis, especially pathway analysis, to identify the “CNS signature.” We performed such a study and revealed new biologic insights.
Methods
Study subjects
Fresh frozen samples of PCNSL, nDLBCL, and enDLBCL from immunocompetent patients were obtained under the protocol approved by the Institutional Review Board of the Mayo Clinic. These samples were surplus tissues after the establishment of definitive pathologic diagnosis. The pathologic diagnosis was confirmed by central pathology review (D.M.M.). Totals of 13 PCNSL, 11 nDLBCL, and 19 enDLBCL were used in this study. For CNS tumors, 11 of 13 were stereotactic needle biopsies; the other 2 were from resections. The quality of the samples was ascertained by CD20 immunohistochemical stain; generally an estimated 80% or more content of B cells was observed. The enDLBCL samples originated from spleen (1), tonsil (4), adenoid (1), skin (1), bone (3), stomach (1), liver (1), testes (2), ovary (1), epidural (1), pericardium (1), thyroid (1), and pleural (1).
Microarray protocols
Total RNA was extracted from the dissected lymphoma tissue using a kit from QIAGEN (Rneasy mini kit; Valencia, CA). A fraction of the total RNA was used to perform a quality check for RNA integrity using the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Only samples yielding profiles of intact total RNA (retention of both ribosomal bands and the broad central peak of mRNA) were used for the microarray analyses reported in this paper. The mRNA in the sample was amplified with RiboAmp HS RNA Amplification Kit (Arcturus Engineering, Sunnyvale, CA). The resulting amplified RNA (aRNA) preparations were labeled either with Alexa Fluor 555 (lymphoma sample) or Alexa Fluor 647 (reference RNA) from Invitrogen (Carlsbad, CA). The reference RNA was the “Universal” human RNA from Stratagene (Santa Clara, CA). The Alexa dyes have been shown to have reduced labeling bias.8 The labeled samples were hybridized to Agilent Human Genomic Oligo 60-mer microarrays (41 061 probes) in Agilent microarray chambers (G2534A) at 60°C for 17 hours. After washing and drying, the array was scanned for analysis in a confocal laser scanner (ScanArray Express, PerkinElmer Life and Analytical Sciences, Waltham, MA) and Imagene software (version 6; BioDiscovery, El Segundo, CA) was used to process the images.
Validation of microarray results
Validation of microarray results was accomplished using quantitative real-time polymerase chain reaction (RT-PCR; detailed protocol is in “Detailed protocol for quantitative real-time PCR,” available on the Blood website; see the Supplemental Materials link at the top of the online article). Briefly, a portion of cDNA that was also used for microarray experiments was used to quantitate 10 genes: ATP5J, BCL-6, CD10, CD44, CHI3L1, COX6B1, IRF4, SPP1, TFPI2, and GAPDH. The level of GAPDH was used as a reference for obtaining the levels of the other mRNAs, and the ratios of CNS/nodal and CNS/extranodal were calculated. These ratios were then plotted vs ratios determined from the microarray data, and the correlation coefficient was determined after linear regression. We also performed immunohistochemistry (IHC) on SPP1 and CHI3L1 (detailed protocol in “Detailed protocol for immunohistochemistry procedures” in the Supplemental Materials).
Bioinformatics methods
Clustering and parametric tests.
The gene list used was approximately 11 500 in number (genes “present” on 35 of 43 arrays). In GeneSpring (version 7.2, Agilent) the LOWESS method of normalization was used, and unsupervised clustering of genome-wide expression profiles of PCNSL and non-CNS nodal and extranodal DLBCL was performed using “standard correlation” metric in GeneSpring. Genes identified by Fisher discriminant analysis (FDA) were also used to cluster the 43 samples, using cluster 3.0.9 The clustering was performed using uncentered correlation and complete linkage using genes identified in the FDA for genes separating the samples into 2 classes (CNS and non-CNS) at P less than .01.
Pathway analysis.
Data were imported into GeneSpring where LOWESS normalization was performed. The data were trimmed to those genes that were present on at least 35 of the 43 arrays, approximately 11 500 in number. In effect, this processing removed most of the weaker signals. Because additional global normalization did not change the SigPathway (Sun Microsystems, Santa Clara, CA) results substantively (not shown), the results presented in this article are from data with only LOWESS normalization performed. In this type of normalization, the ratios of the 2 channels on each array are adjusted to correct for nonlinearity between the ratios and signal intensity. The data were exported into Microsoft Excel (Microsoft, Redmond, WA), then to Star Calc (http://bioconductor.org/bioclite.R), and saved in “comma-separated value” format. For pathway analysis, the data were imported into R (version 2.2.1; http://www.r-project.org) and analyzed with the SigPathway package10 (version 1.1.3, available as a Bioconductor package at http://www.bioconductor.org). Missing values were imputed using the K-nearest neighbor method11 exactly as previously described.12
The SigPathway package performs “pathway analysis” on microarray data by first compiling genes on the microarray into functional (ontologic and pathway associations) categories based on databases searches, producing many gene sets for statistical testing. Assessment of the differential expression of each gene set in pair-wise comparisons of phenotypes in the experimental data is accomplished by calculating a composite t score for each gene set, and then using permutation methods to determine 2 statistical parameters, NT and NE, for each gene set. NT is a measure of the degree to which a given gene set differs from the other gene sets on the array. Rows of the gene X array matrix (where the genes are in rows and the arrays are in columns) are permuted for this calculation (gene labels are permuted). NE is a measure of the degree to which the gene set composite expression is different between phenotypes; columns of the gene X array matrix are permuted for calculation of this parameter (sample labels are permuted). The program ranks gene sets according to the average of the rank-orders of NT and NE; false discovery rate (q value) is calculated to adjust for multiple testing problems. The rank is required to be high in both rank-orders to minimize false positives.
FDA.
Dimensional reduction methods using matrix decomposition have been applied extensively to microarray data. One useful method in this class is Fisher discriminant analysis (FDA), which maximizes separation of phenotypes.13 As these authors describe, FDA is a matrix decomposition method whereby orthonormal dimensions are determined that maximize the separation between classes. Microarray data (∼11 500 genes on at least 35 of 43 arrays) were exported from GeneSpring into Microsoft Excel, in which a text file was composed for analysis by BioSystAnSe. The FDA was performed with the Singular Value Decomposition option at a criterion of P less than .01. The lymphoma samples were classified either by 3 phenotypes (CNS, nodal, extranodal) or by 2 phenotypes (CNS, non-CNS).
Image acquisition and preparation
Pathology slides were viewed with a Leica DMLB optical microscope (Leica Microsystems, Wetzlar, Germany). Cytoseal-60 mounting media (Richard Allen, Kalamazoo, MI) was used. Images were acquired using a SPOT RT Color Camera (Diagnostic Instruments, Sterling Heights, MI), and were processed with SPOT Advanced program version 2.0 (Diagnostic Instruments) and Adobe Photoshop version 6.0 software (Adobe Systems, San Jose, CA).
Results
Initial characterization of the microarray data
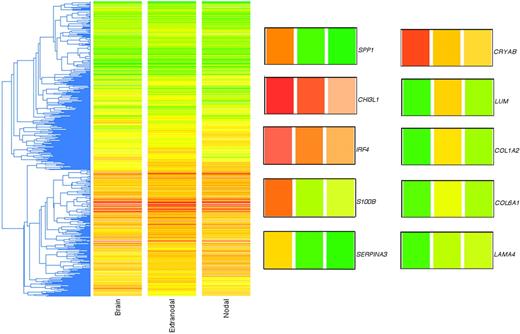
In the final dataset, there were a total of 43 high-quality lymphoma samples that produced reliable microarray data, from which filtering yielded approximately 11 500 genes that were present on at least 35 arrays, a reasonable compromise that optimized both gene numbers and data quality. A standard clustering of these approximately 1l 500 filtered and LOWESS-normalized genes is shown in Figure 1, where the array data are averaged according to the 3 phenotypes. Within the basis list of approximately 11 500 genes, there were 50 genes that were significantly (Student t test; P < .05) expressed at 2-fold or greater difference between the PCNSL and non-CNS DLBCL (Table 1).
Unsupervised clustering of genome-wide expression profiles of PCNSL and non-CNS nodal and extranodal DLBCL. The gene list used was approximately 11 500 in number (genes present on at least 35 of the 43 arrays). The metric used was “standard correlation” in GeneSpring. Because the 2-color array method involved a reference standard, the colors do not represent actual gene expression levels in the tumor samples but rather the ratio of the tumor mRNA to the reference mRNA. The LOWESS method of normalization was used. To the right of the cluster are shown 10 genes of interest enlarged from the cluster; the colored bars correspond to the 3 phenotypes identified at the bottom of the cluster (“Brain,” “Extranodal,” and “Nodal”; left to right). SPP1 (osteopontin), CHI3L1 (chitinase-3 like 1), IRF4 (MUM1), S-100B (S-100 calcium binding protein beta), SERPINA3 (serine proteinase inhibitor, clade A, member 3), CRYAB (crystallin alpha B), LUM (lumican), COL1A2 (collagen type 1 alpha 2), COL6A1 (collagen type 6 alpha 1), and LAMA4 (laminin alpha 4).
