The heterodimeric transcription factor RUNX1/PEBP2-β (also known as AML1/CBF-β) is essential for definitive hematopoiesis. Here, we show that interaction with PEBP2-β leads to the phosphorylation of RUNX1, which in turn induces p300 phosphorylation. This is mediated by homeodomain interacting kinase 2 (HIPK2), targeting Ser249, Ser273, and Thr276 in RUNX1, in a manner that is also dependent on the RUNX1 PY motif. Importantly, we observed the in vitro disruption of this phosphorylation cascade by multiple leukemogenic genetic defects targeting RUNX1/CBFB. In particular, the oncogenic protein PEBP2-β-SMMHC prevents RUNX1/p300 phosphorylation by sequestering HIPK2 to mislocalized RUNX1/β-SMMHC complexes. Therefore, phosphorylation of RUNX1 appears a critical step in its association with and phosphorylation of p300, and its disruption may be a common theme in RUNX1-associated leukemogenesis.
Introduction
The runt-related transcription factor RUNX1 (AML1; PEBP2-α; CBF-α) and its non-DNA binding partner PEBP2-β/CBF-β are frequent targets of leukemogenic genetic changes.1,–3 The heterodimer plays a critical role in definitive hematopoiesis, and disruption of its normal function by chromosomal abnormalities or mutations is collectively the most common cause of acute myeloid leukemia (AML). Their central role in orchestrating proper differentiation of hematopoietic stem cells is underscored by the ablation of definitive hematopoiesis in Runx1 or Pebp2b−/− knockout mice4,–6 and an expanded HSC compartment in conditional Runx1-deficient mice.7,,–10 However, although the overall biologic functions of RUNX1 are becoming clear, how these functions are controlled at the molecular level and the contribution of PEBP2-β remain to be fully determined
The evolutionarily conserved partner protein PEBP2-β is known to enhance DNA binding ability of all 3 mammalian RUNX proteins (RUNX1-3) by inducing allosteric change of DNA binding Runt domain without direct DNA contact. In addition, we have previously reported that PEBP2-β regulates RUNX1 metabolic stability by preventing its ubiquitin-mediated degradation.11 It has also been speculated that PEBP2-β may also function to recruit other proteins to the target gene promoter, although only 2 cytoplasmic proteins, Crl-1 and filamin A, have ever been reported to interact with PEBP2-β.12,,–15 Notwithstanding our current incomplete understanding of molecular interaction between PEBP2-β and RUNX1, it is clear that PEBP2-β is essential for RUNX1 function in vivo, as Pebp2β-deficient mice exhibit a very similar phenotype to Runx1−/− mice.4,–6
Whereas early studies have implicated RUNX1 to be a transcriptional activator, subsequent studies revealed more diverse and intricate functions. It is now clear that RUNX1 can also act as a transcriptional repressor and functions as a molecular scaffold for the organization of partner transcription factors and cofactors at the regulatory regions of target genes. RUNX1 interacts with a growing list of transcription factors, such as Ets1,16 PU.1,17 C/EBPα,17 AP-1,18 and GATA-1.19,20 At the same time, RUNX1 is also shown to associate with coactivators, such as p300/CBP,21 YAP/TAZ,22 and MOZ,23 and corepressors TLE1,24,25 SUV39H1,26 and mSin3A.27 These observations suggest that RUNX1/PEBP2-β possesses multiple facets relating to transcription regulation. However, the molecular mechanism through which RUNX1 switches between its distinct roles, by choosing and activation of specific partner molecule, is poorly addressed. One possible mechanism that may achieve such modulation of RUNX1 function is through covalent modification of the RUNX1 protein, such as phosphorylation. Indeed, the phosphorylation of RUNX1 has been the topic of a number of recent studies, implicating roles for a diverse range of kinases. Of particular relevance to this work, Aikawa et al28 reported the in vitro and in vivo involvement of homeodomain-interacting kinase 2 (HIPK2) in phosphorylating RUNX1 and its coactivator p300. Consequently, mice double-deficient for Hipk1/2 displayed a phenotype subset of those of p300- and CBP-deficient mice.
Here we describe a protein phosphorylation sequence that is initiated by the heterodimerization of RUNX1/PEBP2-β and DNA binding, which leads to the phosphorylation of RUNX1 and p300. We have shown that this is also mediated by HIPK2 and targets proline-directed serine and threonine residues in the C-terminus of RUNX1. We observed that RUNX1 proteins bearing leukemogenic mutations could not be phosphorylated by this mechanism. Similarly, the leukemogenic chimeric protein PEBP2-β/CBF-β-SMMHC (β-SMMHC) also disrupts RUNX1/p300 phosphorylation by sequestration of HIPK2 to large cytoplasmic filaments in a manner dependent on its dimerization with RUNX1. Together, these findings provide novel insights into the regulation of RUNX1/PEBP2-β function at the molecular level and suggest that the targeting of RUNX1-p300 phosphorylation may represent a common mechanism in RUNX1/PEBP2-β–associated leukemogenesis.
Methods
Cell lines and transient transfection
COS-7 and HEK293 cells were maintained as per standard culturing conditions in Dulbecco modified Eagle medium, human colorectal adenoma cell lines HCT-116 in McCoy medium, and human leukemia-derived cell lines HL60, U937, and K562 cells in RPMI medium. All medium was supplemented with 10% fetal calf serum and antibiotics (1000 units/mL penicillin and 1000 μg/mL streptomycin). Typically, 105 cells were plated in 6-well plate and cultured overnight before transiently transfected with expression constructs for RUNX1 (0.1 μg) and PEBP2-β (0.3 μg) and with FLAG-tagged p300 or kinase (0.3 μg), using the FuGENE 6 reagent Roche Diagnostics (Indianapolis, IN) in accordance with the manufacturer's instructions.
Antibodies
Rabbit anti-AML1 and anti-PEBP2-β antibodies were generated as described by Lu et al.29 Mouse M2 anti-FLAG (Sigma-Aldrich, St Louis, MO), rabbit anti-AML1 (Active Motif, Carlsbad, CA) antibodies were obtained commercially as indicated. Secondary horseradish peroxidase-linked donkey anti–rabbit IgG or anti–mouse antibodies used in Western blotting were obtained from GE Healthcare (Little Chalfont, United Kingdom).
Plasmid expression vectors
Expression constructs for pEF-neo-RUNX1 (protein ID NP_001001890.1) -RUNX1S67R, -RUNX1G108R, -RUNX1K83E, -RUNX1R174Q; pEF-bos-FLAG-PEBP2-β1 (NP_071704), -PEBP2-β2, -PEBP2-β3, -PEBP2-β165, -PEBP2-β133, -PEBP2-β110, pEF-bos-PEBP2-β-SMMHC, -SMMHCΔ40-42, -SMMHCΔ38-42, -SMMHCΔ36-42, -SMMHCΔC410, -SMMHCΔC429, -SMMHCΔ33-36, and -SMMHCΔ33-38 were described previously.11,30,,–33 The RUNX1 deletions or point mutation series of constructs were generated by polymerase chain reaction (PCR)-based site-directed mutagenesis (Stratagene, La Jolla, CA). Sequences of oligonucleotide primers for the generation of RUNX1 deletion constructs are as follows: 5′-ATAGGAGCCACCATGAGCGGCGACCGCAGC-3′ for ΔN45; 5′-CAGTCCTACCAATACCTCTCTGCAGAACTT-3′ for Δ260-280; 5′-CGCTACCACACCTACCTGGGCTCGTCGCAAGCGCAG-3′ for ΔPY(355-359). For RUNX1ΔRD deletant (removal of residues 49-173), EcoRV and Mlu1 restriction sites were introduced in flanking the region for deletion for subsequent DNA-binding domain swapping cloning. Briefly, these were constructed by inserting PCR-generated EcoRV-Mlu1 fragments encoding residues 38 to 192 of murine c-Myb, 2 to 147 of yeast Gal4, and 281 to 397 of murine Lef1 into the pEF-Neo-RUNX1ΔRD deletion construct.
