Human T-cell leukemia virus type 1 (HTLV-1) basic leucine zipper factor (HBZ) is dispensable for HTLV-1–mediated cellular transformation in cell culture, but is required for efficient viral infectivity and persistence in rabbits. In most adult T-cell leukemia (ATL) cells, Tax oncoprotein expression is typically low or undetectable, whereas Hbz gene expression is maintained, suggesting that Hbz expression may support infected cell survival and, ultimately, leukemogenesis. Emerging data indicate that HBZ protein can interact with cAMP response element binding protein (CREB) and Jun family members, altering transcription factor binding and transactivation of both viral and cellular promoters. Herein, lentiviral vectors that express Hbz-specific short hairpin (sh)–RNA effectively decreased both Hbz mRNA and HBZ protein expression in transduced HTLV-1–transformed SLB-1 T cells. Hbz knockdown correlated with a significant decrease in T-cell proliferation in culture. Both SLB-1 and SLB-1-Hbz knockdown cells engrafted into inoculated NOD/SCIDγchain−/− mice to form solid tumors that also infiltrated multiple tissues. However, tumor formation and organ infiltration were significantly decreased in animals challenged with SLB-1-Hbz knockdown cells. Our data indicate that Hbz expression enhances the proliferative capacity of HTLV-1–infected T cells, playing a critical role in cell survival and ultimately HTLV-1 tumorigenesis in the infected host.
Introduction
Human T-cell leukemia virus type 1 (HTLV-1) infection causes adult T-cell leukemia (ATL) and is associated with a variety of lymphocyte-mediated disorders.1 Although HTLV-1 can immortalize and ultimately transform primary human T lymphocytes efficiently in culture, disease occurs in only 2% to 5% of infected patients after an extensive latency period. After infection of the T lymphocyte, ATL is thought to arise from a multitude of factors that include both genetic and epigenetic changes in the cell that accumulate over time.2 Although many aspects of HTLV-1 biology have been elucidated, the detailed mechanisms of ATL development remain ill-defined
HTLV-1 is a highly cell-associated virus, and efficient infection requires direct cell-to-cell contact.3 In infected patients, increases in viral progeny occur primarily by clonal expansion of infected T cells and are dependent on the action of the positive regulatory (tax and rex) and accessory genes (ORF I and ORF II) encoded on the plus strand of the genome.2 Although the precise mechanism remains unclear, the accessory gene products (p12, p13, and p30) are dispensable in standard culture assays of virus-mediated cellular transformation, but are required for viral infectivity and persistence in vivo.4,,–7 Tax is the viral transcriptional transactivator that also activates many cellular signaling pathways including nuclear factor kappa-B (NF-κB), cAMP response element binding protein (CREB), activator protein-1 (AP-1), and serum responsive factor (SRF).8,9 In addition, Tax has been shown to inactivate p53, resulting in inhibition of apoptosis. Tax can activate cyclin-dependent kinases, CDK-4 and CDK-6, through direct protein binding leading to the hyperphosphorylation and degradation of retinoblastoma (RB), which then frees the E2F1 transcription factor, thereby accelerating cell-cycle transition from G1 to S.10,–12 Compelling experimental evidence suggests that these pleiotropic effects of Tax on cell biology are required for the transforming or oncogenic capacity of HTLV-1 and thus are critical in the early stage of the malignant process.13,,,–17 However, less clear is the requirement of tax gene expression and ATL. Although most leukemic cells isolated from patients maintain the 3′ portion of the HTLV-1 genome, Tax typically is not expressed, which has been attributed to a deletion of the 5′ LTR/promoter or to gene inactivation by mutations or DNA methylation.18,,,–22 Thus, the loss of Tax expression actually may enable ATL cells to evade host immune surveillance because Tax elicits a strong CTL response in vivo and Tax-expressing cells would rapidly be targeted for elimination.23
The HTLV-1 basic leucine zipper factor gene (Hbz) is located in the 3′ portion of the provirus and is maintained in ATL cells.24,25 Unlike the other HTLV-1 genes, Hbz is encoded in the minus or antisense strand of the genome.24,26 Both spliced and unspliced Hbz transcripts with the potential to encode highly related protein isoforms have been detected in most ATL cells.25,27,28 Exogenously over-expressed HBZ interacts with CREB-2 to down-regulate Tax-mediated HTLV-1 transcription and also interacts with and disrupts the DNA binding activity of JunB and c-Jun (AP-1 components).26,29,30 HBZ also interacts with JunD to activate the transcription of JunD-dependent promoters.31 One recent mutational study provided additional evidence that the Hbz mRNA, not HBZ protein, had the capacity to induce T-cell proliferation by up-regulating cellular E2F1,25 which has lead to the novel hypothesis that Hbz is functional in 2 molecular forms (RNA and protein) and plays an important role in HTLV-1 biology, promoting cell survival by counteracting the effects of Tax-mediated transcription and/or attenuating or activating cellular gene expression. Consistent with this hypothesis, previous studies from our laboratory using an HBZ-deficient infectious molecular clone indicated that HBZ is dispensable for immortalization/transformation of primary T lymphocytes in cell culture, but in a rabbit animal model viruses that produced a severely truncated HBZ were attenuated for viral infectivity, replication, and persistence.32
In this study, we generated and used a panel of lentiviral vectors that express Hbz-specific short hairpin (sh)–RNAs to determine the role of Hbz in T-cell proliferation in vitro and tumor growth in the NOD/SCIDγc−/− (NOG) transplant mouse model.33,–35 We show that Hbz-specific shRNAs effectively decreased both Hbz mRNA and HBZ protein expression, which correlated with significantly decreased cellular proliferation of an established HTLV-1 T-cell line SLB-1, the HTLV-1 leukemic cell line MT-1, and newly HTLV-1–immortalized IL-2–dependent T lymphocytes. Importantly, we show directly that global viral mRNA and protein levels in vector-transduced cells were unaffected by Hbz-specific shRNA expression. In NOG mice, wild-type and Hbz-knockdown SLB-1 cells engrafted to form solid tumor masses that infiltrated multiple organs, but tumor formation and tumor infiltration were reduced significantly in animals challenged with SLB-1 Hbz-knockdown cells. Taken together, our data are consistent with the conclusions that Hbz expression enhances the proliferative capacity of HTLV-1–infected cells and plays a critical role in cell survival and, ultimately, HTLV-1 tumorigenesis.
Methods
Cells
293T cells were maintained in Dulbecco modified Eagle medium (DMEM). SLB-1, 729ACH, MT-1, MT-2, and C8166 (HTLV-1–positive cell lines) and HTLV-1–negative Jurkat T cells were maintained in Iscove medium. All media were supplemented to contain 10% fetal bovine serum (FBS), 2 mM glutamine, penicillin (100 U/mL), and streptomycin (100 μg/mL). In vitro immortalized human peripheral blood T lymphocytes (PBLACH, PBLHTLV-1, and PBLHTLV-1ΔHBZ32 ) were grown in DMEM supplemented with recombinant human IL-2 at 10 U/mL.