Unsupervised clustering of genome-wide expression profiles of PCNSL and non-CNS nodal and extranodal DLBCL. The gene list used was approximately 11 500 in number (genes present on at least 35 of the 43 arrays). The metric used was “standard correlation” in GeneSpring. Because the 2-color array method involved a reference standard, the colors do not represent actual gene expression levels in the tumor samples but rather the ratio of the tumor mRNA to the reference mRNA. The LOWESS method of normalization was used. To the right of the cluster are shown 10 genes of interest enlarged from the cluster; the colored bars correspond to the 3 phenotypes identified at the bottom of the cluster (“Brain,” “Extranodal,” and “Nodal”; left to right). SPP1 (osteopontin), CHI3L1 (chitinase-3 like 1), IRF4 (MUM1), S-100B (S-100 calcium binding protein beta), SERPINA3 (serine proteinase inhibitor, clade A, member 3), CRYAB (crystallin alpha B), LUM (lumican), COL1A2 (collagen type 1 alpha 2), COL6A1 (collagen type 6 alpha 1), and LAMA4 (laminin alpha 4).
Genes at least two-fold different between PCNSL and non-CNS DLBCL at P < 0.05
| Genes . | Fold Change . | P . | Description . |
|---|---|---|---|
| CNS versus non-CNS up-regulated | |||
| SPP1 | 9.73 | < .001 | Secreted phosphoprotein 1 (osteopontin) |
| TF | 8.33 | < .001 | Transferrin |
| DDR1 | 6.08 | < .001 | Discoidin domain receptor family, member 1 |
| NM_178012 | 5.13 | < .001 | Tubulin, beta polypeptide paralog |
| SERPINA3 | 4.67 | < .001 | Serine (or cysteine) proteinase inhibitor, clade A, member 3 |
| S-100B | 4.07 | < .001 | S-100 calcium binding protein, beta (neural) |
| C9orf58 | 4.03 | < .001 | Chromosome 9 open reading frame 58 |
| BC020630 | 3.61 | < .001 | Calcium/calmodulin-dependent protein kinase II |
| CRYAB | 3.36 | < .001 | Crystallin, alpha B (CRYAB) |
| NM_018584 | 3.26 | < .001 | Calcium/calmodulin-dependent protein kinase II |
| AF034208 | 3.24 | .015 | RIG-like 7–1 mRNA, complete cds |
| CXCL13 | 3.22 | .009 | Chemokine (C-X-C motif) ligand 13 (B-cell chemoattractant) |
| NM_152680 | 3.01 | .032 | Hypothetical protein FLJ32028 (FLJ32028) |
| TCL1A | 2.96 | < .001 | T-cell leukemia/lymphoma 1A |
| DKK3 | 2.91 | < .001 | Dickkopf homolog 3 |
| FOXG1B | 2.85 | .003 | Forkhead box G1B |
| UGT2B17 | 2.81 | .041 | UDP glycosyltransferase 2 family, polypeptide B17 |
| TACSTD1 | 2.79 | .006 | Tumor-associated calcium signal transducer 1 |
| FEZ1 | 2.78 | .004 | Fasciculation and elongation protein zeta 1 (zygin I) |
| CHI3L1 | 2.72 | < .001 | Chitinase 3-like 1 (cartilage glycoprotein-39) |
| CA2 | 2.7 | < .001 | Carbonic anhydrase II |
| MGST1 | 2.64 | .037 | Microsomal glutathione S-transferase, transcript variant 1c |
| DNAJC12 | 2.63 | < .001 | DnaJ (Hsp40) homolog, subfamily C, member 12 |
| AK097976 | 2.6 | .026 | cDNA FLJ40657 fis, clone THYMU2019436 |
| NM_018357 | 2.57 | .042 | Acheron (FLJ11196) |
| HBA2 | 2.5 | .019 | Hemoglobin, alpha 2 (HBA2) |
| NM_024897 | 2.44 | .01 | Progestin and adipoQ receptor family member VI (PAQR6) |
| RGS13 | 2.4 | .027 | Regulator of G-protein signalling 13 |
| FCGR3A | 2.37 | .004 | Fc fragment of IgG, low affinity IIIa, receptor for (CD16) |
| CNP | 2.25 | < .001 | 2′,3′-cyclic nucleotide 3′ phosphodiesterase |
| FBP1 | 2.24 | .04 | Fructose-1,6-bisphosphatase 1 |
| BC000525 | 2.21 | .001 | Glutamic-oxaloacetic transaminase 2 |
| BMP7 | 2.11 | .017 | Bone morphogenetic protein 7 |
| NM_013332 | 2.08 | .021 | Hypoxia-inducible protein 2 (HIG2) |
| C1QB | 2.03 | .023 | Complement component 1, q subcomponent, beta polypeptide |
| IRF4 | 2.02 | .004 | Interferon regulatory factor 4 |
| DECR2 | 2.01 | .006 | 2,4-dienoyl CoA reductase 2 |
| CNS versus non-CNS down-regulated | |||
| CX3CR1 | 0.5 | .003 | Chemokine (C-X3-C motif) receptor 1 |
| AK091178 | 0.49 | .042 | cDNA FLJ33859 fis, clone CTONG2006223 |
| D42043 | 0.49 | .002 | KIAA0084 mRNA, partial cds |
| SQLE | 0.49 | .004 | Squalene epoxidase (SQLE) |
| P2RY8 | 0.49 | .006 | Purinergic receptor P2Y, G-protein coupled, 8 |
| LXN | 0.48 | .026 | Latexin |
| LPP | 0.47 | .002 | LIM domain containing preferred translocation partner in lipoma |
| S56205 | 0.47 | .005 | Insulin-like growth factor binding protein 3 {3′ region} |
| Z24727 | 0.47 | .008 | Tropomyosin isoform mRNA, complete CDS |
| STEAP | 0.46 | .015 | Six transmembrane epithelial antigen of the prostate |
| COL6A1 | 0.45 | < .001 | Collagen, type VI, alpha 1 |
| COL12A1 | 0.45 | .002 | Collagen, type XII, alpha 1 |
| AK022110 | 0.44 | .023 | cDNA FLJ12048 fis, clone HEMBB1001990 |
| NR2F2 | 0.43 | .006 | Nuclear receptor subfamily 2, group F, member 2 |
| NNMT | 0.43 | < .001 | Nicotinamide N-methyltransferase |
| AK091153 | 0.42 | .006 | cDNA FLJ33834 fis, clone CTONG2004264 |
| BC040354 | 0.42 | < .001 | Caldesmon 1, transcript variant 3 |
| VEGFC | 0.4 | .001 | Vascular endothelial growth factor C |
| LAMA4 | 0.37 | .008 | Laminin, alpha 4 |
| NM_015714 | 0.32 | < .001 | Putative lymphocyte G0/G1 switch gene (G0S2) |
| CAMKK2 | 0.3 | .026 | Calcium/calmodulin-dependent protein kinase kinase 2, beta |
| FBN1 | 0.29 | .001 | Fibrillin 1 (Marfan syndrome) |
| AJ001381 | 0.28 | .002 | Incomplete cDNA for a mutated allele of a myosin class I, myh-1c |
| LUM | 0.28 | .008 | Lumican (LUM) |
| LOXL1 | 0.27 | .001 | Lysyl oxidase-like 1 |
| COL1A2 | 0.27 | < .001 | Collagen, type I, alpha 2 |
| Genes . | Fold Change . | P . | Description . |
|---|---|---|---|
| CNS versus non-CNS up-regulated | |||
| SPP1 | 9.73 | < .001 | Secreted phosphoprotein 1 (osteopontin) |
| TF | 8.33 | < .001 | Transferrin |
| DDR1 | 6.08 | < .001 | Discoidin domain receptor family, member 1 |
| NM_178012 | 5.13 | < .001 | Tubulin, beta polypeptide paralog |
| SERPINA3 | 4.67 | < .001 | Serine (or cysteine) proteinase inhibitor, clade A, member 3 |
| S-100B | 4.07 | < .001 | S-100 calcium binding protein, beta (neural) |
| C9orf58 | 4.03 | < .001 | Chromosome 9 open reading frame 58 |
| BC020630 | 3.61 | < .001 | Calcium/calmodulin-dependent protein kinase II |
| CRYAB | 3.36 | < .001 | Crystallin, alpha B (CRYAB) |
| NM_018584 | 3.26 | < .001 | Calcium/calmodulin-dependent protein kinase II |
| AF034208 | 3.24 | .015 | RIG-like 7–1 mRNA, complete cds |
| CXCL13 | 3.22 | .009 | Chemokine (C-X-C motif) ligand 13 (B-cell chemoattractant) |
| NM_152680 | 3.01 | .032 | Hypothetical protein FLJ32028 (FLJ32028) |
| TCL1A | 2.96 | < .001 | T-cell leukemia/lymphoma 1A |
| DKK3 | 2.91 | < .001 | Dickkopf homolog 3 |
| FOXG1B | 2.85 | .003 | Forkhead box G1B |
| UGT2B17 | 2.81 | .041 | UDP glycosyltransferase 2 family, polypeptide B17 |