The sequences of primers used for the introduction of point mutations into RUNX1 are as follows: 5′-CAGATCCAACCAgCCCCACCGTGGTC-3′ for S249A; 5′-GGATCCATTGCCgCTCCTTCTGTGCAC-3′ for S266A; and 5′-GTGCACCCAGCAgCGCCCATTgCACCTGGACGTG-3′ for T273A/S276A mutations. All deletions and mutations of p300 (NP_001420) were performed in the context of pcDEF-FLAG-p300. Regions targeted for the deletion of Ser/Thr-Pro rich (STP) region are residues 57 to 327 for ΔSTP1, 828 to 941 for ΔSTP2, 1839 to 1913 for ΔSTP3, and 2269 to 2369 for ΔSTP4. Multiple STP deletants (ie, p300ΔSTP2,3,4, p300ΔSTP1,3,4, p300ΔSTP1,2,4, and p300ΔSTP1,2,3) and point mutants (ie, S831A/S833A, T839A/T841A/T845A, T885A/T887A) were generated by multiple round, sequential site-directed mutageneses.
Human DYRK1A (NP_001387) and HIPK2 (NP_073577) cDNA were amplified by reverse transcription PCR from U937 cells and cloned into pcDNA3-FLAG vector. Kinase-negative (KN) mutants of DYRK1A and HIPK2 were generated by introducing a K188R and K228A mutation, respectively. All new constructs generated for this study were verified by DNA sequencing and Western blotting.
Western blot analysis
Transiently transfected HEK293 and HCT-116 cells were lysed in 20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.5, 450 mM NaCl, 1.0% Triton X-100, 0.1 mM dichlorodiphenyltrichloroethane, 10 mM NaF, 20 mM β-glycerophosphate, 5 mM ethyleneglycoltetraacetic acid, 1 mM ethylenediaminetetraacetic acid, and protease inhibitor cocktail (Roche Diagnostics). Proteins extracts were boiled for 5 minutes, subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA). SDS-PAGE of p300 was performed in 5% polyacrylamide gels to allow better resolution. After probing with appropriate antibodies, protein bands were detected using the enhanced chemiluminescence detection system (GE Healthcare).
Phosphatase treatment of cell lysates
HL60, U937, or K562 leukemia and transiently transfected HEK293 cells were each lysed in 50 μL lysis buffer containing 20 mM Tris, pH 7.9, 400 mM NaCl, 0.3% Triton X-100, and protease inhibitor cocktail (Roche Diagnostics). After incubation on ice for 10 minutes, 3 volumes of 20 mM Tris, pH 7.9, were added to the lysates, before cellular debris were removed by centrifugation. The lysate supernatants were then treated with 10 units of calf intestine alkaline phosphatase (CIAP; Promega, Madison, WI) for 30 minutes at 37°C, before being subjected to SDS-PAGE and Western blot analyses.
Immunofluorescence
COS-7 cells were plated on glass cover slips and cultured for 1 day before being transiently transfected with various expression vectors. Cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) and permeablized with 0.2% Triton X-100, 24 hours after transfection. After blocking with 2% BSA in PBS, the permeabilized cells were incubated with the appropriate primary antibodies in blocking solution for 1 hour at room temperature. Cells were washed 3 times with PBS at room temperature and incubated with Alexa488 or 594 fluorephore-conjugated secondary antibodies for an additional 1 hour at room temperature. Nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI; 10 μg/mL) during mounting with Vectashield medium (Vector Laboratories, Burlingame, CA). Images were captured by an Olympus DP71 digital camera attached to a fluorescence microscope BX51 (Olympus, Tokyo, Japan) through a UPlan SApo 100×/1.4 NA oil objective lens (Olympus).
Results
PEBP2-β protein promotes phosphorylation of RUNX1 on heterodimerization with DNA-bound RUNX1
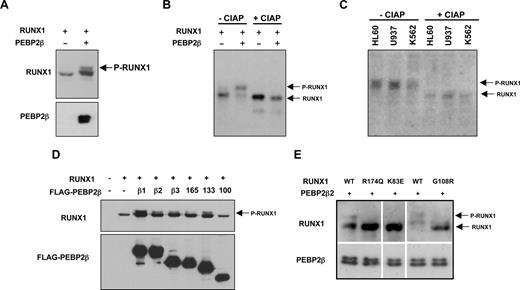
In a previous study, we noted the presence of 2 bands reactive to anti-RUNX1 antibody in Western blot, and it was speculated that the band of lower mobility was a phosphorylated form of RUNX1.34 To further investigate this, we first examined RUNX1 phosphorylation in the presence of PEBP2-β in transiently transfected HEK293 cells. Figure 1A shows that, in the absence of PEBP2-β, RUNX1 was detected as a single band in Western blot. However, coexpression of PEBP2-β induced a less mobile band, suggesting that dimerization with PEBP2-β induces the phosphorylation of RUNX1. Treatment with CIAP specifically abolished the lower mobility band, strongly suggesting that it is phosphorylated RUNX1 and that it is induced in the presence of PEBP2-β (Figure 1B). To demonstrate the phosphorylation of endogenous RUNX1, we examined the human leukemia-derived cell lines, HL60, U937, and K562 that express endogenous RUNX1. As observed in HEK293 cells exogenously expressing RUNX1, treatment with CIAP specifically abolished the low mobility band, indicating that endogenous RUNX1 proteins are phosphorylated in these cell lines under standard growth conditions (Figure 1C lanes 4-6).
Heterodimerization with PEBP2-β/CBF-β promotes phosphorylation of RUNX1 in a DNA binding-dependent manner. (A) PEBP2-β/CBF-β coexpression induces covalent modification of RUNX1 proteins in SDS-PAGE. HEK293 cells were transfected with plasmids for either RUNX1 (100 ng) alone or with PEBP2-β1 (300 ng). Twenty-four hours after transfection, cells were lysed and analyzed by Western blotting using an anti-RUNX1 or anti-PEBP2-β antibody. (B) Transfected HEK293 cell lysates were incubated in the presence or absence of calf intestine alkaline phosphatase (CIAP) followed by Western blotting using an anti-RUNX1 antibody. Phosphorylated RUNX1 proteins (P-RUNX1) and dephosphorylated/unphosphorylated RUNX1 (RUNX1) are indicated accordingly. (C) Ascertaining the phosphorylation status of endogenous RUNX1 in HL60, U937, and K562. Lysates from resting cells were treated with control or CIAP and analyzed by Western blotting using an anti-RUNX1 antibody. (D) Mapping of region in PEBP2-β responsible for the mediation of RUNX1 phosphorylation. HEK293 cells were transfected with RUNX1 (100 ng) and the indicated FLAG-PEBP2-β expression vectors (300 ng). The cells were harvested after 24 hours and analyzed by Western blotting to determine the phosphorylation status of RUNX1 by PAGE electrophoretic mobility. PEBP2-β1, -β2, and -β3 are naturally occurring isoforms of the PEBP2-β subunit. The β110 deletant is unable to dimerize with RUNX1. (E) Effects of RUNX1 point mutants found in AML patients or generated artificially. Wild-type and mutant RUNX1 proteins were coexpressed with PEBP2-β and processed as described in panel A. AML patient-derived mutants RUNX1R174Q and RUNX1K83E lack DNA binding ability, whereas an artificial mutant RUNX1G108R lacks the ability to heterodimerize with PEBP2-β but is able to bind to DNA.