Plasmids
The pME-based HBZ cDNA expression vector (based on the Hbz major transcript27 ) was described previously.32 The third-generation (pRNAT-U6.2/Lenti) lentiviral vector designed to generate short hairpin (sh) RNA from the U6.2 promoter and GFP from the cytomegalovirus (CMV) promoter was purchased from GenScript (Piscataway, NJ). The vector control with no insert was designated V1. The Hbz-specific and scrambled target sequences were cloned into the lentiviral vector using the unique restriction sites BamHI and XhoI. Vectors expressing Hbz transcript target sequences (see Figure 1 for location) were: V2, 5′ 7176AGGACAAGGAGGAGGAGG7159 3′; V3, 5′ 6839ACAGCATAGTGCTAGGAAA6821 3′; and V4, 5′ 8674CGGCCTCAG8666^7265GGCTGTTTCG7257 3′. Sequence and numbering are based on the positive sense nucleotide sequence of the ACH proviral clone.36 V2 and V3 target sequences were previously described (small interfering (si)–RNA4 and siRNA31, respectively25 ) and are expected to target all Hbz transcripts, but also have the potential for off-target effects on HTLV-1 sense strand mRNAs. The target sequence for V4 was designed specifically to span the splice junction and target the Hbz major transcript (Hbz-S1).27 V5 contains a scrambled nucleotide sequence designed to form a nonspecific hairpin shRNA and not target Hbz, other HTLV-1 genes, or cellular genes.
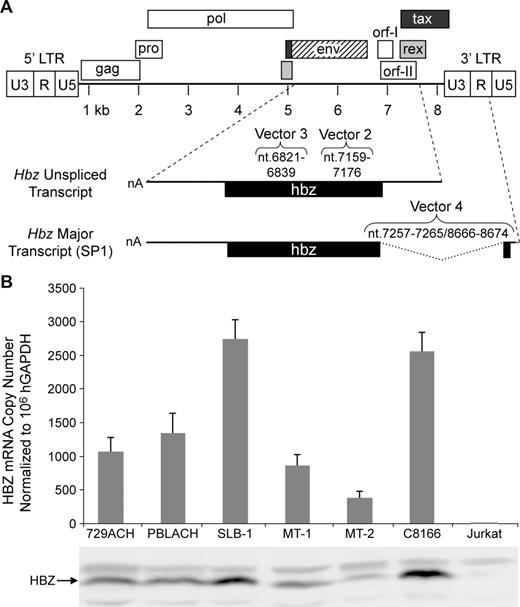
HTLV-1 genome and detection of Hbz mRNA and protein in HTLV-1 cell lines. (A) HTLV-1 proviral genome highlighting the HTLV-1 protein open reading frames (ORFs). Hbz including the unspliced mRNA transcript and ORF and the major spliced transcript (SP1) and ORF are detailed. Black line denotes mRNA and gray line in SP1 denotes multiple transcription start sites.27 The location of the Hbz short hairpin target sequences used in this study (Vector 2-4) relative to positive-strand proviral clone numbering is indicated above the Hbz transcript. (B) Total RNA was isolated from HTLV-1 cell lines and negative control Jurkat T cells and subjected to real-time Taqman RT-PCR to quantify the Hbz major spliced transcript (80%-90% of Hbz-specific mRNA27,32 ). Copy numbers of Hbz mRNA are normalized to 106 copies of the hGAPDH and the average values from 3 independent experiments are presented; error bars denote SD. Western blot analysis of HBZ protein expression (bottom).
HTLV-1 genome and detection of Hbz mRNA and protein in HTLV-1 cell lines. (A) HTLV-1 proviral genome highlighting the HTLV-1 protein open reading frames (ORFs). Hbz including the unspliced mRNA transcript and ORF and the major spliced transcript (SP1) and ORF are detailed. Black line denotes mRNA and gray line in SP1 denotes multiple transcription start sites.27 The location of the Hbz short hairpin target sequences used in this study (Vector 2-4) relative to positive-strand proviral clone numbering is indicated above the Hbz transcript. (B) Total RNA was isolated from HTLV-1 cell lines and negative control Jurkat T cells and subjected to real-time Taqman RT-PCR to quantify the Hbz major spliced transcript (80%-90% of Hbz-specific mRNA27,32 ). Copy numbers of Hbz mRNA are normalized to 106 copies of the hGAPDH and the average values from 3 independent experiments are presented; error bars denote SD. Western blot analysis of HBZ protein expression (bottom).
Transient transfection
To measure the ability of each lentiviral vector plasmid to knock down HBZ protein expression transiently, 1.5 × 105 293T cells were transfected using Lipofectamine reagent (Invitrogen, Carlsbad, CA). The amount of DNA was kept constant and was composed of 0.2 μg of Hbz cDNA expression plasmid along with 1 μg lentiviral vector plasmid (V1-V5) or empty plasmid DNA as negative control. Three independent wells were transfected for each sample and time point. Cells from 3 wells were harvested at 24 and 48 hours and lysed in passive lysis buffer (Promega, Madison, WI). Transfection efficiency was determined by normalization to GFP expression as measured by fluorescence microscopy.
Western blot
Western blot analysis was performed as described37 using lysates from transiently transfected 293T cells, HTLV-1–positive cell lines, and stable vector–infected SLB-1 cell clones and rabbit anti-HBZ polyclonal antiserum32 (1:500) and goat anti–rabbit-conjugated with horseradish peroxidase (Santa Cruz Biotechnology, Santa Cruz, CA). GFP and β-actin were detected using (1:1000) anti-GFP rabbit polyclonal antibody (Santa Cruz Biotechnology) and anti–β-actin rabbit monoclonal antibody, respectively, and goat–anti-rabbit conjugated with horseradish peroxidase (Santa Cruz Biotechnology). HTLV-1 Tax was detected using (1:1000) mouse hybridoma monoclonal antibody and HRP-conjugated goat–anti-mouse secondary antibody. Proteins were visualized using the ECL Western blot analysis system (GE Healthcare, Little Chalfont, United Kingdom).
Virus particle production and cell infection
Plasmid lentiviral vectors (V1-V5) were isolated from bacterial cultures and sent to The Ohio State University Comprehensive Cancer Center/Cincinnati Children's Hospital Medical Center viral vector core for virus particle production. Lentiviruses were produced by transient cotransfection of lentiviral plasmid vectors and packaging plasmids pCDNA3.g/p.4xCTE, pRSV-rev, and M75-VSV-G. 293T cells were transfected using the calcium phosphate method (Sigma-Aldrich, St Louis, MO) in DMEM/10% FBS in the presence of 25 mM chloroquine. At 12 to 18 hours after transfection, the media was changed and supernatants were collected at four 10- to 14-hour intervals, 0.45-μm filtered, and frozen at −80°C. Supernatants were then concentrated by size exclusion using a 10-kDa filter (VivaScience, Hannover, Germany) and infectious titers were determined by expression of GFP on HT1080 cells 4 days after transduction. Cells were infected at a multiplicity of infection (MOI) of 2 in supplemented Iscove media along with 8 μg/mL polybrene (Sigma-Aldrich). At 48 hours after infection, viral inocula were removed and cells were selected in growth medium containing 1 mg/mL Geneticin (Gibco, Carlsbad, CA) for 4 weeks. Each drug-resistant culture was designated by the parental line and the transducing vector (eg, SLB-V1).