| TACSTD1 | 2.79 | .006 | Tumor-associated calcium signal transducer 1 |
| FEZ1 | 2.78 | .004 | Fasciculation and elongation protein zeta 1 (zygin I) |
| CHI3L1 | 2.72 | < .001 | Chitinase 3-like 1 (cartilage glycoprotein-39) |
| CA2 | 2.7 | < .001 | Carbonic anhydrase II |
| MGST1 | 2.64 | .037 | Microsomal glutathione S-transferase, transcript variant 1c |
| DNAJC12 | 2.63 | < .001 | DnaJ (Hsp40) homolog, subfamily C, member 12 |
| AK097976 | 2.6 | .026 | cDNA FLJ40657 fis, clone THYMU2019436 |
| NM_018357 | 2.57 | .042 | Acheron (FLJ11196) |
| HBA2 | 2.5 | .019 | Hemoglobin, alpha 2 (HBA2) |
| NM_024897 | 2.44 | .01 | Progestin and adipoQ receptor family member VI (PAQR6) |
| RGS13 | 2.4 | .027 | Regulator of G-protein signalling 13 |
| FCGR3A | 2.37 | .004 | Fc fragment of IgG, low affinity IIIa, receptor for (CD16) |
| CNP | 2.25 | < .001 | 2′,3′-cyclic nucleotide 3′ phosphodiesterase |
| FBP1 | 2.24 | .04 | Fructose-1,6-bisphosphatase 1 |
| BC000525 | 2.21 | .001 | Glutamic-oxaloacetic transaminase 2 |
| BMP7 | 2.11 | .017 | Bone morphogenetic protein 7 |
| NM_013332 | 2.08 | .021 | Hypoxia-inducible protein 2 (HIG2) |
| C1QB | 2.03 | .023 | Complement component 1, q subcomponent, beta polypeptide |
| IRF4 | 2.02 | .004 | Interferon regulatory factor 4 |
| DECR2 | 2.01 | .006 | 2,4-dienoyl CoA reductase 2 |
| CNS versus non-CNS down-regulated | |||
| CX3CR1 | 0.5 | .003 | Chemokine (C-X3-C motif) receptor 1 |
| AK091178 | 0.49 | .042 | cDNA FLJ33859 fis, clone CTONG2006223 |
| D42043 | 0.49 | .002 | KIAA0084 mRNA, partial cds |
| SQLE | 0.49 | .004 | Squalene epoxidase (SQLE) |
| P2RY8 | 0.49 | .006 | Purinergic receptor P2Y, G-protein coupled, 8 |
| LXN | 0.48 | .026 | Latexin |
| LPP | 0.47 | .002 | LIM domain containing preferred translocation partner in lipoma |
| S56205 | 0.47 | .005 | Insulin-like growth factor binding protein 3 {3′ region} |
| Z24727 | 0.47 | .008 | Tropomyosin isoform mRNA, complete CDS |
| STEAP | 0.46 | .015 | Six transmembrane epithelial antigen of the prostate |
| COL6A1 | 0.45 | < .001 | Collagen, type VI, alpha 1 |
| COL12A1 | 0.45 | .002 | Collagen, type XII, alpha 1 |
| AK022110 | 0.44 | .023 | cDNA FLJ12048 fis, clone HEMBB1001990 |
| NR2F2 | 0.43 | .006 | Nuclear receptor subfamily 2, group F, member 2 |
| NNMT | 0.43 | < .001 | Nicotinamide N-methyltransferase |
| AK091153 | 0.42 | .006 | cDNA FLJ33834 fis, clone CTONG2004264 |
| BC040354 | 0.42 | < .001 | Caldesmon 1, transcript variant 3 |
| VEGFC | 0.4 | .001 | Vascular endothelial growth factor C |
| LAMA4 | 0.37 | .008 | Laminin, alpha 4 |
| NM_015714 | 0.32 | < .001 | Putative lymphocyte G0/G1 switch gene (G0S2) |
| CAMKK2 | 0.3 | .026 | Calcium/calmodulin-dependent protein kinase kinase 2, beta |
| FBN1 | 0.29 | .001 | Fibrillin 1 (Marfan syndrome) |
| AJ001381 | 0.28 | .002 | Incomplete cDNA for a mutated allele of a myosin class I, myh-1c |
| LUM | 0.28 | .008 | Lumican (LUM) |
| LOXL1 | 0.27 | .001 | Lysyl oxidase-like 1 |
| COL1A2 | 0.27 | < .001 | Collagen, type I, alpha 2 |
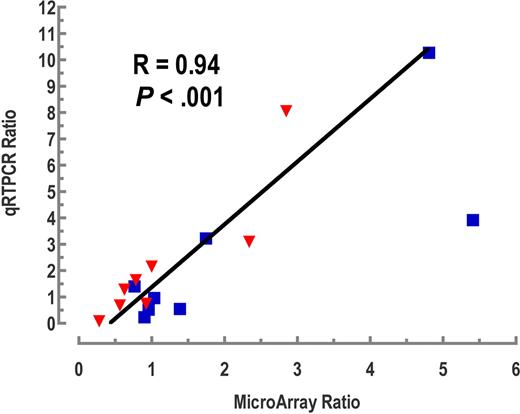
For validation of the microarray data, we performed quantitative RT-PCR for a set of 10 genes, which included several extracellular matrix (ECM)-related genes (which will be shown to be important under “Pathway analysis of the DLBCL gene expression dataset”), several others of interest, and GAPDH. There was excellent agreement between the microarray and quantitative RT-PCR data. Figure 2 shows a plot of the averages of quantitative RT-PCR values for multiple samples of the CNS and non-CNS phenotypes versus their corresponding values on the microarrays. There was one outlier; without this point, the linear correlation coefficient was 0.94 and highly significant (P < .001); the correlation was still statistically significant when the outlier was included (R = 0.79; P < .02).
Validation of selected genes using quantitative RT-PCR. The blue squares represent CNS/nodal sample ratios; the red inverted triangles are the CNS/extranodal sample ratios. The ratios obtained using quantitative RT-PCR are plotted along the y-axis, whereas the ratios calculated from the microarray data are plotted on the x-axis. The genes analyzed were ATP5J, BCL-6, CD10, CD44, CHI3L1, COX6B1, IRF4, SPP1, TFPI2, and GAPDH. The correlation coefficient shown is that calculated without the non-CNS outlier at the right (CHI3L1 CNS/nodal ratio). The correlation remains significant when including this outlier (R = 0.79; P < .02).
Validation of selected genes using quantitative RT-PCR. The blue squares represent CNS/nodal sample ratios; the red inverted triangles are the CNS/extranodal sample ratios. The ratios obtained using quantitative RT-PCR are plotted along the y-axis, whereas the ratios calculated from the microarray data are plotted on the x-axis. The genes analyzed were ATP5J, BCL-6, CD10, CD44, CHI3L1, COX6B1, IRF4, SPP1, TFPI2, and GAPDH. The correlation coefficient shown is that calculated without the non-CNS outlier at the right (CHI3L1 CNS/nodal ratio). The correlation remains significant when including this outlier (R = 0.79; P < .02).
Pathway analysis of the DLBCL gene expression dataset
The bioinformatics program SigPathway was used to identify those gene sets that were most powerful in contrasting phenotypes.10 The PCNSL was contrasted pair-wise with non-CNS DLBCL (nDLBCL + enDLBCL), nDLBCL, and enDLBCL for a total of 3 comparisons (Tables 2,Table 3–4; these tables are abbreviated in the main text; full versions are included in Tables S1Table S3. Unabbreviated version of Table 4 (PDF, 59.8 KB)–S3). Table 2 shows the pathway analysis results for the contrast between the 13 CNS samples versus 30 “non-CNS” samples (nDLBCL and enDLBCL combined). This contrast led to discoveries of primary importance in this study: assessment of biologic pathways unique to PCNSL. Table 3 contrasts CNS with nDLBCL samples; Table 4 contrasts CNS with enDLBCL samples. Our results in Tables 2 to 4 show numerous gene sets for which there are high values of NT and NE along with corresponding low q values (as shown numerically in the full-length Tables S1Table S3. Unabbreviated version of Table 4 (PDF, 59.8 KB)–S3). This indicates statistically strong differential expression of these biologic pathways between the phenotypes. In each table, there are as many as 20 ranked gene sets exhibiting high NT and NE parameters associated with q values less than 0.0001, indicating very high statistical reliability (shown in Tables S1Table S3. Unabbreviated version of Table 4 (PDF, 59.8 KB)–S3).