Heterodimerization with PEBP2-β/CBF-β promotes phosphorylation of RUNX1 in a DNA binding-dependent manner. (A) PEBP2-β/CBF-β coexpression induces covalent modification of RUNX1 proteins in SDS-PAGE. HEK293 cells were transfected with plasmids for either RUNX1 (100 ng) alone or with PEBP2-β1 (300 ng). Twenty-four hours after transfection, cells were lysed and analyzed by Western blotting using an anti-RUNX1 or anti-PEBP2-β antibody. (B) Transfected HEK293 cell lysates were incubated in the presence or absence of calf intestine alkaline phosphatase (CIAP) followed by Western blotting using an anti-RUNX1 antibody. Phosphorylated RUNX1 proteins (P-RUNX1) and dephosphorylated/unphosphorylated RUNX1 (RUNX1) are indicated accordingly. (C) Ascertaining the phosphorylation status of endogenous RUNX1 in HL60, U937, and K562. Lysates from resting cells were treated with control or CIAP and analyzed by Western blotting using an anti-RUNX1 antibody. (D) Mapping of region in PEBP2-β responsible for the mediation of RUNX1 phosphorylation. HEK293 cells were transfected with RUNX1 (100 ng) and the indicated FLAG-PEBP2-β expression vectors (300 ng). The cells were harvested after 24 hours and analyzed by Western blotting to determine the phosphorylation status of RUNX1 by PAGE electrophoretic mobility. PEBP2-β1, -β2, and -β3 are naturally occurring isoforms of the PEBP2-β subunit. The β110 deletant is unable to dimerize with RUNX1. (E) Effects of RUNX1 point mutants found in AML patients or generated artificially. Wild-type and mutant RUNX1 proteins were coexpressed with PEBP2-β and processed as described in panel A. AML patient-derived mutants RUNX1R174Q and RUNX1K83E lack DNA binding ability, whereas an artificial mutant RUNX1G108R lacks the ability to heterodimerize with PEBP2-β but is able to bind to DNA.
To further study the apparent dependence on PEBP2-β for RUNX1 phosphorylation, we tested a series of FLAG-tagged PEBP2-β deletions and isoforms for their ability to induce phosphorylation. The results show that cotransfection of RUNX1 with all but the dimerization-defective PEBP2-β110 induced RUNX1 phosphorylation in HEK293 cells (Figure 1D). The same observation was made when a separate set of untagged PEBP2-β constructs was used (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Together, these observations indicate that heterodimerization with PEBP2-β is needed for the induction of RUNX1 phosphorylation.
We next asked whether the phosphorylation of RUNX1 requires a direct interaction with DNA because the best-known function of PEBP2-β is in increasing the DNA binding affinity of RUNX proteins. To address this, we adopted the AML patient-derived mutants RUNX1R174Q and RUNX1K83E, which are capable of dimerization with PEBP2-β but lack DNA binding ability.35 At the same time, we also tested RUNX1G108R, a DNA-binding but heterodimerization-defective mutant.11,35 As shown in Figure 1E, neither the 2 DNA binding-defective mutants (RUNX1R174Q and RUNX1K83E) nor the dimerization-defective mutant RUNX1G108R could be phosphorylated. These results suggest that, in addition to dimerization with PEBP2-β, the phosphorylation of RUNX1 is also dependent on its binding to DNA, hence implying that the phosphorylation of RUNX1 occurs within a chromatin context and probably involves a nuclear kinase.
Induction of p300 phosphorylation by RUNX1/PEBP2-β complex
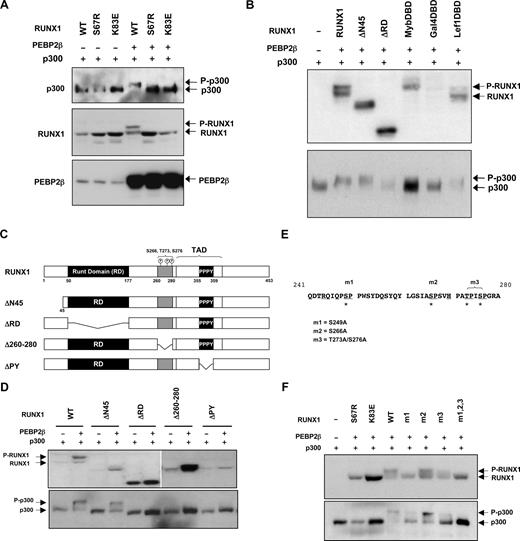
As RUNX1 is known to physically interact with a number of transcriptional cofactors, we studied the impact of RUNX1 phosphorylation on the phosphorylation status of p300, Yap1, SMRT, and Brg proteins. This showed that phosphorylation of RUNX1 resulted in a similar shift in electrophoretic mobility in p300 that could also be abolished by CIAP treatment (data not shown). We further demonstrated that the detected phosphorylation of p300 is dependent on the presence of both RUNX1 and PEBP2-β (Figure 2A). Interestingly, both the DNA-binding mutant RUNX1K83E and another dimerization-defective mutant RUNX1S67R were incapable of inducing p300 phosphorylation. These observations suggest that the formation of the RUNX1/PEBP2-β/DNA ternary complex is prerequisite to the induction of p300 phosphorylation. The similarities shared between the phosphorylation of RUNX1 and p300 also imply that these 2 processes may be coupled as components of a phosphorylation cascade.
DNA bound RUNX1/ PEBP2-β complex promotes RUNX1 and p300 phosphorylation. (A) Coupling of RUNX1 and p300 phosphorylation. HEK293 cells were transfected with RUNX1 or phosphorylation-defective point mutant constructs (100 ng) together with FLAG-tagged p300 (300 ng) in the absence or presence of PEBP2-β (300 ng). Cells were lysed 24 hours after transfection, and lysates were analyzed by Western blotting with antibodies against FLAG (top panel) and RUNX1 (middle panel) and PEPB2β (bottom panel), respectively. (B) HEK293 cells were cotransfected with wild-type RUNX1 or heterologous DNA-binding domain (DBD) mutants (100 ng), FLAG-tagged p300 (300 ng), and PEBP2-β (300 ng) as indicated. Cells were lysed 36 hours after transfection and analyzed by Western blotting using antibodies against RUNX1 (top panel) or FLAG (bottom panel). (C) Schematic representation of domain structure and phosphorylation sites of RUNX1 and mutants used in panels B and E. (D) Mapping of functional domains necessary for the phosphorylation of RUNX1 and p300. Wild-type or mutant RUNX1 were coexpressed with FLAG-p300 and PEBP2-β and analyzed by Western blotting 24 hours after transfection as described in panel A. (E) Mutation of putative phosphorylation target sites on RUNX1. Proline (P)-directed serine (S) and threonine (T) residues targeted in mutations m1 (S249A), m2 (S266A), and m3 (T273A/276A) are denoted by asterisks and mutated to alanine (A). (F) Transient transfection and Western blot analyses of RUNX1 point mutants in HEK293 cells. Wild-type, leukemogenic, and engineered point mutants of RUNX1 were coexpressed with p300 and PEBP2-β and analyzed by Western blotting as described in panel A.