RT-PCR and quantitative Taqman real-time RT-PCR
Total RNA from 107 HTLV-1 positive cell lines, stable lentivirus-infected SLB-1 cell lines (V1-V5), and uninfected Jurkat control cells was extracted using the RNeasy kit and DNase treated column (QIAGEN, Valencia, CA). Two micrograms of total RNA were subjected to reverse transcriptase–polymerase chain reaction (RT-PCR) using 1 uM oligo DT primer for first-strand synthesis to generate cDNA libraries (Invitrogen). Forty cycles of real-time Taqman PCR were conducted to quantitate mRNA transcripts from cDNA libraries. cDNA was amplified in duplicate using the primers and probes for Hbz, tax/rex, and gag/pol sequences as described.38 The final numbers were averaged and transcript quantity was determined based on a standard curve generated from duplicate samples of log10 dilutions of a plasmid containing the Hbz, tax/rex, and gag/pol DNA sequences. The copy of each transcript was normalized to 106 copies of hGAPDH. hGAPDH was amplified using primers previously described.39
Proliferation assay and cell viability
Cell Titer 96 Aqueous One Solution Cell Proliferation Assays (Promega, Madison, WI) were performed on mock-infected and stable lentivirus-infected cell lines according to the manufacturer's protocol. Briefly, cells were counted and plated at 1000 cells/well in 96-well round-bottom plates on day 0 and monitored over a 5-day time course. Cell Titer 96 reagent was added to each well, agitated slightly, and incubated at 37°C, 5% CO2 for 2 hours. The optical density absorbance at 490 nm was collected on an enzyme-linked immunosorbent assay (ELISA) plate reader. To verify cellular viability over time, the same cell lines were plated in 96-well round-bottom plates and counted by trypan blue exclusion over the same 5-day time course. For each cell line, data represent 3 independent experiments performed in triplicate.
Mouse inoculation procedure and IL-2rα ELISA
Fifteen 7-week-old female NOD.Cg-PrkdcSCIDIL2rgtm1Wjl/SzJ (NOG) mice (pathogen-free) were obtained from The Jackson Laboratory (Bar Harbor, ME). The mice were inoculated subcutaneously in the lumbar region with 107 parental SLB-1, SLB-1 V4 lentivirus-infected (HBZ knockdown), SLB-1 V5 lentivirus-infected (scrambled), or Jurkat T-cell lines. Serum was drawn from the anterior facial vein at days 0, 7, 15, and 22. Mice were monitored daily, weight was recorded 3 times per week, and tumor size was measured manually using hand calipers. Mice were killed and necropsied at day 22. At necropsy, tumor was isolated from the underlying tissue and weighed. Tumors were flash frozen and remaining mouse tissues were fixed in 10% neutral buffered formalin, trimmed, paraffin-embedded, sectioned, and stained using routine hematoxylin and eosin. Representative tissue sections were stained with Ki67 (KiS5 clone) monoclonal antibody. This study protocol was approved by the University Laboratory Animal Resources (ULAR) of The Ohio State University. Because SLB-1 cells are known to produce interleukin-2 receptor alpha (IL-2rα), mouse serum reactivity to IL-2rα was used as a biomarker for SLB-1 cell proliferation in vivo. IL-2Rα serum levels were detected using a commercial Quantikine IL-2Rα ELISA (R&D Systems, Minneapolis, MN) using mouse serum diluted (1:10) to obtain absorbance values in the linear range of the assay. Western blot analysis was performed on homogenized tumor tissue lysates using antibodies as described in “Western blot.”
Results
Hbz mRNA and HBZ protein expression in HTLV-1 cell lines
Both spliced (major, SP1; minor, SP2) and unspliced Hbz transcripts with the potential to encode highly related protein isoforms have been detected in most ATL cells (Figure 1A).25,27,28 The spliced SP1 HBZ protein isoform previously was detected in HTLV-1–positive cell lines MT-4, C8166, SLB-1, and 729Ach.26,32 Satou et al also reported the successful knockdown of Hbz mRNA in HTLV-1 cell lines using an siRNA approach.25 Overall, they concluded from their studies that the Hbz mRNA had a proliferative effect on T cells independent of Tax. However, these studies did not evaluate HBZ protein or the potential for off-target effects of Hbz siRNA on other viral mRNAs or proteins. Our first objective was to quantitate Hbz expression in a panel of HTLV-1 cell lines and correlate mRNA levels to HBZ protein expression. Hbz-specific mRNA expression was detected, but variable, in all HTLV-1–positive cell lines (Figure 1B). We observed the highest Hbz mRNA copy number (normalized to hGAPDH) in established SLB-1 and C8166 cell lines with the next highest level displayed by newly HTLV-1–immortalized IL-2–dependent T lymphocytes (PBLACH). Furthermore, HBZ protein expression directly correlated with the Hbz mRNA levels in all positive samples (Figure 1B). However, we were surprised to detect HBZ protein in MT-2 cells because a previous study reported the detection of Hbz mRNA, but not protein, in MT-2.26
shRNA lentiviral vector design and transient knockdown of HBZ protein expression
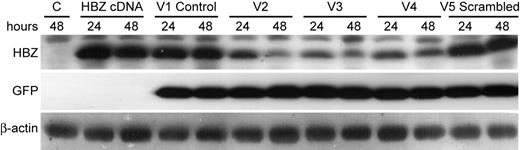
To address the role of Hbz in HTLV-1 immortalized/transformed cells both in cell culture and in vivo, we used an shRNA approach to knock down Hbz mRNA and protein expression. Lentiviral vectors generating shRNAs against various sequences of Hbz were generated. V2 and V3 target Hbz sequences identical to previously described siRNA4 and siRNA31, respectively, and have the potential to target all known Hbz transcripts.25 V4 was designed specifically to target the Hbz major spliced transcript; this target sequence spans the splice junction. Two additional negative controls including the empty vector (V1) and a vector expressing a scrambled nonspecific sequence (V5) also were generated. To determine whether the lentiviral vector plasmid DNAs encoded the capacity to knock down Hbz gene expression, we initially tested them in transient transfection. 293T cells were cotransfected in duplicate with an HBZ cDNA expression vector and lentiviral vectors (V1-V5). HBZ protein expression was measured by Western blot at 24 and 48 hours after transfection. GFP and β-actin protein expression were used as internal controls for loading and transfection efficiency. The 3 Hbz-specific target vectors (V2-V4) had the capacity to significantly knock down HBZ protein expression at 24 hours and, to a greater extent, at 48 hours after transfection (Figure 2). Our results with V2 are in contrast to those previously reported in which the identical target sequence was found not to suppress Hbz mRNA expression.25
Hbz-specific shRNA lentiviral plasmid vectors knock down HBZ protein expression transiently. 293T cells (105) were cotransfected with 0.2 μg HBZ cDNA expression vector and 1.0 μg shRNA lentiviral vector as indicated (V1-V5). Cellular lysates were harvested at 24- and 48-hour time points and subjected to Western blot analysis to detect HBZ, GFP, and β-actin. β-actin and GFP levels were used as internal loading and transfection normalization controls, respectively.