SigPathway results: PCNSL versus non-CNS DLBCL
| . | Gene set category . | Pathway . | Set size . | % up . | NTk rank . | NEk rank . |
|---|---|---|---|---|---|---|
| 1 | KEGG:04512 | ECM-receptor interaction | 47 | 19 | 3 | 1 |
| 2 | GO:0005604 | Basement membrane | 23 | 9 | 5 | 2 |
| 3 | KEGG:04510 | Focal adhesion | 135 | 30 | 6 | 8 |
| 4 | GO:0005201 | Extracellular matrix structural constituent | 40 | 18 | 1 | 20 |
| 5 | GO:0004857 | Enzyme inhibitor activity | 128 | 28 | 19 | 3 |
| 6 | KEGG:01430 | Cell communication | 53 | 23 | 7 | 24 |
| 7 | GO:0016126 | Sterol biosynthesis | 22 | 27 | 11 | 31 |
| 8 | GO:0005605 | Basal lamina | 14 | 7 | 10 | 38 |
| 9 | GO:0007265 | Ras protein signal transduction | 29 | 34 | 33 | 15 |
| 10 | GO:0001817 | Regulation of cytokine production | 16 | 50 | 39 | 13 |
| 11 | GO:0042035 | Regulation of cytokine biosynthesis | 16 | 50 | 39 | 13 |
| 12 | GO:0009064 | Glutamine family amino acid metabolism | 23 | 65 | 48 | 12 |
| 13 | KEGG:00220 | Urea cycle and metabolism of amino groups | 18 | 72 | 44 | 17 |
| 14 | KEGG:00330 | Arginine and proline metabolism | 38 | 66 | 46 | 16 |
| 15 | GO:0007588 | Excretion | 16 | 63 | 22 | 44 |
| 16 | GO:0050954 | Sensory perception of mechanical stimulus | 39 | 31 | 74 | 7 |
| 17 | GO:0007605 | Sensory perception of sound | 39 | 31 | 74 | 7 |
| 18 | GO:0042089 | Cytokine biosynthesis | 17 | 47 | 62 | 22 |
| 19 | GO:0043062 | Extracellular structure organization and biogenesis | 11 | 9 | 8 | 81 |
| 20 | GO:0030198 | Extracellular matrix organization and biogenesis | 11 | 9 | 8 | 81 |
| 21 | GO:0007059 | Chromosome segregation | 25 | 12 | 9 | 81 |
| 22 | GO:0006937 | Regulation of muscle contraction | 17 | 24 | 14 | 81 |
| 23 | KEGG:04060 | Cytokine-cytokine receptor interaction | 79 | 30 | 21 | 81 |
| 24 | GO:0005578 | Extracellular matrix (sensu Metazoa) | 102 | 21 | 2 | 105 |
| 25 | GO:0031012 | Extracellular matrix | 102 | 21 | 2 | 105 |
| 26 | GO:0005581 | Collagen | 15 | 13 | 4 | 108 |
| 27 | GO:0043161 | Proteasomal ubiquitin-dependent protein catabolism | 11 | 18 | 108.5 | 4 |
| 28 | GO:0042087 | Cell-mediated immune response | 11 | 45 | 108.5 | 5 |
| 29 | GO:0042088 | T-helper 1 type immune response | 11 | 45 | 108.5 | 5 |
| 30 | BioCarta | ALK in cardiac myocytes | 14 | 21 | 108.5 | 6 |
| . | Gene set category . | Pathway . | Set size . | % up . | NTk rank . | NEk rank . |
|---|---|---|---|---|---|---|
| 1 | KEGG:04512 | ECM-receptor interaction | 47 | 19 | 3 | 1 |
| 2 | GO:0005604 | Basement membrane | 23 | 9 | 5 | 2 |
| 3 | KEGG:04510 | Focal adhesion | 135 | 30 | 6 | 8 |
| 4 | GO:0005201 | Extracellular matrix structural constituent | 40 | 18 | 1 | 20 |
| 5 | GO:0004857 | Enzyme inhibitor activity | 128 | 28 | 19 | 3 |
| 6 | KEGG:01430 | Cell communication | 53 | 23 | 7 | 24 |
| 7 | GO:0016126 | Sterol biosynthesis | 22 | 27 | 11 | 31 |
| 8 | GO:0005605 | Basal lamina | 14 | 7 | 10 | 38 |
| 9 | GO:0007265 | Ras protein signal transduction | 29 | 34 | 33 | 15 |
| 10 | GO:0001817 | Regulation of cytokine production | 16 | 50 | 39 | 13 |
| 11 | GO:0042035 | Regulation of cytokine biosynthesis | 16 | 50 | 39 | 13 |
| 12 | GO:0009064 | Glutamine family amino acid metabolism | 23 | 65 | 48 | 12 |
| 13 | KEGG:00220 | Urea cycle and metabolism of amino groups | 18 | 72 | 44 | 17 |
| 14 | KEGG:00330 | Arginine and proline metabolism | 38 | 66 | 46 | 16 |
| 15 | GO:0007588 | Excretion | 16 | 63 | 22 | 44 |
| 16 | GO:0050954 | Sensory perception of mechanical stimulus | 39 | 31 | 74 | 7 |
| 17 | GO:0007605 | Sensory perception of sound | 39 | 31 | 74 | 7 |
| 18 | GO:0042089 | Cytokine biosynthesis | 17 | 47 | 62 | 22 |
| 19 | GO:0043062 | Extracellular structure organization and biogenesis | 11 | 9 | 8 | 81 |
| 20 | GO:0030198 | Extracellular matrix organization and biogenesis | 11 | 9 | 8 | 81 |
| 21 | GO:0007059 | Chromosome segregation | 25 | 12 | 9 | 81 |
| 22 | GO:0006937 | Regulation of muscle contraction | 17 | 24 | 14 | 81 |
| 23 | KEGG:04060 | Cytokine-cytokine receptor interaction | 79 | 30 | 21 | 81 |
| 24 | GO:0005578 | Extracellular matrix (sensu Metazoa) | 102 | 21 | 2 | 105 |
| 25 | GO:0031012 | Extracellular matrix | 102 | 21 | 2 | 105 |
| 26 | GO:0005581 | Collagen | 15 | 13 | 4 | 108 |
| 27 | GO:0043161 | Proteasomal ubiquitin-dependent protein catabolism | 11 | 18 | 108.5 | 4 |
| 28 | GO:0042087 | Cell-mediated immune response | 11 | 45 | 108.5 | 5 |
| 29 | GO:0042088 | T-helper 1 type immune response | 11 | 45 | 108.5 | 5 |
| 30 | BioCarta | ALK in cardiac myocytes | 14 | 21 | 108.5 | 6 |
SigPathway results: PCNSL versus nodal DLBCL
| . | Gene set category . | Pathway . | Set size . | % up . | NTk rank . | NEk rank . |
|---|---|---|---|---|---|---|
| 1 | KEGG:00330 | Arginine and proline metabolism | 38 | 68 | 11 | 2 |
| 2 | GO:0016066 | Cellular defense response (sensu Vertebrata) | 12 | 75 | 10 | 6 |
| 3 | KEGG:04512 | ECM-receptor interaction | 47 | 19 | 3 | 16 |
| 4 | GO:0042087 | Cell-mediated immune response | 11 | 73 | 16 | 3 |
| 5 | GO:0042088 | T-helper 1 type immune response | 11 | 73 | 16 | 3 |
| 6 | BioCyc | Fatty acid oxidation pathway | 14 | 71 | 17 | 10 |
| 7 | GO:0005604 | Basement membrane | 23 | 4 | 2 | 28 |
| 8 | KEGG:04510 | Focal adhesion | 135 | 38 | 7 | 25 |
| 9 | GO:0016126 | Sterol biosynthesis | 22 | 23 | 30 | 8 |
| 10 | GO:0001817 | Regulation of cytokine production | 16 | 75 | 20 | 26 |
| 11 | GO:0019888 | Protein phosphatase regulator activity | 32 | 22 | 36 | 12 |
| 12 | GO:0050954 | Sensory perception of mechanical stimulus | 39 | 38 | 48 | 4 |
| 13 | GO:0007605 | Sensory perception of sound | 39 | 38 | 48 | 4 |
| 14 | GO:0019208 | Phosphatase regulator activity | 33 | 24 | 54 | 9 |
| 15 | GO:0001816 | Cytokine production | 17 | 76 | 18 | 48 |
| 16 | GO:0042089 | Cytokine biosynthesis | 17 | 76 | 18 | 48 |
| 17 | GO:0042995 | Cell projection | 36 | 19 | 47 | 19 |
| 18 | GO:0042157 | Lipoprotein metabolism | 25 | 68 | 68 | 5 |
| 19 | GO:0016836 | Hydrolyase activity | 29 | 76 | 6 | 77 |
| 20 | KEGG:04662 | B-cell receptor signaling pathway | 45 | 73 | 9 | 80 |
| 21 | GO:0005201 | Extracellular matrix structural constituent | 40 | 23 | 4 | 93 |
| 22 | GO:0007588 | Excretion | 16 | 69 | 96 | 7 |
| 23 | GO:0042158 | Lipoprotein biosynthesis | 16 | 75 | 96 | 14 |
| 24 | GO:0006497 | Protein amino acid lipidation | 16 | 75 | 96 | 14 |
| 25 | GO:0007409 | Axonogenesis | 18 | 61 | 96 | 15 |
| 26 | GO:0050900 | Immune cell migration | 10 | 60 | 96 | 17 |
| 27 | GO:0030595 | Immune cell chemotaxis | 10 | 60 | 96 | 17 |
| 28 | GO:0048468 | Cell development | 34 | 59 | 96 | 18 |
| 29 | GO:0006631 | Fatty acid metabolism | 80 | 60 | 5 | 124 |
| 30 | GO:0005605 | Basal lamina | 14 | 0 | 12 | 124 |
| . | Gene set category . | Pathway . | Set size . | % up . | NTk rank . | NEk rank . |
|---|---|---|---|---|---|---|
| 1 | KEGG:00330 | Arginine and proline metabolism | 38 | 68 | 11 | 2 |
| 2 | GO:0016066 | Cellular defense response (sensu Vertebrata) | 12 | 75 | 10 | 6 |