DNA bound RUNX1/ PEBP2-β complex promotes RUNX1 and p300 phosphorylation. (A) Coupling of RUNX1 and p300 phosphorylation. HEK293 cells were transfected with RUNX1 or phosphorylation-defective point mutant constructs (100 ng) together with FLAG-tagged p300 (300 ng) in the absence or presence of PEBP2-β (300 ng). Cells were lysed 24 hours after transfection, and lysates were analyzed by Western blotting with antibodies against FLAG (top panel) and RUNX1 (middle panel) and PEPB2β (bottom panel), respectively. (B) HEK293 cells were cotransfected with wild-type RUNX1 or heterologous DNA-binding domain (DBD) mutants (100 ng), FLAG-tagged p300 (300 ng), and PEBP2-β (300 ng) as indicated. Cells were lysed 36 hours after transfection and analyzed by Western blotting using antibodies against RUNX1 (top panel) or FLAG (bottom panel). (C) Schematic representation of domain structure and phosphorylation sites of RUNX1 and mutants used in panels B and E. (D) Mapping of functional domains necessary for the phosphorylation of RUNX1 and p300. Wild-type or mutant RUNX1 were coexpressed with FLAG-p300 and PEBP2-β and analyzed by Western blotting 24 hours after transfection as described in panel A. (E) Mutation of putative phosphorylation target sites on RUNX1. Proline (P)-directed serine (S) and threonine (T) residues targeted in mutations m1 (S249A), m2 (S266A), and m3 (T273A/276A) are denoted by asterisks and mutated to alanine (A). (F) Transient transfection and Western blot analyses of RUNX1 point mutants in HEK293 cells. Wild-type, leukemogenic, and engineered point mutants of RUNX1 were coexpressed with p300 and PEBP2-β and analyzed by Western blotting as described in panel A.
To demonstrate that RUNX1 phosphorylation is not dependent on DNA binding per se, a series of chimeric proteins were generated, in which the DNA-binding Runt domain in RUNX1 was replaced with DNA-binding domains of Myb, gal4, or Lef1. As shown in Figure 2B, the attachment of RUNX1 to DNA via Myb, gal4, and Lef1 DNA- binding domains resulted in much lower levels of RUNX1 phosphorylation. In comparison, the RUNX1ΔRD lacking the DNA-binding Runt domain could not be phosphorylated, unlike the full-length protein or the RUNX1ΔN45 variant. Consistent with the observation in Figure 2A, the ability of the RUNX1 mutants in inducing p300 phosphorylation was also disrupted (Figure 2B bottom panel). Collectively, these results suggest that, although the binding to DNA by RUNX1 is necessary for the induction of RUNX1 and p300 phosphorylation, it is insufficient. Therefore, it is likely that conformational alteration in RUNX1 on dimerization with PEBP2-β is critical to the promotion of phosphorylation, probably through the recruitment of specific kinases. Importantly, these results also indicate a novel and separate function for PEBP2-β in addition to its known function in enhancing RUNX1 DNA binding.
Delineation of RUNX1 protein domains essential for PEBP2-β–induced phosphorylation
To further characterize the molecular mechanism involved in RUNX1 and p300 phosphorylation, we sought to determine the protein domains necessary and phosphorylation sites within each protein. In addition to RUNX1ΔRD and RUNX1ΔN45, we generated a series of truncated RUNX1 variants (Figure 2C) and tested these in HEK293 cells by transient transfection and Western blot analyses. We observed that, along with RUNX1ΔRD, the RUNX1Δ260-280 and RUNX1ΔPY variants were resistant to PEBP2-β–mediated phosphorylation (Figure 2D top panel). The Δ260-280 and PY motifs were specifically targeted in these experiments because the former contains several proline-directed serine and threonine residues reported as ERK phosphorylation targets, whereas the latter resides within the transactivation domain of RUNX1 and is known to mediate protein-protein interaction.22,36,37
We further examined the ability of these RUNX1 variants in inducing p300 phosphorylation and found that all 3 phosphorylation-defective RUNX1 variants were unable to promote p300 phosphorylation in the presence of PEBP2-β (Figure 2D bottom panel). These observations are consistent with the notion that phosphorylation of RUNX1 and p300 are coupled events.
Identification of PEBP2-β–dependent phosphorylation sites on RUNX1 and their effect on p300 phosphorylation
To identify the specific amino acid residues targeted in PEBP2-β–induced phosphorylation, we made a series of alanine mutants of proline-directed serine or threonine residues as illustrated in Figure 2E. The results show that mutant RUNX1 proteins in which Ser249 and Thr273/Ser276 were substituted with alanine residues (RUNX1S249A and RUNX1T273A/S276A, respectively) were resistant to phosphorylation (Figure 2F top panel). However, substitution of Ser266 with alanine had little or no effect on RUNX1 phosphorylation status. These data suggest that Ser249, Thr273, and Ser276 are probable target sites for PEBP2-β–mediated phosphorylation.
We further examine whether the phosphorylation of these RUNX1 residues is prerequisite for p300 phosphorylation. The bottom panel of Figure 2F shows that the 2 phosphorylation-defective mutants RUNX1S249A and RUNX1T273A/S276A caused a significant decrease in phosphorylation-induced mobility shift of p300 compared with wild-type RUNX1 or RUNX1S266A. Importantly, a compound RUNX1 mutant containing alanines at residue position 249, 266, 273, and 276 completely abolished p300 mobility shift, suggesting that phosphorylation of these RUNX1 residues is essential for promoting p300 phosphorylation.
Identification of phosphorylation regions and sites in p300
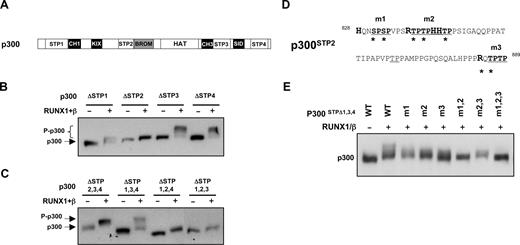
To identify the phosphorylation sites within p300, we inspected 53 proline-directed serines and threonines in p300 and found that most of them are located on 4 regions as indicated in Figure 3A. We next targeted these serine-, threonine-, and proline-rich regions, termed STP region, and generated a set of deletion constructs to examine their effects on p300 phosphorylation. Western blot analyses show that, with the deletion of STP2, the phosphorylation-mediated shift induced by RUNX1/PEBP2-β was markedly reduced. In the case of p300ΔSTP1, a minor change in band shift was observed, but the deletion of STP3 and STP4 had no observable effect (Figure 3B). In addition, through a separate series of domain-targeting deletion constructs, we observed that the SRC1-interacting domain and bromodomain are also necessary for RUNX1/PEBP2-β–induced phosphorylation (Figure S2).
Determination of phosphorylation targets within p300. (A) Schematic representation of domain structure of wild-type p300 and deletion mutants used in panels B and C. Four regions rich in proline-directed serine/threonine-rich residues are denoted STP1 to STP4. (B,C) Deletion variants of FLAG-p300 lacking single STP regions (B) or 3 STP regions (C) were transiently expressed in HEK293 cells in the presence or absence of exogenous RUNX1/PEBP2-β, as indicated. Cells were lysed 24 hours after transfection and the relative phosphorylation efficiencies of the p300 variants were analyzed by Western blotting using anti-FLAG antibodies. (D) The amino acid sequences of STP2 region (residues 828-889 of p300). Proline (P)-directed serine (S) and threonine (T) residues targeted in mutants p300Δ1,3,4/m1-m3 are changed to alanine, as denoted by asterisks. (E) Transient transfection and Western blot analyses of p300 mutants in HEK293 cells. Wild-type and point mutants of p300ΔSTP1,3,4 proteins were coexpressed with RUNX1/PEBP2-β as illustrated and analyzed by Western blotting as described in panel B.