Hbz-specific shRNA lentiviral plasmid vectors knock down HBZ protein expression transiently. 293T cells (105) were cotransfected with 0.2 μg HBZ cDNA expression vector and 1.0 μg shRNA lentiviral vector as indicated (V1-V5). Cellular lysates were harvested at 24- and 48-hour time points and subjected to Western blot analysis to detect HBZ, GFP, and β-actin. β-actin and GFP levels were used as internal loading and transfection normalization controls, respectively.
Stable shRNA suppression of Hbz in SLB-1 cells does not alter tax/rex or gag/pol expression
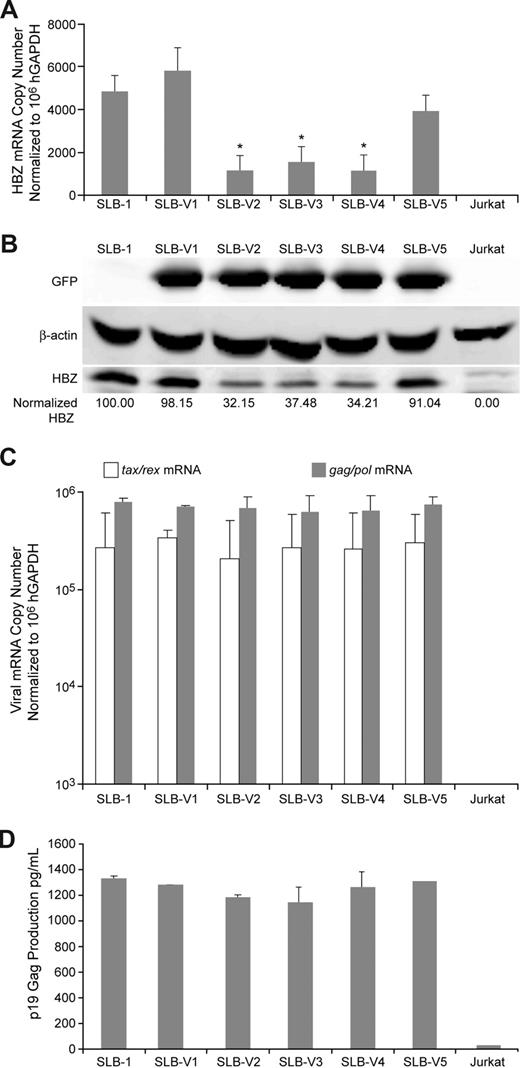
Lentivirus particles encoding the shRNAs against Hbz were generated and used to transduce HTLV-1 SLB-1–transformed T cells. We focused our initial studies on the SLB-1 cell line due to high Hbz expression levels relative to the other cell lines and the previous successful use of this cell line in the mouse tumorigenicity model.33 SLB-1 cells were infected with lentiviruses and stably transduced cells were selected and expanded for 4 weeks in geneticin-containing growth media. To determine whether a stable knockdown of Hbz expression was achieved, total RNA was isolated from each transduced line and subjected to real-time RT-PCR to quantitate Hbz mRNA copy number relative to cellular hGAPDH control. Hbz mRNA expression was significantly knocked down (approximately 3-fold) in SLB-1 cells transduced by lentiviruses V2, V3, or V4 as compared with wild-type SLB-1 and SLB-1 transduced by control lentiviruses V1 or V5 (Figure 3A). Western blot analysis indicated that HBZ protein expression was reduced, which correlated with the Hbz mRNA suppression (Figure 3B). Next, we wanted to be sure that V2-, V3-, and V4-generated shRNAs targeted Hbz specifically and did not have any off-target effects on other key viral genes. Total RNA was subjected to real time RT-PCR to quantitate both tax/rex and gag/pol mRNA copy number. Our results showed that Hbz knockdown had no significant effect on tax/rex or gag/pol mRNA expression (Figure 3C). Moreover, p19 Gag production in the viral supernatant was also unaffected (Figure 3D). Taken together, these results indicate that Hbz mRNA and protein are significantly and specifically suppressed by Hbz-targeting shRNAs.
Characterization of HTLV-1 gene expression in SLB-1 cells stably infected with shRNA lentiviral vectors. (A) Total RNA was isolated from parental SLB-1, stably transduced SLB-1 (V1-V5), and control Jurkat cells and subjected to real-time RT-PCR to quantify Hbz transcripts. Numbers are normalized to 106 copies of hGAPDH. Cells transduced with Hbz-specific shRNA vectors (V2-V4) have a significant 3- to 3.5-fold decrease in Hbz mRNA levels compared with controls (*). (B) Western blot analysis on total cellular lysates from indicated cells shows a significant 3-fold decrease in HBZ protein expression (normalized HBZ densitometry numbers) in cells transduced with Hbz-specific shRNA vectors (V2-V4). Protein reduction correlates directly to mRNA reduction. (C) Quantitation of tax/rex and gag/pol mRNA expression in total cellular RNA by real-time RT-PCR (normalized to 106 hGAPDH) and (D) p19 Gag expression in culture supernatant by ELISA. The data presented in panels A, C, and D are the average values from 3 independent experiments; error bars denote SD. Statistical significance was determined by analysis of variance (ANOVA) followed by Tukey test.
Characterization of HTLV-1 gene expression in SLB-1 cells stably infected with shRNA lentiviral vectors. (A) Total RNA was isolated from parental SLB-1, stably transduced SLB-1 (V1-V5), and control Jurkat cells and subjected to real-time RT-PCR to quantify Hbz transcripts. Numbers are normalized to 106 copies of hGAPDH. Cells transduced with Hbz-specific shRNA vectors (V2-V4) have a significant 3- to 3.5-fold decrease in Hbz mRNA levels compared with controls (*). (B) Western blot analysis on total cellular lysates from indicated cells shows a significant 3-fold decrease in HBZ protein expression (normalized HBZ densitometry numbers) in cells transduced with Hbz-specific shRNA vectors (V2-V4). Protein reduction correlates directly to mRNA reduction. (C) Quantitation of tax/rex and gag/pol mRNA expression in total cellular RNA by real-time RT-PCR (normalized to 106 hGAPDH) and (D) p19 Gag expression in culture supernatant by ELISA. The data presented in panels A, C, and D are the average values from 3 independent experiments; error bars denote SD. Statistical significance was determined by analysis of variance (ANOVA) followed by Tukey test.