| 3 | KEGG:04512 | ECM-receptor interaction | 47 | 19 | 3 | 16 |
| 4 | GO:0042087 | Cell-mediated immune response | 11 | 73 | 16 | 3 |
| 5 | GO:0042088 | T-helper 1 type immune response | 11 | 73 | 16 | 3 |
| 6 | BioCyc | Fatty acid oxidation pathway | 14 | 71 | 17 | 10 |
| 7 | GO:0005604 | Basement membrane | 23 | 4 | 2 | 28 |
| 8 | KEGG:04510 | Focal adhesion | 135 | 38 | 7 | 25 |
| 9 | GO:0016126 | Sterol biosynthesis | 22 | 23 | 30 | 8 |
| 10 | GO:0001817 | Regulation of cytokine production | 16 | 75 | 20 | 26 |
| 11 | GO:0019888 | Protein phosphatase regulator activity | 32 | 22 | 36 | 12 |
| 12 | GO:0050954 | Sensory perception of mechanical stimulus | 39 | 38 | 48 | 4 |
| 13 | GO:0007605 | Sensory perception of sound | 39 | 38 | 48 | 4 |
| 14 | GO:0019208 | Phosphatase regulator activity | 33 | 24 | 54 | 9 |
| 15 | GO:0001816 | Cytokine production | 17 | 76 | 18 | 48 |
| 16 | GO:0042089 | Cytokine biosynthesis | 17 | 76 | 18 | 48 |
| 17 | GO:0042995 | Cell projection | 36 | 19 | 47 | 19 |
| 18 | GO:0042157 | Lipoprotein metabolism | 25 | 68 | 68 | 5 |
| 19 | GO:0016836 | Hydrolyase activity | 29 | 76 | 6 | 77 |
| 20 | KEGG:04662 | B-cell receptor signaling pathway | 45 | 73 | 9 | 80 |
| 21 | GO:0005201 | Extracellular matrix structural constituent | 40 | 23 | 4 | 93 |
| 22 | GO:0007588 | Excretion | 16 | 69 | 96 | 7 |
| 23 | GO:0042158 | Lipoprotein biosynthesis | 16 | 75 | 96 | 14 |
| 24 | GO:0006497 | Protein amino acid lipidation | 16 | 75 | 96 | 14 |
| 25 | GO:0007409 | Axonogenesis | 18 | 61 | 96 | 15 |
| 26 | GO:0050900 | Immune cell migration | 10 | 60 | 96 | 17 |
| 27 | GO:0030595 | Immune cell chemotaxis | 10 | 60 | 96 | 17 |
| 28 | GO:0048468 | Cell development | 34 | 59 | 96 | 18 |
| 29 | GO:0006631 | Fatty acid metabolism | 80 | 60 | 5 | 124 |
| 30 | GO:0005605 | Basal lamina | 14 | 0 | 12 | 124 |
SigPathway results: PCNSL versus extranodal DLBCL
| . | Gene set category . | Pathway . | Set size . | % up . | NTk rank . | NEk rank . |
|---|---|---|---|---|---|---|
| 1 | GO:0004857 | Enzyme inhibitor activity | 128 | 30 | 14 | 2 |
| 2 | KEGG:01430 | Cell communication | 53 | 28 | 5 | 15 |
| 3 | KEGG:04510 | Focal adhesion | 135 | 33 | 7 | 19 |
| 4 | KEGG:04512 | ECM-receptor interaction | 47 | 21 | 3 | 27 |
| 5 | GO:0031497 | Chromatin assembly | 54 | 20 | 13 | 18 |
| 6 | KEGG:04210 | Apoptosis | 51 | 29 | 15 | 16 |
| 7 | GO:0000785 | Chromatin | 94 | 32 | 12 | 21 |
| 8 | GO:0005604 | Basement membrane | 23 | 17 | 8 | 26 |
| 9 | GO:0006695 | Cholesterol biosynthesis | 15 | 27 | 32 | 8 |
| 10 | GO:0007059 | Chromosome segregation | 25 | 12 | 6 | 38 |
| 11 | GO:0043161 | Proteasomal ubiquitin-dependent protein catabolism | 11 | 9 | 38 | 9 |
| 12 | GO:0001501 | Skeletal development | 47 | 23 | 16 | 32 |
| 13 | GO:0006334 | Nucleosome assembly | 52 | 19 | 17 | 40 |
| 14 | BioCarta | Ceramide signaling pathway | 14 | 21 | 49 | 13 |
| 15 | GO:0043062 | Extracellular structure organization and biogenesis | 11 | 9 | 9 | 56 |
| 16 | GO:0030198 | Extracellular matrix organization and biogenesis | 11 | 9 | 9 | 56 |
| 17 | BioCarta | Caspase cascade in apoptosis | 17 | 29 | 63 | 11 |
| 18 | GO:0016126 | Sterol biosynthesis | 22 | 32 | 73 | 10 |
| 19 | KEGG:00220 | Urea cycle and metabolism of amino groups | 18 | 56 | 72 | 20 |
| 20 | GO:0005201 | Extracellular matrix structural constituent | 40 | 23 | 1 | 116 |
| 21 | GO:0005694 | Chromosome | 190 | 35 | 10 | 110 |
| 22 | GO:0005581 | Collagen | 15 | 13 | 4 | 137 |
| 23 | GO:0016810 | Hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | 38 | 61 | 138.5 | 5 |
| 24 | GO:0000786 | Nucleosome | 39 | 18 | 21 | 137 |
| 25 | GO:0009064 | Glutamine family amino acid metabolism | 23 | 65 | 138.5 | 22 |
| 26 | GO:0005605 | Basal lamina | 14 | 14 | 19 | 166 |
| 27 | GO:0006928 | Cell motility | 130 | 38 | 18 | 176 |
| 28 | GO:0040011 | Locomotion | 130 | 38 | 18 | 176 |
| 29 | GO:0051674 | Localization of cell | 130 | 38 | 18 | 176 |
| 30 | GO:0005578 | Extracellular matrix (sensu Metazoa) | 102 | 24 | 2 | 212.5 |
| . | Gene set category . | Pathway . | Set size . | % up . | NTk rank . | NEk rank . |
|---|---|---|---|---|---|---|
| 1 | GO:0004857 | Enzyme inhibitor activity | 128 | 30 | 14 | 2 |
| 2 | KEGG:01430 | Cell communication | 53 | 28 | 5 | 15 |
| 3 | KEGG:04510 | Focal adhesion | 135 | 33 | 7 | 19 |
| 4 | KEGG:04512 | ECM-receptor interaction | 47 | 21 | 3 | 27 |
| 5 | GO:0031497 | Chromatin assembly | 54 | 20 | 13 | 18 |
| 6 | KEGG:04210 | Apoptosis | 51 | 29 | 15 | 16 |
| 7 | GO:0000785 | Chromatin | 94 | 32 | 12 | 21 |
| 8 | GO:0005604 | Basement membrane | 23 | 17 | 8 | 26 |
| 9 | GO:0006695 | Cholesterol biosynthesis | 15 | 27 | 32 | 8 |
| 10 | GO:0007059 | Chromosome segregation | 25 | 12 | 6 | 38 |
| 11 | GO:0043161 | Proteasomal ubiquitin-dependent protein catabolism | 11 | 9 | 38 | 9 |
| 12 | GO:0001501 | Skeletal development | 47 | 23 | 16 | 32 |
| 13 | GO:0006334 | Nucleosome assembly | 52 | 19 | 17 | 40 |
| 14 | BioCarta | Ceramide signaling pathway | 14 | 21 | 49 | 13 |
| 15 | GO:0043062 | Extracellular structure organization and biogenesis | 11 | 9 | 9 | 56 |
| 16 | GO:0030198 | Extracellular matrix organization and biogenesis | 11 | 9 | 9 | 56 |
| 17 | BioCarta | Caspase cascade in apoptosis | 17 | 29 | 63 | 11 |
| 18 | GO:0016126 | Sterol biosynthesis | 22 | 32 | 73 | 10 |
| 19 | KEGG:00220 | Urea cycle and metabolism of amino groups | 18 | 56 | 72 | 20 |
| 20 | GO:0005201 | Extracellular matrix structural constituent | 40 | 23 | 1 | 116 |
| 21 | GO:0005694 | Chromosome | 190 | 35 | 10 | 110 |
| 22 | GO:0005581 | Collagen | 15 | 13 | 4 | 137 |
| 23 | GO:0016810 | Hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | 38 | 61 | 138.5 | 5 |
| 24 | GO:0000786 | Nucleosome | 39 | 18 | 21 | 137 |
| 25 | GO:0009064 | Glutamine family amino acid metabolism | 23 | 65 | 138.5 | 22 |
| 26 | GO:0005605 | Basal lamina | 14 | 14 | 19 | 166 |
| 27 | GO:0006928 | Cell motility | 130 | 38 | 18 | 176 |
| 28 | GO:0040011 | Locomotion | 130 | 38 | 18 | 176 |
| 29 | GO:0051674 | Localization of cell | 130 | 38 | 18 | 176 |
| 30 | GO:0005578 | Extracellular matrix (sensu Metazoa) | 102 | 24 | 2 | 212.5 |
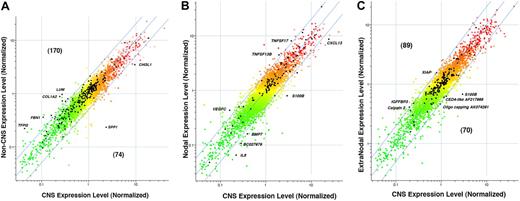