Determination of phosphorylation targets within p300. (A) Schematic representation of domain structure of wild-type p300 and deletion mutants used in panels B and C. Four regions rich in proline-directed serine/threonine-rich residues are denoted STP1 to STP4. (B,C) Deletion variants of FLAG-p300 lacking single STP regions (B) or 3 STP regions (C) were transiently expressed in HEK293 cells in the presence or absence of exogenous RUNX1/PEBP2-β, as indicated. Cells were lysed 24 hours after transfection and the relative phosphorylation efficiencies of the p300 variants were analyzed by Western blotting using anti-FLAG antibodies. (D) The amino acid sequences of STP2 region (residues 828-889 of p300). Proline (P)-directed serine (S) and threonine (T) residues targeted in mutants p300Δ1,3,4/m1-m3 are changed to alanine, as denoted by asterisks. (E) Transient transfection and Western blot analyses of p300 mutants in HEK293 cells. Wild-type and point mutants of p300ΔSTP1,3,4 proteins were coexpressed with RUNX1/PEBP2-β as illustrated and analyzed by Western blotting as described in panel B.
To confirm the observations in Figure 3B, a reciprocal experiment was performed. A set of deletion constructs, each possessing only one STP region while lacking the remaining STP regions, was generated and tested (Figure 3C). Consistent with the data presented in Figure 3B, deletion constructs p300ΔSTP2,3,4 and p300ΔSTP1,3,4 showed significant degree of phosphorylation, indicating that STP1 and STP2 contain the necessary phosphorylation targets (Figure 3D). Having observed that phosphorylation target residues in RUNX1 are invariably preceded by basic amino acids, we reasoned that, if the same kinase were involved, it would recognize similar signatures in p300. To test this, we specifically targeted proline-directed serines or threonines in the STP2 region to generate a series of mutant constructs in the context of the p300ΔSTP1,3,4 variant (Figure 3E). As shown in Figure 3E, all single mutants showed modest effects on phosphorylation-induced mobility shift, but phosphorylation was significantly reduced in the compound mutants, p300ΔSTP1,3,4/m1,2 and p300ΔSTP1,3,4/m1,2,3. From these results, we conclude that p300 is phosphorylated at multiple sites primarily within STP2 and STP1 by a kinase that is recruited to chromatin-bound RUNX1/PEBP2-β complex. Importantly, the kinase involved also targeted proline-directed serines and threonines residues in p300, as in the case of RUNX1 phosphorylation, indicating the involvement of a single, common kinase.
Kinase-negative HIPK2 protein inhibits RUNX1/PEBP2-β–induced p300 phosphorylation
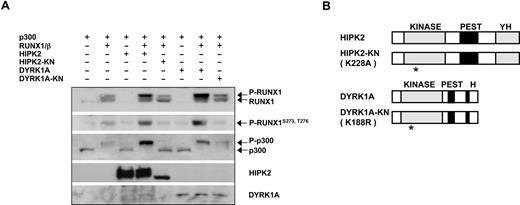
Analyses of amino acid signatures at the phosphorylation sites of RUNX1 and p300 revealed a number of candidate kinases. On the basis that RUNX1 and p300 are phosphorylated constitutively and all phosphorylated residues are downstream of basic amino acids, we tested 2 YAK family members, HIPK2 and DYRK1A.38,39 As shown in Figure 4A, wild-type HIPK2 or DYRK1A significantly enhanced RUNX1 and p300 phosphorylation. The phosphorylation of RUNX1 was further confirmed by the use of a polyclonal antibody specific for phosphorylated RUNX1 (Figures 4A, S3). Importantly, HIPK2 and DYRK1A increased phosphorylation of p300 greatly only in the presence of RUNX1/PEBP2-β, consistent with the notion that RUNX1/PEBP2-β complex is involved in the recruitment of these kinases. To identify the endogenous kinase responsible, we generated and tested kinase-negative forms of HIPK2 and DYRK1A (HIPK2-KN and DYRK1A-KN, respectively). Strong inhibition of RUNX1 and p300 phosphorylation was observed only when HIPK2-KN was overexpressed, whereas DYRK1A-KN had only marginal effects (Figure 4A). Collectively, these results suggest that, under physiologic conditions, HIPK2, rather than DYRK1A, is involved in the phosphorylation of PEBP2-β–induced RUNX1 and p300.
HIPK2 is involved in RUNX1/ PEBP2-β–dependent p300 phosphorylation. (A) FLAG-p300 was transfected with RUNX1 and PEBP2-β in the presence or absence of wild-type or kinase-negative FLAG-tagged HIPK2 or DYRK1A, as indicated. Immunoblotting was performed on transiently transfected HEK293 cell lysates using the indicated antibodies. (B) Schematic representation of FLAG-tagged HIPK2 and DYRK1A proteins with functional domains used in panel A. KINASE indicates kinase catalytic domain; PEST, domain rich in proline, glutamate, serine, and threonine; YH, domain rich in tyrosine and histidine; H, histidine repeat; KN, catalytically inactive mutant.
HIPK2 is involved in RUNX1/ PEBP2-β–dependent p300 phosphorylation. (A) FLAG-p300 was transfected with RUNX1 and PEBP2-β in the presence or absence of wild-type or kinase-negative FLAG-tagged HIPK2 or DYRK1A, as indicated. Immunoblotting was performed on transiently transfected HEK293 cell lysates using the indicated antibodies. (B) Schematic representation of FLAG-tagged HIPK2 and DYRK1A proteins with functional domains used in panel A. KINASE indicates kinase catalytic domain; PEST, domain rich in proline, glutamate, serine, and threonine; YH, domain rich in tyrosine and histidine; H, histidine repeat; KN, catalytically inactive mutant.
Inhibition of p300 phosphorylation by oncogenic chimeric protein PEBP2-β/CBF-β-SMMHC
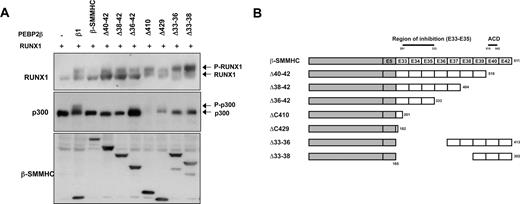
Chimeric RUNX1 and PEBP2-β proteins arising from leukemogenic chromosomal translocation are considered dominant inhibitors of RUNX1/PEBP2-β functions.33,40,41 To investigate whether these proteins disrupt the RUNX1/p300 phosphorylation cascade, we studied the effects of PEPB2-β/CBF-β-SMMHC (β-SMMHC).42,43 We coexpressed RUNX1, p300 and full-length β-SMMHC in COS-7 cells and observed a clear disruption of RUNX1 and p300 phosphorylation (Figure 5A). Because the PEBPβ moiety of β-SMMHC is capable of dimerizing with RUNX1, this observation implies that the disruption of RUNX1-p300 phosphorylation is mediated through the SMMHC moiety. To further investigate the role of the SMMHC moiety, we examined the inhibitory properties of a series of β-SMMHC deletants (Figure 5A). Interestingly, with the exception of the full-length β-SMMHC chimeric protein, various levels of RUNX1 phosphorylation were observed for all deletants (Figure 5A). Indeed, the removal of exons 40 to 42 at the carboxyl terminus, which contains the previously described assembly competent domain, readily restored RUNX1 phosphorylation.44,45 In addition, we observed a significant restoration of RUNX1 phosphorylation when a region between exons 33 and 36 (residues 201-333) was removed (Figure 5A). Interestingly, here we observed an apparent uncoupling of RUNX1 and p300 phosphorylation as several β-SMMHC deletants (Δ40-42, Δ38-42, Δ33-36, and Δ33-38) that only partially inhibit RUNX1 phosphorylation continue to suppress p300 phosphorylation (Figure 5A, middle panel). This is consistent with the notion that the phosphorylation of RUNX1 is upstream of p300 phosphorylation. In support of these observations, we observed also the disruption of p300 phosphorylation by RUNX1-ETO, another prominent leukemogenic chimeric protein targeting RUNX1/PEBP2-β (Figure S4).