Knockdown of Hbz gene expression significantly suppresses T-lymphocyte proliferation
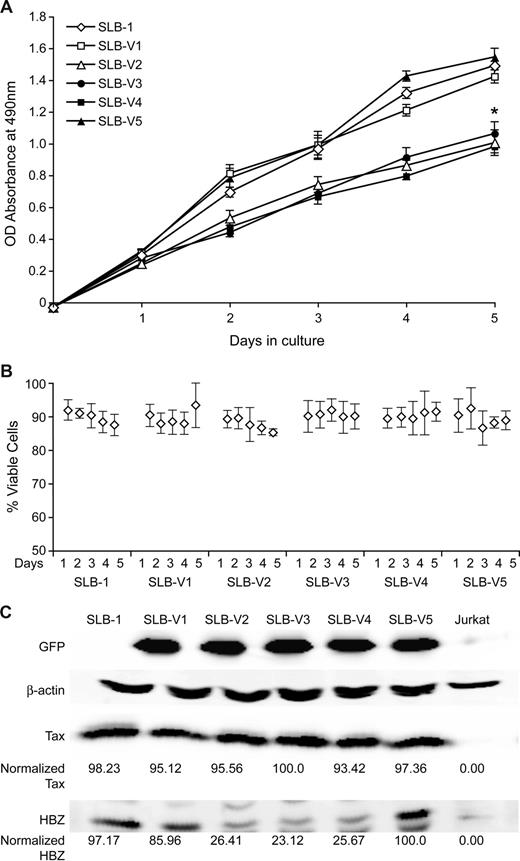
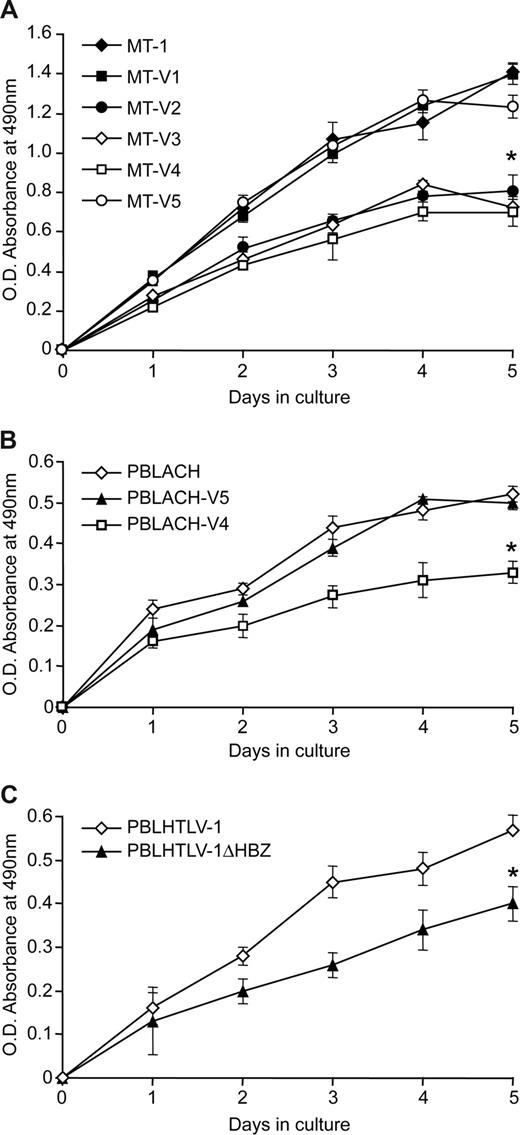
We next evaluated whether Hbz knockdown altered the proliferation of our stable lentivirus-transduced SLB-1 cells. Cells were plated at low concentrations and monitored daily for 5 days by standard MTS assay. The data indicate that knockdown of Hbz results in a significant decrease in proliferation (∼ 33%), compared with mock and transduced controls (Figure 4A). In addition, cells also were monitored and enumerated by trypan blue exclusion to verify that the decrease in proliferation rate was not attributed to increased cell death (Figure 4B). Furthermore, we examined cell-cycle progression of propidium iodide–stained SLB-1 and SLB-1 Hbz knockdown cells by fluorescence-activated cell sorting analysis. Consistent with the 5-day proliferation curves, SLB-1 Hbz knockdown cells showed a 7% decrease in cells in S phase as compared with parental SLB-1 cells (data not shown). We confirmed that the proliferating cells evaluated in the growth assay maintained the stable knockdown of HBZ protein. As presented in Figure 4C, HBZ protein expression was consistently and significantly knocked down 4-fold, whereas Tax protein expression was not affected. Together, the data indicate that Hbz expression significantly supported the proliferation of SLB-1 cells and that the decrease in proliferation in the Hbz knockdown cells was not attributable to apoptosis. It should be noted that the growth-promoting activity of Hbz has been previously reported in MT-1 and TL-Om1 in which tax gene expression is extremely low or silenced.25 Using our vector system we confirmed that shRNA suppression of Hbz in MT-1 cells resulted in growth suppression similar to SLB-1 (Figure 5A). More importantly, we extended this observation to newly HTLV-1–immortalized T lymphocytes. Knockdown of Hbz in PBLACH results in a significant decrease in proliferation compared with the parental and mock-transduced controls (Figure 5B). Although HBZ protein is not required for the immortalization of primary T lymphocytes in culture,32 we show here that cells newly immortalized by HTLV-1ΔHBZ display a reduced capacity to proliferate as compared with wild-type HTLV-1–immortalized cells (Figure 5C). Although these proliferation results are consistent with SLB-1, MT-1, and PBLACH in comparison to their Hbz-specific shRNA-expressing counterparts, it is important to note that because PBLHTLV-1 and PBLHTLV-1ΔHBZ are independent cell isolates it is possible that their distinct growth properties are independent of their HBZ status. However, taken together our data clearly show that in the context of an integrated provirus, Hbz promotes T-lymphocyte proliferation.
Hbz supports SLB-1 cell proliferation in culture. (A) 103 SLB-1 or stable SLB-1 lentiviral-infected cells (V1-V5) were plated in normal growth medium in 96-well plates and MTS assays were performed on triplicate wells at 24-hour intervals for a total of 5 days. The average absorbance numbers are plotted and error bars denote SD. * denotes the significant difference in proliferation in Hbz-specific shRNA-transduced cells compared with controls. (B) The same cell lines as in panel A were subjected to trypan blue exclusion to assess cell viability. A total of 5 wells per cell line were enumerated per day and presented as average percent of viable cells with error bars denoting SD. (C) Western blot analysis was performed on total cell lysates as indicated. HBZ and Tax levels were quantified and normalized to β-actin control by densitometry. Statistical significance was determined by ANOVA followed by Tukey test.
Hbz supports SLB-1 cell proliferation in culture. (A) 103 SLB-1 or stable SLB-1 lentiviral-infected cells (V1-V5) were plated in normal growth medium in 96-well plates and MTS assays were performed on triplicate wells at 24-hour intervals for a total of 5 days. The average absorbance numbers are plotted and error bars denote SD. * denotes the significant difference in proliferation in Hbz-specific shRNA-transduced cells compared with controls. (B) The same cell lines as in panel A were subjected to trypan blue exclusion to assess cell viability. A total of 5 wells per cell line were enumerated per day and presented as average percent of viable cells with error bars denoting SD. (C) Western blot analysis was performed on total cell lysates as indicated. HBZ and Tax levels were quantified and normalized to β-actin control by densitometry. Statistical significance was determined by ANOVA followed by Tukey test.