Examination of these 3 tables reveals that the PCNSL phenotype differentially expresses 2 major types of ontologic gene sets: one type that primarily sets apart the PCNSL phenotype from both nDLBCL and enDLBCL combined and other gene sets that differentiate PCNSL from each non-CNS group, either nDLBCL or enDLBCL separately. Gene sets of the first type will appear in all 3 tables. For example, in the PCNSL versus non-CNS contrast (Table 2), there are several gene sets that exhibit biologic associations with the ECM and adhesion: gene set 1 (ECM-receptor interaction, gene set 2 (basement membrane), gene set 3 (focal adhesion), gene set 4 (ECM structural constituent), gene set 8 (basal lamina), gene set 19 (extracellular structure organization and biogenesis), gene set 20 (ECM organization and biogenesis), gene set 24 (ECM [sensu Metazoa]), gene set 25 (ECM), gene set 26 (collagen), and gene set 33 (ECM/adhesion molecules). Nine of these 11 gene sets also appear in the PCNSL versus nDLBCL contrast (Table 3), whereas 10 of these 11 gene sets also appear in the PCNSL versus enDLBCL contrast (Table 4). After removing duplicate listings, there were 244 unique genes in these 11 gene sets listed from Table 2. Their expression levels are plotted (in black) overlying the approximately 11 500 total genes (shown in color) in the scatter-plot in Figure 3A. The normalized levels for non-CNS samples (n = 30) were averaged and plotted against averages of the normalized levels for the PCNSL samples (n = 13). Of these 244 ECM and adhesion-related genes (plotted in black), 170 lie above the line of equivalent expression, indicating relatively higher expression in non-CNS samples; 74 genes are expressed at higher levels in the PCNSL samples. Several of these ECM and adhesion-related genes are labeled in Figure 3A. Notably, SPP1 (secreted phosphoprotein 1, osteopontin; NM_000582) and CHI3L1 (chitinase-3 like 1, cartilage glycoprotein-39, ECM structural hydrolase; NM_001276) are expressed at much higher levels in PCNSL (9.7-fold and 2.7-fold, PCNSL > non-CNS, respectively). Several other ECM-related genes labeled in Figure 3A are expressed higher in the non-CNS samples: TFP12 (tissue factor pathway inhibitor 2; NM_006528; 5.1-fold non-CNS > PCNSL), FBN1 (fibrillin 1; NM_000138; 3.4-fold non-CNS > PCNSL), COL1A2 (collagen type 1 alpha2; NM_000089; 3.7-fold non-CNS > PCNSL), and LUM (lumican, collagen binding protein; NM_002345; 3.6-fold non-CNS > PCNSL). Differential expression of genes in sterol biosynthesis pathway also appears to be part of the CNS pathway signature as this pathway is found to be significant in all 3 contrasts.
Expression of selected gene sets. (A) Expression of a set of 244 ECM and adhesion-related genes that distinguish PCNSL from non-CNS DLBCL. LOWESS normalization was performed using genes present on at least 35 of the 43 arrays. The colored points are normalized gene ratios for these approximately 11 500 genes, whereas the black points are the ECM and adhesion-related genes. (B) Expression of a set of 92 cytokine genes that distinguish PCNSL and nodal DLBCL. LOWESS normalization was performed using genes present on at least 35 of the 43 arrays. The colored points are normalized gene ratios for these approximately 11 500 genes, whereas the black points are the cytokine-related genes. (C) Expression of a set of 159 apoptosis-related genes that distinguish PCNSL from extranodal DLBCL. LOWESS normalization was performed using genes present on at least 35 of the 43 arrays. The colored points are normalized gene ratios for these approximately 11 500 genes, whereas the black points are the apoptosis-related genes. The range of colors in these panels reflects range of gene levels in the CNS phenotype in panel A, or the Nodal and Extranodal phenotype in panels B and C, respectively. Specifically, the gene level refers to the ratio formed by dividing the gene level in the tumor by the level of the universal reference. Red indicates levels more than 1.0, whereas green indicates fractions less than 1.0.
Expression of selected gene sets. (A) Expression of a set of 244 ECM and adhesion-related genes that distinguish PCNSL from non-CNS DLBCL. LOWESS normalization was performed using genes present on at least 35 of the 43 arrays. The colored points are normalized gene ratios for these approximately 11 500 genes, whereas the black points are the ECM and adhesion-related genes. (B) Expression of a set of 92 cytokine genes that distinguish PCNSL and nodal DLBCL. LOWESS normalization was performed using genes present on at least 35 of the 43 arrays. The colored points are normalized gene ratios for these approximately 11 500 genes, whereas the black points are the cytokine-related genes. (C) Expression of a set of 159 apoptosis-related genes that distinguish PCNSL from extranodal DLBCL. LOWESS normalization was performed using genes present on at least 35 of the 43 arrays. The colored points are normalized gene ratios for these approximately 11 500 genes, whereas the black points are the apoptosis-related genes. The range of colors in these panels reflects range of gene levels in the CNS phenotype in panel A, or the Nodal and Extranodal phenotype in panels B and C, respectively. Specifically, the gene level refers to the ratio formed by dividing the gene level in the tumor by the level of the universal reference. Red indicates levels more than 1.0, whereas green indicates fractions less than 1.0.
Other groupings of ontologic gene sets differentiate the PCNSL from either enDLBCL or nDLBCL separately. For example, The gene sets 10, 15, and 16 in Table 3, where CNS and nDLBCL are contrasted, are involved in cytokine production; these groups do not appear in Table 4, where CNS and enDLBCL are contrasted. Only gene set 41 in Table 4 is linked to cytokines, and it concerns receptor functions, not cytokine production. There are 92 unique genes in gene sets 10, 15, and 16 combined; they are plotted in black overlying all approximately 11 500 genes used in the analysis (plotted in color) in Figure 3B. Several individual genes in this composite are labeled: S-100B, IRF4, CXCL13, BMP7, BC 027979, and IL8 are elevated 2-fold or more in the PCNSL; TNFSF17, TNFSF13B, and VEGFC are elevated in nDLBCL at least 2-fold with respect to CNS samples. Another grouping of gene sets in Table 3 concerns the immunologic functions and responses of T cells and B cells: gene sets 4, 5, 20, 26, 27, and 41. None of the gene sets appear in the contrast between CNS and EN (Table 4). The contrast PCNSL versus nDLBCL also showed significant differential expression of many metabolic pathways. Of these, lipoprotein-related pathways are absent in PCNSL versus enDLBCL contrast.
The PCNSL versus enDLBCL contrast (Table 4) exhibits several gene sets associated with apoptosis (6, 17, 32, 33, 34, 35) that do not appear in the PCNSL versus nDLBCL contrast. These 7 gene sets contain 159 unique genes, which are plotted (in black) overlying all approximately 11 500 genes in the basis (color) in Figure 3C. Several genes of interest in this composite are labeled: S-100B, AF 217966 (CED4-like death effector filament forming), AK074291 (Oligo capping), which are relatively higher in expression in the PCNSL; U45880 (X-linked inhibitor of apoptosis protein, XIAP), S56204 (insulin-like growth factor binding protein 3), and CAPN2 (calpain type 2), which are expressed 2-fold or more higher in the enDLBCL relative to PCNSL. Moreover, certain chromatin and chromosome-related pathways (5, 7, 10, 21, 24) showed significant differential expression between PCNSL and enDLBCL. They are conspicuously absent from PCNSL versus nDLBCL contrast. The contrast results for PCNSL versus enDLBCL also reveal that certain aspects of amine metabolism separate these 2 phenotypes (gene sets 19 and 25).