Domain-dependent impairment of RUNX1 and p300 phosphorylation by leukemogenic PEBP2-β-SMMHC fusion protein. (A) Determination of regions within PEBP2-β-SMMHC necessary for the repression of p300 phosphorylation. Wild-type PEBP2-β or deletion variants of PEBP2-β-SMMHC fusion proteins were coexpressed in HEK293 cells with RUNX1 and FLAG-p300. Western blot analyses were performed using the indicated antibodies. (B) Schematic diagrams depicting different PEBP2-β-SMMHC deletion constructs used in panel A. The region delineated to be important in the inhibition of RUNX1 phosphorylation is also indicated.
Domain-dependent impairment of RUNX1 and p300 phosphorylation by leukemogenic PEBP2-β-SMMHC fusion protein. (A) Determination of regions within PEBP2-β-SMMHC necessary for the repression of p300 phosphorylation. Wild-type PEBP2-β or deletion variants of PEBP2-β-SMMHC fusion proteins were coexpressed in HEK293 cells with RUNX1 and FLAG-p300. Western blot analyses were performed using the indicated antibodies. (B) Schematic diagrams depicting different PEBP2-β-SMMHC deletion constructs used in panel A. The region delineated to be important in the inhibition of RUNX1 phosphorylation is also indicated.
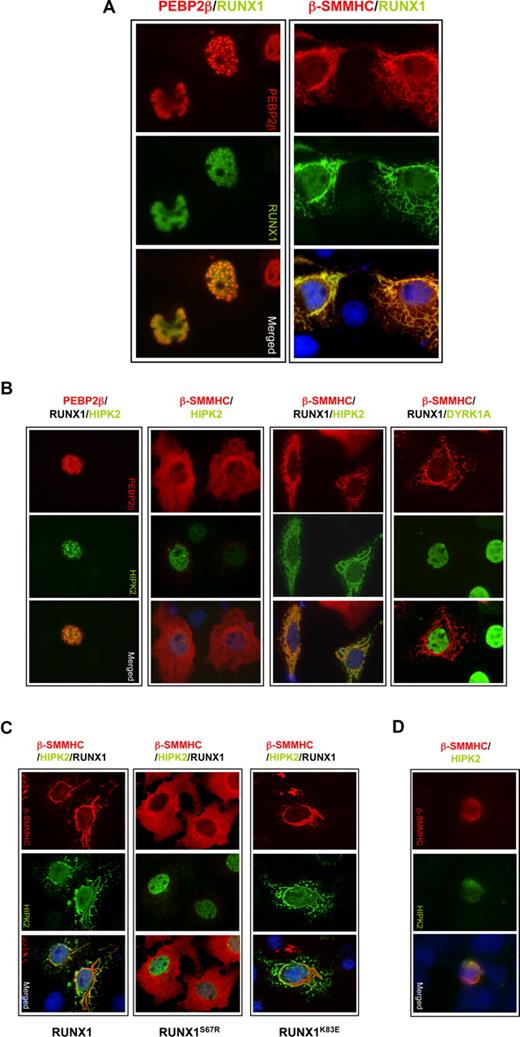
Coexpression of RUNX1 and β-SMMHC chimeric protein result in cytoplasmic sequestration of HIPK2
To better understand the functional relationship between RUNX1, PEBP2-β, and HIPK2, and how β-SMMHC may disrupt RUNX1 and p300 phosphorylation, we performed cellular localization experiments in COS-7 cells. These show that wild-type RUNX1 and PEBP2-β colocalized within the nucleus (Figure 6A). In contrast, ectopic expression of RUNX1 and β-SMMHC induced the formation of striking, filamentous structures in the cytoplasm that showed positivity for anti–PEBP2-β antibody. In this in vitro protein overexpression system, interaction with these filaments prominently resulted in the targeting of RUNX1 to the cytoplasm. Consistent with our biochemical data, in the presence of wild-type RUNX1 and PEBP2-β, HIPK2 is targeted to the nucleus where it colocalizes with PEBP2-β and, presumably, RUNX1 (Figure 6B first column). This localization pattern of HIPK2 was greatly disrupted with the introduction of β-SMMHC, where it became targeted to the β-SMMHC–related filaments (Figure 6B third column). Importantly, our data indicate that this cytoplasmic sequestration is dependent on RUNX1, as HIPK2 remained targeted to the nucleus in the presence of exogenous β-SMMHC alone (Figure 6B second column). In addition, we observed that this interaction is specific for HIPK2, as coexpression of DYRK1A with RUNX1 and β-SMMHC showed no such colocalization (Figure 6B fourth column). We further investigated the involvement of RUNX1 by testing the ability of dimerization-defective RUNX1S67R and DNA binding-defective RUNX1K83E mutants to mediate the sequestration of HIPK2 to β-SMMHC (Figure 6C). This revealed that RUNX1S67R, but not RUNX1K83E, was incapable of sequestering HIPK2. This observation provides strong evidence that the sequestration of HIPK2 is dependent on the ability of RUNX1 to directly interact with β-SMMHC, but not on its DNA binding. Finally, to establish that these observations are reproducible in relevant human leukemia-derived cells, the experiment was repeated on U937 premonocytic leukemic cells that express endogenous RUNX1. We observed clear cytoplasmic colocalization of HIPK2 to β-SMMHC filaments despite the relatively scant cytoplasm of U937 cells (Figure 6D). Together, these observations provide a compelling mechanism that accounts for the inhibition of RUNX1and p300 phosphorylation by the oncogenic β-SMMHC protein, namely, through the sequestration, and possible inactivation, of HIPK2.
Cytoplasmic sequestration of HIPK2 in filamentous structures formed by interaction between RUNX1 and PEBP2-β-SMMHC. (A) PEBP2-β-SMMHC forms filament-like cytoplasmic structures in the presence of RUNX1. COS-7 cells were transiently transfected with RUNX1 along with PEBP2-β or PEBP2-β-SMMHC and analyzed for RUNX1 (green) and PEBP2-β or PEBP2-β-SMMHC (red) by indirect immunofluorescence with anti-RUNX1 or -PEBP2-β antibodies accordingly, followed by Alex 488- or 594-conjugated secondary antibodies, respectively. Merged images showing overlapping localization are shown in the third panel. The nuclei were counterstained with DAPI (blue). All images: original magnification ×1000. (B) HIPK2 is specifically sequestrated in pronounced cytoplasmic RUNX1/PEBP2-β-SMMHC complexes. FLAG-HIPK2 is coexpressed together with wild-type PEBP2-β and RUNX1 (first column), β-SMMHC alone (second column), β-SMMHC and RUNX1 (third column), in COS-7 cells. Negative control experiment was performed using FLAG-DYRK1A (fourth column). FLAG-HIPK2 (green), FLAG-DYRK1A (green), PEBP2-β (red), or β-SMMHC (red) proteins were visualized by indirect immunofluorescence as described for panel A using anti-FLAG or -PEBP2-β antibodies, respectively. Merged images showing nuclei counterstained with DAPI (blue) are shown in the lowest panels. (C) Sequestration of HIPK2 to RUNX1/β-SMMHC complex is dependent on RUNX1 dimerization and not DNA-binding activity. FLAG-HIPK2 is coexpressed together with β-SMMHC and wild-type RUNX1 (first column), dimerization-defective RUNX1S67R (second column), or DNA binding-defective RUNX1K83E (third column) in COS-7 cells. Ectopically expressed β-SMMHC (red) and FLAG-HIPK2 (green) were visualized by indirect immunofluorescence, and in merged images nuclei were counterstained with DAPI (blue). (D) U937 cells coexpressing ectopic FLAG-HIPK2 and β-SMMHC proteins were stained and visualized by indirect immunofluorescence microscopy to reveal the sequestration of HIPK2 to prominent cytoplasmic β-SMMHC filamentous structures.