Hbz supports immortalized T-lymphocyte proliferation in culture. (A) 103 MT-1 and stable MT-1 lentiviral- (V1-V5) infected cells, (B) PBLACH and stable PBLACH lentiviral- (V4 or V5) infected cells, and (C) PBLHTLV-1 and PBLHTLV-1ΔHBZ cells were plated in normal growth medium in 96-well plates and MTS assays were performed on triplicate wells at 24-hour intervals for a total of 5 days. The average absorbance numbers are plotted and error bars denote SD. * denotes the significant difference in proliferation in Hbz-specific shRNA transduced cells or HTLV-1ΔHBZ–infected cells compared with controls or wild-type infected cells. Statistical significance was determined by ANOVA followed by Tukey test.
Hbz supports immortalized T-lymphocyte proliferation in culture. (A) 103 MT-1 and stable MT-1 lentiviral- (V1-V5) infected cells, (B) PBLACH and stable PBLACH lentiviral- (V4 or V5) infected cells, and (C) PBLHTLV-1 and PBLHTLV-1ΔHBZ cells were plated in normal growth medium in 96-well plates and MTS assays were performed on triplicate wells at 24-hour intervals for a total of 5 days. The average absorbance numbers are plotted and error bars denote SD. * denotes the significant difference in proliferation in Hbz-specific shRNA transduced cells or HTLV-1ΔHBZ–infected cells compared with controls or wild-type infected cells. Statistical significance was determined by ANOVA followed by Tukey test.
Knockdown of Hbz gene expression decreases tumor size in NOG mice
To evaluate the role that Hbz plays in tumor cell growth and organ infiltration in vivo, NOG mice were inoculated subcutaneously with wild-type SLB-1, SLB-V4 (Hbz knockdown), SLB-V5 control (scrambled), or Jurkat cell control (n = 5 each). After inoculation, mice were observed on a daily basis for tumor engraftment, and weighed every other day.
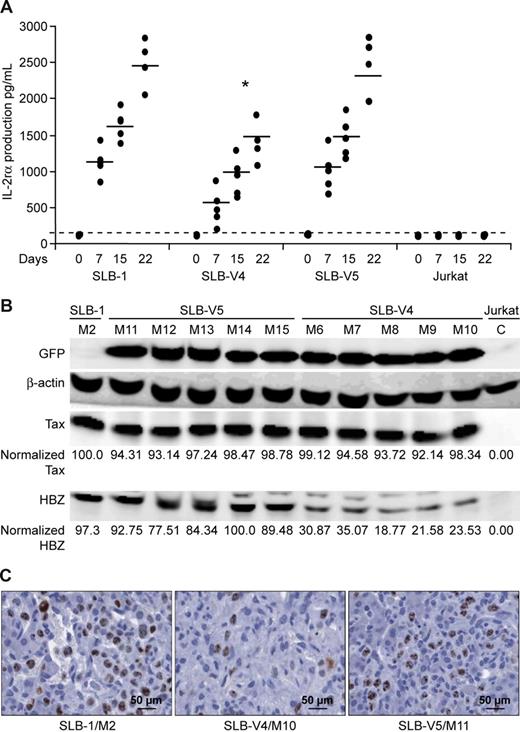
Because IL-2Rα is secreted by SLB-1 cells, it can be used as a biomarker for cellular proliferation in vivo. Blood was drawn from each mouse just before inoculation (day 0) and at days 7, 15, and 22 after inoculation and IL-2Rα was quantitated in the serum by ELISA. We observed a statistically significant decrease (∼ 35%-40%) of IL-2Rα in the serum of mice inoculated with SLB-V4 Hbz knockdown cells (Figure 6A). Consistent with the ELISA data, mice inoculated with SLB-V4 also displayed a significant 49% to 58% average decrease in overall tumor size and volume on the day they were killed (day 22) as compared with SLB-V5 control and parental SLB-1 (Table 1). Western blot on tumor tissue lysates from SLB-V4–inoculated mice revealed 3- to 5-fold less HBZ protein as compared with tumor material from a representative SLB-1–inoculated mouse (M2) or SLB-V5–inoculated mice (Figure 6B). Tax protein expression was similar in tumors of all inoculated mice consistent with the conclusion that tumor cell growth directly correlated with Hbz expression and not the expression level of the Tax oncoprotein. In addition, the decrease in HBZ protein expression in the tumors from SLB-V4–inoculated mice was consistent with the level of HBZ protein knockdown observed in culture experiments. At death, mouse tissues were examined histologically by routine H&E staining and Ki67 expression for cell proliferation and tumor cell infiltration. Table 2 presents an average score for tumor cell infiltration in select tissues from each of the inoculated groups. Tumor infiltration to liver, heart, lung, kidney, and pancreas tissue was dramatically lower in SLB-V4–inoculated mice. Figure 6C shows representative Ki67 staining of liver from mice inoculated with SLB-1, SLB-V4, or SLB-V5. Results demonstrated a dramatic reduction in proliferation and infiltration of liver tissue by SLB-V4 (Hbz knockdown) cells as compared with SLB-1 or SLB-V5 controls. Taken together, our results showed that knockdown in Hbz gene expression significantly decreases proliferation of SLB-1 cells in vivo. The decrease in proliferation of the SLB-1 Hbz knockdown cells in vivo correlated with a decrease in tumor size and infiltration of tumor cells to surrounding tissue.
SLB-1 Hbz knockdown cell proliferation is significantly reduced in NOG mice. (A) NOG mice were inoculated with 107 parental SLB-1, stable lentiviral-infected SLB-1 derivatives (V4, V5), or Jurkat cell control (5 mice each). Because SLB-1 secretes IL-2Rα, mouse serum levels of IL-2Rα were measured by ELISA as a biomarker for SLB-1 proliferation in vivo. Each dot represents the average absorbance value of a single inoculated mouse at 0, 7, 15, and 22 days after inoculation within each group. The horizontal line represents the average of the mouse group at each weekly time point and the dotted line represents negative absorbance values. * denotes significant differences from controls. Statistical significance was determined by ANOVA followed by Tukey test. (B) Western blot analysis was performed on total tumor cell lysates of individual mice as indicated. HBZ and Tax levels were quantified and normalized to β-actin control by densitometry. Statistical significance was determined by ANOVA followed by Tukey test. (C) Immunohistochemistry was performed on representative mouse liver tissue stained for Ki67 human antigen, a marker for proliferation. The glass slides were digitally scanned and captured at 40× magnification using the Aperio ScanScope and ImageScope Software (Aperio Technologies. Vista, CA). Average liver counts for each group are: SLB-1, 389.25 (n = 4); SLB-V4, 97.75 (n = 4); and SLB-V5, 358.6 (n = 5).