FDA of the DLBCL gene expression dataset
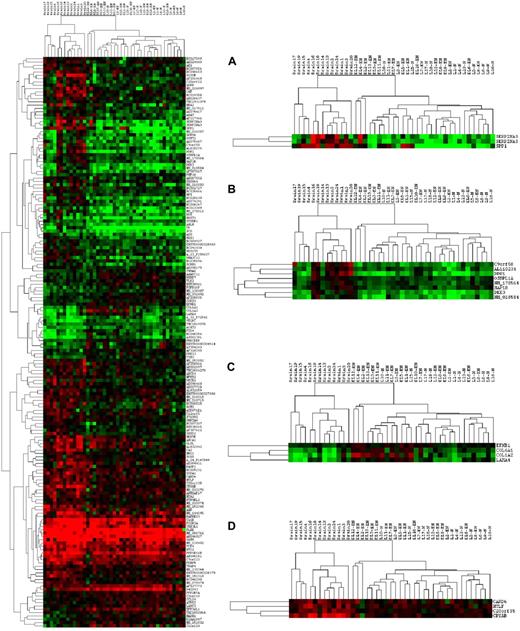
When FDA was applied to the discrimination of 2 classes, CNS from non-CNS, with the criterion of P less than .01 reliability, 172 genes were identified, and these included 7 of the ECM and adhesion-related group, 3 of the cytokine group, and 4 of the apoptosis group. Clustering (using cluster 3.0) with these 172 genes completely separated the CNS samples from the non-CNS samples (Figure 4; the ratio data were log-transformed and clustered, with cluster 3.0 using uncentered correlation and complete linkage. The right-hand portion of Figure 4 shows enlarged views of several gene clusters of interest, including SPP1 (Figure 4A), DDR1 and DKK1 (Figure 4B), a group of ECM-related genes COL6A1, COL1A2, and LAM4 (Figure 4C), and a cluster containing CRYAB (Figure 4D). Careful examination of the main clustering results reveals patterns of expression that separate most of the samples according to the 3 main phenotypes (“brain” [CNS], nodal, and extranodal), as well as patterns that subdivide the CNS phenotype into 2 subclasses.
Clustering results using FDA genes separating 2 classes: CNS versus non-CNS. The left-hand plot shows the complete tree, whereas 4 regions within the tree are shown at right. Shades of red indicate ratios more than 1.0; shades of green indicate ratios less than 1.0; black indicates a ratio of 1.0.
Clustering results using FDA genes separating 2 classes: CNS versus non-CNS. The left-hand plot shows the complete tree, whereas 4 regions within the tree are shown at right. Shades of red indicate ratios more than 1.0; shades of green indicate ratios less than 1.0; black indicates a ratio of 1.0.
When FDA was performed to identify genes that separated 3 classes (CNS, nodal, extranodal) at P less than .01, 144 genes were identified. The gene list was similar to that from the 2-class analysis and contained many of the genes in Table 1 and in the SigPathway analyses (not shown). In this analysis, the CNS phenotypes were separated into 2 adjacent groups, one of which contained only CNS samples (n = 8) and the other of which contained the other 5 CNS samples and one EN sample (not shown).
Immunohistochemical studies of DLBCL
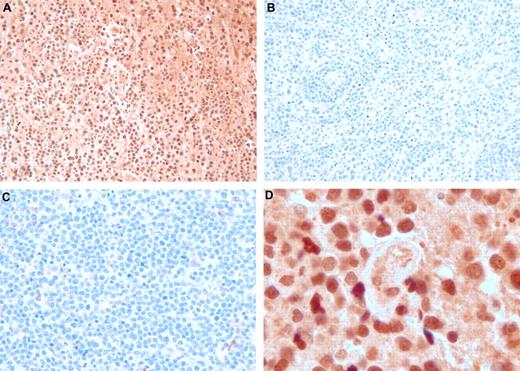
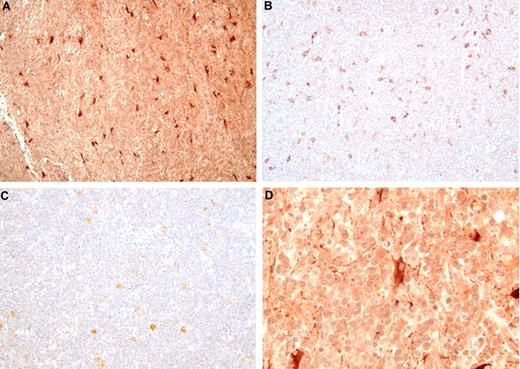
Two genes that were strongly and reliably up-regulated in PCNSL were selected for IHC. Expression of osteopontin (SPP1) and chitinase-3 like1 (CHI3L1) proteins was studied in DLBCL samples from PCNSL (n = 15, which made up the 5 used in the microarray study plus 10 additional), nDLBCL (n = 10 with 5 samples used in the microarray study plus 5 additional), and enDLBCL (n = 7 with 5 samples used in the microarray study plus 2 additional). Figure 5 shows examples of osteopontin IHC from PCNSL (Figure 5A,D), nDLBCL (Figure 5B), and enDLBCL (skin, Figure 5C). The 2 views of the PCNSL sample show that most of the tumor cells express moderate to high levels of osteopontin, whereas tumor cells from the 2 samples of non-CNS DLBCL contain little or no osteopontin. The staining pattern for SPP1 was predominantly nuclear, but cytoplasmic staining was also seen. At least some positive staining for SPP1 was seen in 100% of PCNSL and 80% of non-CNS DLBCL. Heavy staining was present in 92% of PCNSL and 26% of non-CNS DLBCL. Figure 6 contains images of CHI3L1 IHC in PCNSL (Figure 6A,D), nDLBCL (Figure 6B), and enDLBCL (spleen, Figure 6C). The 2 views of the PCNSL sample show that most tumor cells express moderate levels of CHI3L1 immunoreactivity with some heavy staining of astrocyte-like cells; the nodal and extranodal sample exhibit some moderate immunoreactivity in nontumor cells (probably macrophages). The staining pattern for CHI3L1 was both cytoplasmic and nuclear, but the nuclear staining was more predominant. Positive staining for CHI3L1 was seen in 73% of PCNSL and 41% of non-CNS DLBCL. Heavy staining was present in 40% of PCNSL and 18% of non-CNS DLBCL.
Osteopontin immunohistochemistry in DLBCL. The immunoperoxidase complexes were visualized with diaminobenzidine (brown), and the sections were counterstained with hematoxylin. (A) PCNSL: original magnification ×200. Nearly every tumor cell of this brain biopsy is immunoreactive. (B) Nodal DLBCL: original magnification ×200. Essentially no tumor cell contains immunoreactivity. (C) Extranodal DLBCL (skin): original magnification ×200. Essentially no tumor cell contains immunoreactivity. (D) PCNSL: original magnification ×1000 oil. Cross section of a small vessel, probably a vein, surrounded by osteopontin-positive tumor cells.
Osteopontin immunohistochemistry in DLBCL. The immunoperoxidase complexes were visualized with diaminobenzidine (brown), and the sections were counterstained with hematoxylin. (A) PCNSL: original magnification ×200. Nearly every tumor cell of this brain biopsy is immunoreactive. (B) Nodal DLBCL: original magnification ×200. Essentially no tumor cell contains immunoreactivity. (C) Extranodal DLBCL (skin): original magnification ×200. Essentially no tumor cell contains immunoreactivity. (D) PCNSL: original magnification ×1000 oil. Cross section of a small vessel, probably a vein, surrounded by osteopontin-positive tumor cells.
Chitinase-3-like 1 immunohistochemistry in DLBCL. The immunoperoxidase complexes were visualized with diaminobenzidine (brown), and the sections were counterstained with hematoxylin. (A) PCNSL: original magnification ×200. Most tumor cells express moderate levels of CHI3L1 with a minority expressing strong levels. The largest profiles with heavy immunoreactivity are possibly astrocytes (see also panel D). (B) Nodal DLBCL: original magnification ×200. Most of the tumor cells contain low levels of immunoreactivity. The larger, strongly positive cells may be macrophages. (C) Extranodal DLBCL (spleen): original magnification ×200. (D) PCNSL: original magnification ×400. This is a higher power view of the PCNSL shown in panel A, showing astrocyte-like profiles with moderate to strong levels of immunoreactivity.
Chitinase-3-like 1 immunohistochemistry in DLBCL. The immunoperoxidase complexes were visualized with diaminobenzidine (brown), and the sections were counterstained with hematoxylin. (A) PCNSL: original magnification ×200. Most tumor cells express moderate levels of CHI3L1 with a minority expressing strong levels. The largest profiles with heavy immunoreactivity are possibly astrocytes (see also panel D). (B) Nodal DLBCL: original magnification ×200. Most of the tumor cells contain low levels of immunoreactivity. The larger, strongly positive cells may be macrophages. (C) Extranodal DLBCL (spleen): original magnification ×200. (D) PCNSL: original magnification ×400. This is a higher power view of the PCNSL shown in panel A, showing astrocyte-like profiles with moderate to strong levels of immunoreactivity.