Cytoplasmic sequestration of HIPK2 in filamentous structures formed by interaction between RUNX1 and PEBP2-β-SMMHC. (A) PEBP2-β-SMMHC forms filament-like cytoplasmic structures in the presence of RUNX1. COS-7 cells were transiently transfected with RUNX1 along with PEBP2-β or PEBP2-β-SMMHC and analyzed for RUNX1 (green) and PEBP2-β or PEBP2-β-SMMHC (red) by indirect immunofluorescence with anti-RUNX1 or -PEBP2-β antibodies accordingly, followed by Alex 488- or 594-conjugated secondary antibodies, respectively. Merged images showing overlapping localization are shown in the third panel. The nuclei were counterstained with DAPI (blue). All images: original magnification ×1000. (B) HIPK2 is specifically sequestrated in pronounced cytoplasmic RUNX1/PEBP2-β-SMMHC complexes. FLAG-HIPK2 is coexpressed together with wild-type PEBP2-β and RUNX1 (first column), β-SMMHC alone (second column), β-SMMHC and RUNX1 (third column), in COS-7 cells. Negative control experiment was performed using FLAG-DYRK1A (fourth column). FLAG-HIPK2 (green), FLAG-DYRK1A (green), PEBP2-β (red), or β-SMMHC (red) proteins were visualized by indirect immunofluorescence as described for panel A using anti-FLAG or -PEBP2-β antibodies, respectively. Merged images showing nuclei counterstained with DAPI (blue) are shown in the lowest panels. (C) Sequestration of HIPK2 to RUNX1/β-SMMHC complex is dependent on RUNX1 dimerization and not DNA-binding activity. FLAG-HIPK2 is coexpressed together with β-SMMHC and wild-type RUNX1 (first column), dimerization-defective RUNX1S67R (second column), or DNA binding-defective RUNX1K83E (third column) in COS-7 cells. Ectopically expressed β-SMMHC (red) and FLAG-HIPK2 (green) were visualized by indirect immunofluorescence, and in merged images nuclei were counterstained with DAPI (blue). (D) U937 cells coexpressing ectopic FLAG-HIPK2 and β-SMMHC proteins were stained and visualized by indirect immunofluorescence microscopy to reveal the sequestration of HIPK2 to prominent cytoplasmic β-SMMHC filamentous structures.
Discussion
Protein phosphorylation in the nucleus is thought to be a fundamental regulatory mechanism for transcription, together with other covalent modifications, such as ubiquitination, sumoylation, acetylation, methylation, which allow nuclear factors to be dynamic with regard to their activity, subcellular localization, molecular interactions, and stability. Activation cascades in response to external signals, such as growth factors or DNA damage, or internal cues, such as the cell cycle, are extensively investigated along with their nuclear targets in a vast number of studies. However, many phosphorylation sites found in transcription factors or transcription-related factors remained to be characterized. Within these sites, phosphorylations of proline-directed serine or threonine are frequently observed. A recent proteomic study demonstrated that approximately 60% of phosphorylation sites identified in HeLa nuclear proteins under normal growth condition are represented by these Pro-directed Ser or Thr residues. However, little is currently known of the molecular mechanisms behind these phosphorylations, even though the involvement of protein-protein interactions has been speculated.46
Here we present results demonstrating that the heterodimerization between RUNX1 and PEBP2-β leads to the phosphorylation of RUNX1 and one of its coactivators, p300. Furthermore, these are dependent on the DNA binding activity of RUNX1 and mediated through the chromatin-associated protein kinase HIPK2. In addition, the involvement of the proline-rich PY motif of RUNX1 is also implicated. Together, these observations fit best a model in which the attachment of RUNX1/PEBP2-β to the chromatin provides a stable interaction between the PY-motif of RUNX1 and HIPK2.
We observed that RUNX1Δ260-280 was resistant to phosphorylation and turned our attention to the proline-directed serine and threonine residues within this region, as previous studies have identified phosphorylation targets within this region.35,47,48 Site-directed mutation study elucidates Ser249, Thr273, and Ser276 as the targets of phosphorylation. Computer software analysis of the signature motifs returned several candidate kinases that could be responsible for this phosphorylation, including MAPK, GSK3, and members of the CDK family. However, we noted that these residues are downstream of basic amino acids and that RUNX1 phosphorylation was observed in the absence of extracellular stimuli. These led us to the identification of HIPK2 as the kinase responsible for the phosphorylation of RUNX1 in our system. In a recent study, Ser276 was reported to be also a target of Cdk1 and Cdk2 phosphorylation, which marks RUNX1 for Cdc20-dependent anaphase-promoting complex degradation.49 Interestingly, in their system, the authors reported residual phosphorylation of RUNX1 persisted in the presence of multiple Cdk-specific inhibitors, suggesting that Ser276 can serve concurrently as the target for several kinases.49
In a related study, Aikawa et al recently reported the phosphorylation of RUNX1 and p300 by HIPK2 and HIPK1.28 Using a proteomic approach, they observed the phosphorylation of Ser249 and Ser276 in phosphorylated RUNX1 proteins isolated from myeloid cells.28 Although the p300 mapping data are largely complimentary, significant differences between the 2 experimental approaches should be noted. In their study, Aikawa et al performed immunoprecipitation experiments to delineated p300 regions interacting with RUNX1 and HIPK2, 49 whereas we chose a functional readout in our mapping experiments, namely, the detection of phosphorylation-induced band shift. The strength in this approach is that it takes into account the participation of other, yet undetermined upstream events and interactions. Indeed, we have further delineated the involvement of p300's bromo- and SRC1-interacting domains in the overall phosphorylation process through this approach (Figure S2).
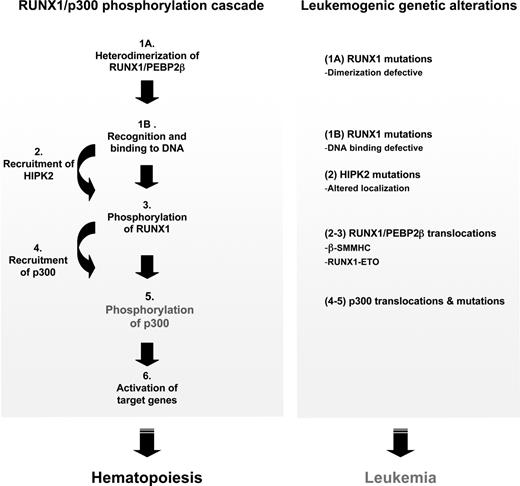
Importantly, the phosphorylation of the identified RUNX1 residues appears prerequisite to p300 phosphorylation, suggesting a tight coupling between these phosphorylation events. Phosphorylation of these RUNX1 residues has previously been linked to both RUNX1 and p300 transcriptional activities.28,36 These give rise to a model in which RUNX1 orchestrates its downstream genetic program by directing p300 through its binding to key promoters and activating p300's HAT activity by phosphorylation. The data presented here extend this model by demonstrating that heterodimerization with PEBP2-β and DNA binding are critical requirements leading to RUNX1 phosphorylation (Figure 7), hence implicating the binding of RUNX1/PEBP2-β to its cognate site as an initiating event in the RUNX1/p300 phosphorylation cascade.