SLB-1 Hbz knockdown cell proliferation is significantly reduced in NOG mice. (A) NOG mice were inoculated with 107 parental SLB-1, stable lentiviral-infected SLB-1 derivatives (V4, V5), or Jurkat cell control (5 mice each). Because SLB-1 secretes IL-2Rα, mouse serum levels of IL-2Rα were measured by ELISA as a biomarker for SLB-1 proliferation in vivo. Each dot represents the average absorbance value of a single inoculated mouse at 0, 7, 15, and 22 days after inoculation within each group. The horizontal line represents the average of the mouse group at each weekly time point and the dotted line represents negative absorbance values. * denotes significant differences from controls. Statistical significance was determined by ANOVA followed by Tukey test. (B) Western blot analysis was performed on total tumor cell lysates of individual mice as indicated. HBZ and Tax levels were quantified and normalized to β-actin control by densitometry. Statistical significance was determined by ANOVA followed by Tukey test. (C) Immunohistochemistry was performed on representative mouse liver tissue stained for Ki67 human antigen, a marker for proliferation. The glass slides were digitally scanned and captured at 40× magnification using the Aperio ScanScope and ImageScope Software (Aperio Technologies. Vista, CA). Average liver counts for each group are: SLB-1, 389.25 (n = 4); SLB-V4, 97.75 (n = 4); and SLB-V5, 358.6 (n = 5).
Knockdown of Hbz gene expression significantly decreased tumor size in vivo
| Animal inoculum . | Tumor size, mm . | Average . | Standard . | Tumor volume . | Average . | Standard . | Tumor weight, g . | Average . | Standard . |
|---|---|---|---|---|---|---|---|---|---|
| SLB-1 | |||||||||
| M1 | 16 × 21 | 268.8 | 1.82 | ||||||
| M2 | 16 × 18 | 227.0 | 1.56 | ||||||
| M3 | 16 × 21 | 268.8 | 2.00 | ||||||
| M4 | 21 × 22 | 363.1 | 1.75 | ||||||
| M5 | − | 355.5 | 74.5 | − | 281.9 | 57.6 | − | 1.79 | .18 |
| SLB-V4 | |||||||||
| M6 | 12 × 14 | 132.7 | 1.04 | ||||||
| M7 | 14 × 15 | 165.2 | 0.93 | ||||||
| M8 | − | − | − | ||||||
| M9 | 12 × 14 | 132.7 | 0.94 | ||||||
| M10 | 12 × 13 | 175.5* | 88.9 | 122.7 | 138.3* | 18.5 | 1.12 | 1.01* | .09 |
| SLB-V5 | |||||||||
| M11 | 18 × 19 | 268.8 | 1.42 | ||||||
| M12 | 15 × 21 | 254.5 | 1.72 | ||||||
| M13 | 15 × 17 | 201.1 | 1.29 | ||||||
| M14 | − | − | − | ||||||
| M15 | 16 × 18.5 | 302.0 | 44.5 | 233.7 | 239.5 | 29.4 | 1.58 | 1.50 | .19 |
| Jurkat, M16-M20 | N/A | N/A | N/A |
| Animal inoculum . | Tumor size, mm . | Average . | Standard . | Tumor volume . | Average . | Standard . | Tumor weight, g . | Average . | Standard . |
|---|---|---|---|---|---|---|---|---|---|
| SLB-1 | |||||||||
| M1 | 16 × 21 | 268.8 | 1.82 | ||||||
| M2 | 16 × 18 | 227.0 | 1.56 | ||||||
| M3 | 16 × 21 | 268.8 | 2.00 | ||||||
| M4 | 21 × 22 | 363.1 | 1.75 | ||||||
| M5 | − | 355.5 | 74.5 | − | 281.9 | 57.6 | − | 1.79 | .18 |
| SLB-V4 | |||||||||
| M6 | 12 × 14 | 132.7 | 1.04 | ||||||
| M7 | 14 × 15 | 165.2 | 0.93 | ||||||
| M8 | − | − | − | ||||||
| M9 | 12 × 14 | 132.7 | 0.94 | ||||||
| M10 | 12 × 13 | 175.5* | 88.9 | 122.7 | 138.3* | 18.5 | 1.12 | 1.01* | .09 |
| SLB-V5 | |||||||||
| M11 | 18 × 19 | 268.8 | 1.42 | ||||||
| M12 | 15 × 21 | 254.5 | 1.72 | ||||||
| M13 | 15 × 17 | 201.1 | 1.29 | ||||||
| M14 | − | − | − | ||||||
| M15 | 16 × 18.5 | 302.0 | 44.5 | 233.7 | 239.5 | 29.4 | 1.58 | 1.50 | .19 |
| Jurkat, M16-M20 | N/A | N/A | N/A |
Mouse tumor size was measured in millimeters (length × width) with the average and standard deviation indicated. Mouse tumor volumes were calculated by the formula 3.142 × radius2 . Mouse solid tumor weights (in grams) were taken at day of sacrifice. M5 and M8 were sacrificed on day 17 due to necrosis and are not included in the final averaged numbers (−). M14 died on day 22 due to complications from peritoneal injection and also was not included in the final average numbers (−). N/A; not applicable, because no tumor mass at day 22 in any of the Jurkat cell–inoculated animals.
indicates statistically significant difference from SLB-1– and SLB-V5–inoculated mice.
Mouse tissues examined histologically by routine H&E and Ki67 staining
| Tissue . | SLB-1 . | SLB-V4 . | SLB-V5 . | Jurkat . |
|---|---|---|---|---|
| Liver | +++ | + | +++ | − |
| Heart | ++ | ++ | ++++ | − |
| Lung | +++ | + | ++ | − |
| Kidney | +++ | + | ++ | − |
| Pancreas | ++++ | + | ++++ | − |
| Tissue . | SLB-1 . | SLB-V4 . | SLB-V5 . | Jurkat . |
|---|---|---|---|---|
| Liver | +++ | + | +++ | − |
| Heart | ++ | ++ | ++++ | − |
| Lung | +++ | + | ++ | − |
| Kidney | +++ | + | ++ | − |
| Pancreas | ++++ | + | ++++ | − |
Tissues were fixed in 10% neutral buffered formalin, trimmed, paraffin-embedded, sectioned, and stained using routine hematoxylin and eosin. Scores represent the average values of each animal group. All tissues were scored using the following scale: no neoplastic cells (−); minimal (+) corresponds to an infiltrate of 0 to 5 cells in radius; mild (++) corresponds to an infiltrate of 6 to 10 cells in radius; moderate (+++) corresponds to an infiltrate of 11 to 15 cells in radius; maximal (++++) corresponds to an infiltrate of 16 or more cells in radius. Data represent the averages from the mouse groups of tumor cell infiltration in each tissue.
Discussion
The role of the HTLV-1 negative strand Hbz gene in viral replication and/or pathogenesis remains to be fully defined. In the context of an HTLV-1 molecular clone, HBZ protein is dispensable for transformation/immortalization of T lymphocytes in culture, but disruption of HBZ dramatically reduced viral infectivity and persistence in inoculated rabbits.32 Exogenously overexpressed HBZ interacts with cellular transcription factors of the Jun and CREB family of proteins and negatively regulates Tax-mediated and AP-1–mediated transcription. Interestingly, in addition to the functional properties of the HBZ protein, the secondary structure of the Hbz mRNA has been reported to support the proliferation of T cells.25 Both spliced and unspliced Hbz transcripts with the potential to encode highly related protein isoforms have been described.25,27,28 The Hbz major spliced transcript (SP1) is the most abundant and is detected in multiple HTLV-1–infected cell lines and most all ATL cells.25,27,28 In addition, our group and Murata et al recently demonstrated by Western blot analyses and immunochemistry that in vitro–transformed HTLV-1 and ATL cells predominantly express the protein isoform of the major spliced transcript.28,32 In the current study, we detected Hbz mRNA and protein in all the HTLV-1–positive cell lines tested. We noted 1 inconsistency from a previous report in which HBZ protein was not detectable in MT-2 cells.26 Differential detection between the 2 studies could be attributed to the different HBZ antibodies used or to the use of variants of the MT-2 cell line. SLB-1 became the cell line of choice for our studies for several reasons. (1) SLB-1 is a well characterized and established HTLV-1–transformed T-cell line that consistently expresses the highest level of Hbz mRNA and protein. (2) SLB-1 secretes IL-2Rα, thus providing a biomarker for cellular proliferation that can be measured easily by quantitative ELISA. (3) Previous experiments indicated that SLB-1 cells form solid subcutaneous tumors in NOG mice, whereas other cells such as MT-1 form a dispersed tumor pattern that is difficult to monitor, and newly HTLV-1–immortalized PBLACH cells do not engraft (data not shown).33
We infected HTLV-1 positive SLB-1 cells with lentiviruses expressing shRNAs targeting Hbz sequences to determine the role of Hbz in global viral gene expression and cell proliferation. We clearly showed that the lentiviral plasmid vectors targeting Hbz sequences significantly knocked down Hbz gene expression both transiently and long-term in stably transduced SLB-1 cells. SLB-1 cells with suppressed Hbz gene expression showed a significant decrease in cellular proliferation with no apparent loss of viability or increase in cell death. Furthermore, we extended the direct correlation of Hbz expression and proliferation to additional cell lines including MT-1 and more relevant newly HTLV-1–immortalized T lymphocytes, PBLACH and PBLHTLV-1ΔHBZ. It is interesting to speculate that Hbz V2 and V3 target sequences could have the potential for off-target effects on viral-positive strand mRNAs that might include gag/pol, p12, or p30. Importantly, we show for the first time that suppressing Hbz gene expression has no significant effect on the levels of gag/pol and tax/rex mRNA and p19 Gag and Tax protein. The fact that tax and other quantified viral mRNAs are not altered in SLB-1 Hbz knockdown cells may be contradictory to the current dogma for the function of the HBZ protein. For example, the loss of HBZ expression in the context of a molecular clone resulted in an increase in Tax transactivation and overall viral gene transcription.32 However, in those studies, HBZ protein was completely knocked out, whereas in the present study, Hbz was suppressed, but not knocked out. Thus, we speculate that there could be a threshold level of HBZ protein required for its repressive effects on Tax-mediated transactivation.
We used the immune-compromised NOG mouse to determine the role of Hbz gene expression in cell growth and tumor infiltration in vivo.40,41 Our results demonstrated that the high level of Hbz expression in SLB-1 cells was not required for tumor cell engraftment, but Hbz expression significantly contributed to tumor growth and organ infiltration. Moreover, we showed that serum levels of IL-2Rα in animals challenged with SLB-V4 Hbz knockdown cells was significantly decreased throughout the study relative to controls. These results indicated that Hbz expression contributes significantly to the proliferation of cells in the NOG mouse, thereby facilitating tumor growth as measured by increased tumor weight and volume (Table 1). Western blots on tumor lysates confirmed that the Hbz repression was maintained and HBZ protein levels directly correlated with tumor growth. Furthermore, tumor growth appears to be independent of the Tax oncoprotein, because Tax expression was not significantly affected in any mouse tumors, irrespective of HBZ levels. The animals challenged with the SLB-V4 Hbz knockdown cells also displayed a reduced pattern of infiltrating tumor cells (Table 2, Figure 6C).
Based on the data that HBZ protein represses Tax-mediated transcription and attenuates AP-1 activity, we hypothesized that HBZ might function with other viral gene products to tightly regulate viral replication and/or promote infected cell survival. Because Tax is known to elicit a strong cytotoxic T-lymphocyte (CTL) response in vivo, the down-regulation of Tax clearly would be beneficial to the infected cell by enabling it to evade host immune surveillance. However, if Tax expression is down-regulated, the infected cell would lose key growth and survival signals critical during the early stages of viral infectivity, replication, transformation, and persistence.13,14,17 One such pathway involves Tax-mediated degradation of the tumor suppressor retinoblastoma (Rb), which leads to the release and activation of E2F1, a key regulator of cell progression from G1 to S phase.10,–12 A recent finding showed that the Hbz mRNA enhances E2F1 gene expression.25 This observation suggests an alternate mechanism by which HTLV-1–infected cells, independent of Tax, can maintain the deregulation of E2F1 leading to cell-cycle transition and continued cell proliferation. HBZ protein is detectable in all HTLV-1–positive cell lines tested in this study as wells as in most all ATL cells, suggesting that at least 1 of the activities of Hbz is required late in the leukemogenic process. In this regard, the cellular protein hTERT has been shown to be up-regulated in ATL cells and many cancers. A recent publication reported that HBZ protein enhances transcription of hTERT to its highest levels most notably in the absence of Tax,42 providing further support for both the mRNA and protein forms of Hbz in the dysregulation of cellular activities. Current studies are under way to determine whether the Hbz mRNA and the HBZ protein synergistically affect cellular genes within the infected cell and to assess the overall relationship between Hbz and tax gene expression. Taken together, it seems likely that Tax and Hbz function cooperatively—Tax early and Hbz late—in the process of HTLV-1 tumorigenesis.
In summary, we show that knockdown of Hbz gene expression in SLB-1 cells had no effect on Tax expression but significantly altered the growth properties of these cells in culture and their ability to develop solid tumors and infiltrate/spread to other tissues in the NOG mouse. These results are consistent with the maintenance and expression of Hbz in most all ATL cells and support an important role for Hbz in leukemogenesis. It appears that HTLV-1 has evolved to use HBZ (protein and mRNA) in unique ways to exploit the host cell machinery and enhance the growth of the infected cell. It is possible that the Hbz mRNA and HBZ protein function synergistically in infected cells to support leukemogenesis. Further studies may provide new targets for therapy to disrupt virus replication and the proliferation and survival of infected cells.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank C. Machado-Parrulla and M. Kesic for technical assistance, K. Hayes-Ozello for editorial comments on the manuscript, and T. Vojt for figure preparation.
This work was supported by grants from the National Institutes of Health (AI064440 and CA077566).
National Institutes of Health
Authorship
Contribution: J.A. performed research, analyzed data, and wrote the paper; B.Z. performed research and analyzed data; M.L. performed research and analyzed data; M.D.L. helped design research and analyzed data; and P.L.G. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Patrick L. Green, The Ohio State University, 1925 Coffey Road, Columbus, OH 43210; e-mail: green.466@osu.edu.