Discussion
We have identified alterations in gene expression signature of DLBCL that correlate with anatomic locations, using a statistically powerful method, SigPathway, which makes use of the fact that many, if not most, genes are coregulated according to activation or repression of particular pathways. Most important in the present study was the finding that the PCNSL expresses a unique set of ECM and adhesion-related pathways and genes when contrasted with non-CNS DLBCL. This “CNS signature” for PCNSL was readily attained by contrasting PCNSL with the all the non-CNS DLBCL combined (nDLBCL + enDLBCL). Similar findings were also seen when PCNSL was contrasted with either nDLBCL or enDLBCL, further indicating that these findings are uniquely important for PCNSL. The most significant gene set found in pathway analysis was the ECM-receptor pathway, suggesting that the interaction between the CNS microenvironment and lymphoma cells is of great importance for PCNSL. At the single gene expression level, we also found significant, differential expression of numerous ECM and adhesion-related genes. In addition, we demonstrated the up-regulation in PCNSL of 2 important ECM-related genes, SPP1 and CHI3L1, at the protein level.
The differential expression of ECM-related genes, especially adhesion genes, has been long suspected in PCNSL but has not been proven in previous studies.14-16 The reason for this may be that most of the genes in these pathways do not differentially express at a statistically significant level when tested individually. However, when they are analyzed in groups by SigPathway, many ECM-related pathways, including focal adhesion pathways, are found to be significantly implicated in PCNSL biology. Thus, the SigPathway results demonstrate the power of pathway analysis to infer statistically reliable biologic mechanisms in DLBCL, in particular indicating the existence of a unique expression profile for PCNSL. The results of the FDA analyses also provide support for the idea of a unique PCNSL signature. FDA is a matrix decomposition method that identifies those genes whose composite expression patterns are most powerful in distinguishing the phenotypes, and is mathematically very different from the methods of SigPathway. From the basis dataset of approximately 11 500 genes, subsets of fewer than 200 genes were identified by FDA that have expression patterns that can completely, or nearly completely, classify a DLBCL sample according to one of either 2 or 3 phenotypes. Many of the genes identified by FDA were also present in the SigPathway results.
By several measures, the most significant up-regulated gene in PCNSL is an ECM-related gene, SPP1 (osteopontin; OPN). The SigPathway results implicate SPP1 in numerous cellular functions, including cell communication, focal adhesion, immune cell activation, and immune cell migration. This multiplicity of function is consistent with the literature, which has shown SPP1 involvement in various aspects of cancer biology, including cellular proliferation, invasion, metastasis, and regulation of cytokine expression and angiogenesis.17,18 A high level of expression of SPP1 has been associated with aggressive cancers and poor prognosis.17 Our immunohistochemical finding of predominantly nuclear staining pattern in PCNSL cells is quite unique, as SPP1 staining is usually cytoplasmic in other malignant tumors.19 The nuclear localization of SPP1 has been linked to cellular proliferation.20 SPP1 has been found to be up-regulated in other CNS diseases, such as multiple sclerosis,21 and glioblastoma multiforme, and astrocytomas.17 It appears that SPP1 plays an important role in pathogenesis of CNS diseases. To our knowledge, this is the first report of significant up-regulation of SPP1 in PCNSL. It is noteworthy that SPP1 has not been previously reported to be expressed significantly in B cells.
Our IHC experiments show that CHI3L1 expression is higher in PCNSL compared with non-CNS DLBCL. CHI3L1 (YKL-40) is an ECM-related gene widely implicated in the biology of several types of cancer. It has a role in cancer cell proliferation, differentiation, survival, invasiveness, metastasis, angiogenesis, and remodeling of ECM surrounding the tumor.22 Highest serum levels of CHI3L1 are found in patients with metastatic cancer with the shortest recurrence-free interval and shortest overall survival.22 The presence of immunohistochemically detected CHI3L1 in breast cancer is associated with a poor prognosis.23 Other significantly up-regulated ECM/adhesion genes in our study include DDR1 and TACSTD1 (EpCAM). DDR1 is a member of a novel family of receptor tyrosine kinases thought to play a role in cell adhesion.24 It has been shown to be consistently and selectively expressed in human brain tumors.25 TACSTD1 (EpCAM) is a cell adhesion molecule expressed by a variety of carcinomas.26 It is also expressed in normal retina and retinoblastoma.27
Several previous studies of PCNSL are relevant to other findings in our experiments. Up-regulated CXCL13, a B cell attracting–chemokine, has been shown previously to occur in PCNSL.28 We also found that RGS13, which regulates germinal center B-cell responsiveness to CXCL13,29 is up-regulated. Another up-regulated gene, MUM1, a marker of ABC subtype of nDLBCL, has been reported as expressed in more than 90% of PCNSL.5
Other up-regulated genes in our dataset have been implicated previously in cancer biology. TCL1A has been implicated in lymphatic leukemias and lymphomas.30 CRYAB has been found to be up-regulated in cancers and correlated with risk of cancer recurrence.31,32 DKK3 has been implicated in cancer, although its exact role has not been clarified.33 We found that MGST1, one of the glutathione S-transferases, is up-regulated in PCNSL. Glutathione S-transferases have been implicated in lymphomas34 and chemotherapy resistance.35 S-100B has been implicated in neoplasia, especially in melanoma.36,37 TF was the second most up-regulated gene for PCNSL. It has been shown to act as an autocrine regulator of cellular proliferation.38 The receptor for transferrin is frequently expressed in non-Hodgkin lymphomas.39
We have interpreted gene expression alterations within the context of signaling or regulatory pathways to develop hypotheses of biologic mechanisms in DLBCL. In doing so, it is important to keep in mind that the tumors are heterogeneous. In PCNSL samples, malignant B cells are admixed with infiltrating immune cells and cells from the CNS microenvironment. These non-B cells may express some of the implicated genes or indirectly influence B-cell gene expression. Thus, it remains possible that some cell type other than a B cell is the site of expression of a given gene of interest. It is our opinion that the contribution of surrounding tissue is probably minor but that the presence of non-B cells may in some instances be functionally relevant to some aspects of the expression profiles. Sorting out complexities like these will require extensive follow-up experiments with immunohistochemical procedures and microdissection combined with microarray or quantitative RT-PCR approaches.
We think that our findings have significant biologic relevance for lymphoma research and development of novel treatments. The findings indicate that the CNS microenvironment is of great importance for PCNSL. The ECM and adhesion-related pathways may determine some of the biologic characteristics of PCNSL, such as CNS tropism. Individual genes discovered in our study may have roles in different aspects of the biology of PCNSL. SPP1 and DDR1 may play a role in CNS tropism of PCNSL. CXCL13 and SPP1 are probably relevant to the B-cell migration involved in the pathogenesis of PCNSL. B-cell proliferation may be associated with increased expression of SPP1, TCL1A, and CHI3L1. Elevated MUM1 expression indicates that PCNSL is of activated B-cell subtype as previously reported. The coordinate up-regulation of SPP1, CHI3L1, and MUM1 is consistent with the known aggressive clinical behavior of PCNSL. SPP1 and CHI3L1 have been associated with aggressive metastatic cancers, suggesting that PCNSL has an aggressive metastatic cancer phenotype. Because our approach contrasted PCNSL with a wide spectrum of non-CNS DLBCL on a genomic scale with in-depth bioinformatic analysis, the gene expression signature identified in our study may represent a true “CNS signature.”
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Kathleen Roberson for expert secretarial help.
This work was supported by the Mayo Foundation (research program, M.M.; internal grant, H.T.), the University of Iowa/Mayo Clinic Lymphoma Specialized Programs of Research Excellence (SPORE; P50 CA97274), the Mayo SPORE in Brain Cancer (P50 CA108961), and the Immunochemistry Core at the Mayo Clinic Jacksonville (MCJ) Cancer Center, a National Cancer Institute-designated Comprehensive Cancer Center (P30 CA15083).
National Institutes of Health
Authorship
Contribution: H.W.T. designed the study, performed the statistical analysis and interpretation, and wrote the manuscript. D.P. and K.A.B. performed bench work. D.M.M. performed a pathology review. K.A.J. obtained samples. P.K., B.E., and A.C.Z. performed the immunohistochemistry work. B.P.O. obtained samples. W.R.L. and P.J.P. performed statistical analysis and interpretation. M.M. designed the study, performed the statistical analysis and interpretation, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Han W. Tun, Department of Hematology and Oncology, Mayo Clinic Jacksonville, 4500 San Pablo Road, Jacksonville, FL 32224; e-mail: Tun.Han@mayo.edu; or Michael McKinney, Department of Molecular Pharmacology and Therapeutics, 4500 San Pablo Road, Jacksonville, FL 32224; e-mail: mckinney@mayo.edu.