Dimerization-induced RUNX1/p300 phosphorylation cascade and its proposed central role in hematopoiesis and leukemia. A schematic that summarizes the findings reported in this study. The left panel illustrates the initiation of a phosphorylation cascade after the dimerization of RUNX1 with PEBP2-β at functional binding sites. This leads to the activation of specific target genes and enables RUNX1/PEBP2-β to act as master regulators of hematopoiesis. The right panel shows known leukemogenic genetic alterations in the components of this proposed cascade, indicating its central importance. The targeting of this phosphorylation cascade may represent a common mechanism in the genesis of RUNX1-related leukemia.
Dimerization-induced RUNX1/p300 phosphorylation cascade and its proposed central role in hematopoiesis and leukemia. A schematic that summarizes the findings reported in this study. The left panel illustrates the initiation of a phosphorylation cascade after the dimerization of RUNX1 with PEBP2-β at functional binding sites. This leads to the activation of specific target genes and enables RUNX1/PEBP2-β to act as master regulators of hematopoiesis. The right panel shows known leukemogenic genetic alterations in the components of this proposed cascade, indicating its central importance. The targeting of this phosphorylation cascade may represent a common mechanism in the genesis of RUNX1-related leukemia.
In evaluating the functional relevance of this phosphorylation cascade, we noted that all leukemogenic RUNX1 mutations tested in our experiments inhibited p300 phosphorylation, which further indicates its central importance. To further address this connection, we studied the effects of 2 leukemogenic chimeric proteins, PEBP2-β-SMMHC and RUNX1-ETO, products of the highly frequent inv(16) and t(8;21) chromosomal translocations, respectively. These leukemogenic proteins are known to disrupt normal RUNX1/PEBP2-β functions as dominant-negative proteins that also possess additional gains in function. Our results clearly demonstrate the disruption of RUNX1/p300 phosphorylation cascade by β-SMMHC. In the case of RUNX1-ETO, which lacks the C-terminal region of RUNX1 containing the target residues and the PY motif, we expected it to be ineffective in inducing p300 phosphorylation. Consistent with this prediction, RUNX1-ETO did not induce p300 phosphorylation in the presence of PEBP2-β (Figure S4). These observations shed new light on the molecular mechanisms by which chimeric proteins interrupt normal RUNX1/PEBP2-β functions to initiate leukemia. Moreover, that multiple genetic alterations targeting RUNX1/PEBP2-β resulted in the impairment of RUNX1/p300 phosphorylation strongly implicates the targeting of dimerization-initiated RUNX1/p300 phosphorylation cascade as part of a common mechanism in RUNX1-related leukemogeneses.
In the case of β-SMMHC, we further observed that coexpression with RUNX1 resulted in conspicuous multimerization in the cytoplasm, which coincides with the mislocalization of RUNX1 and HIPK2. Several previous studies have reported that β-SMMHC altered subcellular localizsation of RUNX1, either as large subnuclear inclusion bodies, or as cytoplasmic deposits on cytoskeletal filaments.29,50,51 Here we showed that the multimerized filamentous structure formed by β-SMMHC, when cotransfected with RUNX1, specifically sequesters HIPK2 to the cytoplasm. Although it is true that our observations are made in an in vitro, overexpression system, it is clear that the observed interactions are highly reproducible and specific for RUNX1 and HIPK2: (1) RUNX1 does not colocalize in the cytoplasm unless β-SMMHC is cotransfected (Figure 6A); (2) unless RUNX1 is cotransfected, HIPK2 does not colocalize with β-SMMHC and RUNX1 in the cytoplasm (Figure 6B); (3) only HIPK2, and not the related DYRK1A, can be colocalized with β-SMMHC and RUNX1 (these presumed ternary complexes discriminate HIPK2 from DYRK1A; Figure 6B); (4) a single amino acid substitution mutant of RUNX1 that eliminates its heterodimerization ability loses the ability to form ternary complex (Figure 6C); and (5) a DNA-binding negative form of RUNX1 retains the ability to form the ternary complex (Figure 6C). Although the expression levels and distribution of these proteins may be different in leukemic cells expressing β-SMMHC, the remarkable specificities of these interactions would argue strongly that the function of RUNX1 and HIPK2 would be impaired by β-SMMHC in leukemic cells.
Consistent with its role in RUNX1 and p300 phosphorylation, double-deficient hipk1−/−hipk2−/− mice show various abnormalities, including impaired hematopoiesis and blood vessel formation, contributing to prenatal death.28,52 It has been proposed that HIPK2 fulfills its many different functions at least in part through its highly structured subcellular localization.53 Indeed, 2 mutations at the speckle-retention signal domain of HIPK2, resulting in abnormal subcellular localization, have recently been reported in AML patients.54 Therefore, the profound alteration in HIPK2 cellular distribution induced by RUNX1/β-SMMHC would, in all probability, disrupt normal HIPK2 functions.
Prominent among HIPK2's many reported functions is its role in phosphorylating p53 at Ser46 after DNA damage, thereby activating p53-mediated apoptosis.55,56 Notably, it has been reported that β-SMMHC attenuates the induction of p53 after DNA damage, resulting in reduced apoptosis in hematopoietic cells.57 Therefore, our observation of HIPK2 sequestration by RUNX1/β-SMMHC oligomers provides a key missing link between these earlier observations. Furthermore, these observations suggest that, in addition to its disruption of RUNX1 functions, β-SMMHC confers an additional, and important, gain of function to preneoplastic hematopoietic cells, namely, the attenuation of HIPK2/p53 apoptotic pathway.
In conclusion, our data have shed new light on the important question of how RUNX1 directs p300 to its target genes as it coordinates definitive hematopoiesis and other functions. Our elucidation of a phosphorylation cascade that is initiated by the heterodimerization and DNA binding of RUNX1/PEBP2-β adds to an emerging picture in which key regulators of hematopoiesis, such as RUNX1, C/EBPβ, and Myb, use a common mechanism to direct downstream genetic program, ie, recruitment of HIPK2 for the phosphorylation of p300. True to its integral nature, mutations in each component in this pathway have now been reported in human AML (Figure 7), highlighting the possibility that impairment of RUNX1/p300 phosphorylation is a common mechanism to most, if not all, RUNX1-related leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Motomi Osato and Gang Huang for critical reading of the manuscript.
This work was supported by A*STAR (the Agency for Science, Technology and Research), Singapore.
Authorship
Contribution: H.-J.W. designed and performed research, analyzed data, and wrote the paper; D.C.-C.V. analyzed data and wrote the paper; S.-C.B. provided part of the funding; and Y.I. designed research, analyzed data, and provided funding.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yoshiaki Ito, Genomics and Genetics Division, Institute of Molecular and Cell Biology, Agency for Science, Technology and Research, 61 Biopolis Drive, Proteos, Singapore 138673; e-mail: itoy@imcb.a-star.edu.sg.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal