Abstract
The exact prognostic role of TP53 mutations (without 17p deletion) and any impact of the deletion without TP53 mutation in CLL are unclear. We studied 126 well-characterized CLL patients by direct sequencing and DHPLC to detect TP53 mutations (exons 2-11). Most patients with 17p deletions also had TP53 mutations (81%). Mutations in the absence of 17p deletions were found in 4.5%. We found a shorter survival for patients with TP53 mutation (n = 18; P = .002), which was more pronounced when analyzed from the time point of mutation detection (6.8 vs 69 months, P < .001). The survival was equally poor for patients with deletion 17p plus TP53 mutation (7.6 months, n = 13), TP53 mutation only (5.5 months, n = 5), and 17p deletion only (5.4 months, n = 3). The prognostic impact of TP53 mutation (HR 3.71) was shown to be independent of stage, VH status, and 11q and 17p deletion in multivariate analysis. Serial samples showed evidence of clonal evolution and increasing clone size during chemotherapy, suggesting that there may be patients where this treatment is potentially harmful. TP53 mutations are associated with poor sur-vival once they occur in CLL. The de-monstration of clonal evolution under selective pressure supports the biologic significance of TP53 mutations in CLL.
Introduction
Chronic lymphocytic leukemia (CLL) is the most frequent type of leukemia in the Western world and is characterized by a highly variable clinical course.1,2 Treatment of early-stage patients has not been shown to prolong survival.3 Traditionally, therapy has been used for advanced-stage or symptomatic disease and consisted of chemotherapy with alkylating agents, purine analogs, or a combination. Over recent years, highly effective and potentially curative approaches, such as the combination of antibodies with chemotherapy and autologous or allogeneic stem cell transplantation, have been developed.
The deletion 17p affecting the TP53 gene has been associated with failure after treatment with alkylating agents, purine analogs, and rituximab.4-8 In a chromosome banding study of patients treated in a prospective trial based on alkylating agents, 17p aberrations were the only chromosomal aberration of prognostic relevance.6 An interphase-fluorescence in situ hybridization (FISH) study also showed that patients whose CLL cells showed a 17p deletion had significantly shorter survival times than patients without this aberration, and a relationship was found between the deletion and the response to treatment.5 Although 56% of patients without 17p deletion went into remission after treatment with purine analogs, none of the patients with 17p deletion showed a response. Similarly, the monoclonal anti-CD20 antibody rituximab did not show efficacy in CLL with 17p deletion.7 Results from the Eastern Cooperative Oncology Group 2997 trial, the German CLL4 trial and the United Kingdom Leukaemia Research Fund CLL4 confirm the results showing that the deletion of 17p is associated with poor response to fludarabine and alkylators as well as poor event-free and overall survival.9-11 Although the group of patients with deletion 17p is quite small, the detection of this chromosome aberration is of paramount importance because these patients are unlikely to respond to current standard chemotherapy.
There is growing evidence that durable response can be achieved in CLL with 17p deletion using, for example, the monoclonal anti-CD52 antibody alemtuzumab.8,12 In a retrospective series of CLL cases mostly refractory to fludarabine therapy treatment with intravenous alemtuzumab, resulted in a complete or partial response in 11 of 36 (31%) and in 6 of 15 (40%) of patients with TP53 mutations or deletions.12 Similar results have been reported from a larger prospective trial.13 Other treatment approaches for patients with impaired p53 may include high-dose steroids with alemtuzumab, lenalidomide, or flavopiridol.14
Mutations of TP53 have been found in 4% to 37% of patients with CLL.9,12,15-22 The highest incidence is seen in patients with fludarabine-refractory CLL.22 The presence of mutations in TP53 has been associated with poor prognosis in a number of retrospective studies, but the association of 17p deletion and TP53 mutation has led to the pooling of mutations with 17p deletion (usually the majority) and cases without 17p deletion.15,18 Therefore, the exact prognostic relevance of TP53 mutations (in relation to 17p deletion) has not been documented, and most previous studies did not provide comprehensive molecular genetic characterizations, including FISH and VH mutational status. A recent analysis from a prospective trial failed to show a prognostic significance of monoallelic TP53 mutations, but there are some fundamental differences in this study compared with the data presented here, including the limitation to p53 exons 5 to 9, denaturing gradient gel electrophoresis, and, most importantly, the grouping of unconfirmed mutations (DGGE profile abnormal but no mutation detected on sequencing) and polymorphisms in the “mutation” group.9 In addition to the deletion and mutation of TP53, functional defects of p53 have been reported, and there is growing interest in how to best assess p53 defects.23,24
The thorough investigation of the role of TP53 mutations in CLL is particularly important in light of the prognostic role of the deletion 17p and its consequence on patient management. The availability of high-throughput techniques for mutation screening as, for example, denaturing high performance liquid chromatography (DHPLC), will allow using mutation detection in larger cohorts of patients and in a timely fashion.
The aim of the current study was to assess the role of TP53 mutations in a well-characterized patient cohort with mature follow-up and detailed molecular genetic analysis.
Methods
Patients
Between October 1990 and August 1998, 126 CLL patients from a single institution (Med. Klinik und Poliklinik V, University of Heidelberg, Heidelberg, Germany) were enrolled in this study and followed with regard to survival. This study was approved by the institutional review board at the University of Ulm and performed in accordance with the Declaration of Helsinki. Genetic data have been described previously, together with detailed clinical characteristics.25,26 Treatment was not in controlled trials, and standard clinical criteria were used for the initiation of therapy that was usually alkylator based at first line and mostly fludarabine at second line. At the time of mutation analysis, 27 of 126 patients had received chemotherapy (21%). Median time from diagnosis to first mutation analysis was 0.65 months. Seventy-six percent of cases were studied within 12 months from diagnosis. We were particularly interested to analyze serial samples in cases with available material. The selection of the cohort was influenced by the availability of DNA. To obtain adequate numbers of patients with 17p deletion, we selected these patients (17p-) from a larger cohort of unselected patients.25 This cohort is therefore not representative of a particular clinical situation.
At the time of enrollment, ages ranged from 30 to 87 years (median, 62 years): 59 patients were at Binet stage A, 37 were at Binet stage B, and 28 were at Binet stage C. Staging information was incomplete for 2 patients. Estimated median survival time of the cohort was 70 months. The median follow-up was 58.9 months.27 During this study period, 59 patients died.
Genetic analysis
FISH analysis and VH sequencing were performed in all cases as previously described.4,5,25,26 To define the cut-off level for the diagnosis of the p53 gene deletion hybridization experiments of blood specimens from probands were performed. The cut-off level was defined by the mean plus 3 SD of the frequency of control cells exhibiting only one p53 signal.5 To assess genetic complexity, we analyzed a part of the cohort and additional patients with known 17p/TP53 mutation status (n = 60) by array-comparative genomic hybridization (CGH) and single nucleotide polymorphism (SNP) arrays. Array-CGH was performed as previously described.28 The Affymetrix 500k SNP array-set was used on samples separated for CD19 by immunomagnetic beads. The CD19 negative fraction was used as intraindividual reference. DNA was hybridized to the Affymetrix 500k Array Set (Santa Clara, CA), and data were analyzed by the Copy Number Analyser for Gene Chip (CNAG). A germ line homology of 98% was used as the cut-off between VH mutated and VH unmutated cases.26
TP53 sequence analysis
We analyzed all samples by automated fluorescent sequencing using the Big Dye Terminator Kit and ABI 3100 sequencer (exons 2-11, Applied Biosystems, Foster City, CA). The primer sequences are available on request. The primers were designed to cover all coding exons and intron-exon boundaries.
DHPLC
DHPLC was used to identify samples containing mutations in exons 2 to 11 (coding region of p53; n = 80). WAVE analysis is based on the temperature-dependent differences in column-retention time of polymerase chain reaction products generated from homoduplex (wild-type) and heteroduplex (mutated) DNA, resulting in the presence of distorted or additional peaks when mutations are present. To detect homozygous mutations or mutations in samples that had undergone allelic loss, each test sample was mixed with a known wild-type DNA control (20%). Assay conditions were optimized for analysis of each polymerase chain reaction fragment with samples previously characterized by DNA sequencing. All samples were denatured and cooled slowly to room temperature before WAVE analysis to maximize heteroduplex formation (95° 2 minutes, then decrease 1° every 40 seconds, to 45° [30 minutes]). Oligonucleotide primers and DHPLC conditions were chosen as previously described with small modifications (available on request).29,30
Statistical analysis
The primary endpoint was survival from the time of diagnosis and/or genetic testing of the index sample. Survival time distributions were plotted using Kaplan-Meier estimates. Median duration of follow-up was calculated according to the method of Korn.27 Group-wise comparisons of distributions of clinical, laboratory, and genetic data were performed with the Fisher exact test (categorical variables). All tests were 2-sided. An effect was considered statistically significant when P was less than .05. Statistical analyses were performed with GraphPad Prism, version 3.00 (GraphPad Software, San Diego, CA). Prognostic factors for overall survival were analyzed using Cox proportional hazards regression in 113 patients where the full dataset was available. Besides the TP53 mutation (without deletion 17p), the models included Binet stage and prior therapy at the time of mutation analysis, as well as molecular genetics (13q-, 11q-, 17p-, and VH mutation status).
Results
To assess whether the cohort was representative, we analyzed the survival data according to our published hierarchical model and confirmed the previous results. The median survival from time of diagnosis for the groups with 17p deletion, 11q deletion (without 17p-), unmutated VH status (without 11q-, 17p-), and mutated VH status (without 11q-, 17p-) were 36, 63, 73, and 92 months (P = .001).
Incidence of TP53 mutation
We found 19 TP53 mutations in 18 patients (Tables 1,2) by both DHPLC and sequencing. The detection of mutations was associated with more advanced disease (higher Binet stage at study (P = .01)) and a poor risk profile (prior chemotherapy (P = .001), 17p deletion (P < .001), and an unmutated VH status (P = .03)). Although the distribution of Binet stage did not differ between patients with or without TP53 mutations at the time of diagnosis, there was a significant difference at the time point of mutation detection. TP53 mutations in the absence of 17p deletions were found in 4.5% of CLL patients (5 of 110). Of the cases with TP53 mutation without 17p deletion, 4 of 5 cases had an unmutated VH mutation status. No patients with trisomy 12 were found to have a TP53 mutation. Two patients with 11q deletion (without 17p deletion) had TP53 mutations (23 total cases with 11q deletion).
Clinical and genetic characteristics of CLL patients in the cohort stratified by TP53 mutation
| Characteristic/category . | All . | WT TP53 . | Mutation TP53 . | Significance, P . |
|---|---|---|---|---|
| Stage (at diagnosis) | ||||
| Mean age, y | 61.5 | 64.4 | 61.1 | |
| Male, % | 60 | 58 | 67 | |
| Female, % | 40 | 42 | 33 | NS |
| Binet A, % | 56 | 57 | 44 | |
| Binet B, % | 25 | 23 | 39 | |
| Binet C, % | 11 | 12 | 6 | |
| NA, % | 8 | 7 | 11 | NS |
| Stage (at study) | ||||
| Binet A, % | 47 | 53 | 17 | |
| Binet B, % | 29 | 27 | 44 | |
| Binet C, % | 22 | 19 | 39 | |
| NA, % | 2 | 2 | 0 | .015 |
| Chemotherapy prior to mutation analysis | ||||
| None, % | 78 | 83 | 44 | |
| ≥ 1, % | 21 | 16 | 56 | |
| NA, % | 1 | 1 | 0 | < .001 |
| FISH | ||||
| Normal, % | 16 | 18 | 6 | |
| 13q- single, % | 29 | 31 | 11 | |
| 11q-, % | 19 | 20 | 11 | .09 |
| 12q+, % | 16 | 19 | 0 | .07 |
| 17p-, % | 13 | 3 | 72 | < .001 |
| Other, % | 8 | 9 | 0 | |
| VH status | ||||
| Mutated, % | 32 | 35 | 11 | |
| Unmutated, % | 59 | 55 | 83 | |
| NA, % | 10 | 10 | 6 | .03 |
| Total, % | 100 (n = 126) | 100 (n = 108) | 100 (n = 18) |
| Characteristic/category . | All . | WT TP53 . | Mutation TP53 . | Significance, P . |
|---|---|---|---|---|
| Stage (at diagnosis) | ||||
| Mean age, y | 61.5 | 64.4 | 61.1 | |
| Male, % | 60 | 58 | 67 | |
| Female, % | 40 | 42 | 33 | NS |
| Binet A, % | 56 | 57 | 44 | |
| Binet B, % | 25 | 23 | 39 | |
| Binet C, % | 11 | 12 | 6 | |
| NA, % | 8 | 7 | 11 | NS |
| Stage (at study) | ||||
| Binet A, % | 47 | 53 | 17 | |
| Binet B, % | 29 | 27 | 44 | |
| Binet C, % | 22 | 19 | 39 | |
| NA, % | 2 | 2 | 0 | .015 |
| Chemotherapy prior to mutation analysis | ||||
| None, % | 78 | 83 | 44 | |
| ≥ 1, % | 21 | 16 | 56 | |
| NA, % | 1 | 1 | 0 | < .001 |
| FISH | ||||
| Normal, % | 16 | 18 | 6 | |
| 13q- single, % | 29 | 31 | 11 | |
| 11q-, % | 19 | 20 | 11 | .09 |
| 12q+, % | 16 | 19 | 0 | .07 |
| 17p-, % | 13 | 3 | 72 | < .001 |
| Other, % | 8 | 9 | 0 | |
| VH status | ||||
| Mutated, % | 32 | 35 | 11 | |
| Unmutated, % | 59 | 55 | 83 | |
| NA, % | 10 | 10 | 6 | .03 |
| Total, % | 100 (n = 126) | 100 (n = 108) | 100 (n = 18) |
NA indicates not applicable; and NS, not significant.
TP53 mutation profile in CLL
| Patient no. . | Time to study, mo . | 17p-(%) by FISH . | Exon . | Codon . | bp exchange . | AA exchange . | Mutation type . | Transition (Ts)/transversion (Tv) . | CpG . | Mutation . | Mutation . | Frequency according to database* . | Predicted p53 residual activity, %* . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 152 | 59 | 37 | 6 | 193 | cat-cgt | His-Arg | PM | Ts | p.His193Arg | c.578A>G | 86 | 10,15 | |
| 36 | 0 | 55 | 5 | 179 | cat-tat | His - Tyr | PM | Ts | p.His179Tyr | c.535C>T | 131 | 13,27 | |
| 122 | 37 | 65 | 7 | 246 | atg-gtg | Met-Val | PM | Ts | p.Met246Val | c.736A>G | 57 | 0,00 | |
| 62 | 5 | 75 | 5 | 179 | cat-tat | His - Tyr | PM | Ts | p.His179Tyr | c.535C>T | 131 | 13,27 | |
| 17 | 0 | 79 | 8 | 273 | cgt-cat | Arg - His | PM | Ts | Yes | p.Arg273His | c.818G>A | 793 | 1,01 |
| 116 | 0 | 80 | 5 | 141 | tgc-tac | Cys-Tyr | PM | Ts | p.Cys141Tyr | c.422G>A | 105 | 9,84 | |
| 12 | 12 | 87 | 4 | 98 | ccttcc-cc**** | Frameshift | DEL | — | p.Pro98fs* | c.294-297delTTCC | NA | 0 | |
| 61 | 0 | 87 | 6 | 209 | aga-**a | Frameshift | DEL | — | p.Arg209fsX5 | c.626_627delGA | 14 | 0 | |
| 31 | 9 | 89 | 5 | 179 | cat-cgt | His - Arg | PM | Ts | p.His179Arg | c.536A>G | 150 | 13,02 | |
| 9 | 39 | 97 | 5 | 167 | cag-tag | Stopcodon | PM | Ts | p.Gln167X | c.499C>T | 42 | 0 | |
| 150 | 75 | 98 | 9 | 317 | cag-tag | Stopcodon | PM | Ts | p.Gln317X | c.949C>T | 25 | 0 | |
| 3 | 79 | — | 7 | 232 | atc-ttc | Ile-Phe | PM | Tv | p.Ile232Phe | c.694A>T | 11 | 2,46 | |
| 22 | 17 | — | 6 | 214 | cat-cgt | His-Arg | PM | Ts | p.His214Arg | c.641A>G | 69 | 3,12 | |
| 35 | 54 | — | 4 | 62 | gaa-taa | Stopcodon | PM | Tv | p.Glu62X | c.184G>T | 9 | 0 | |
| 54 | 0 | — | 5 | 163 | tac-tgc | Tyr-Cys | PM | Ts | p.Tyr163Cys | c.488A>G | 145 | 18,30 | |
| 149 | 89 | — | 5 | 139 | aag-aat | Lys-Asp | PM | Tv | p.Lys139Asn | c.417G>T | 6 | 10,40 | |
| 149 | 89 | — | 5 | 176 | tgc-ttc | Cys-Phe | PM | Tv | p.Cys176Phe | c.527G>T | 192 | 22,88 | |
| 7 | 122 | Yes | 5 | 181 | cgc-cac | Arg-His | PM | Ts | p.Arg181His | c.542G>A | 35 | 34,07 | |
| 26 | 8 | Yes | Intron5 | 187-1 | tag-tac | pot. Splice site | PM | Tv | c.560-1G>C | NA | 0 |
| Patient no. . | Time to study, mo . | 17p-(%) by FISH . | Exon . | Codon . | bp exchange . | AA exchange . | Mutation type . | Transition (Ts)/transversion (Tv) . | CpG . | Mutation . | Mutation . | Frequency according to database* . | Predicted p53 residual activity, %* . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 152 | 59 | 37 | 6 | 193 | cat-cgt | His-Arg | PM | Ts | p.His193Arg | c.578A>G | 86 | 10,15 | |
| 36 | 0 | 55 | 5 | 179 | cat-tat | His - Tyr | PM | Ts | p.His179Tyr | c.535C>T | 131 | 13,27 | |
| 122 | 37 | 65 | 7 | 246 | atg-gtg | Met-Val | PM | Ts | p.Met246Val | c.736A>G | 57 | 0,00 | |
| 62 | 5 | 75 | 5 | 179 | cat-tat | His - Tyr | PM | Ts | p.His179Tyr | c.535C>T | 131 | 13,27 | |
| 17 | 0 | 79 | 8 | 273 | cgt-cat | Arg - His | PM | Ts | Yes | p.Arg273His | c.818G>A | 793 | 1,01 |
| 116 | 0 | 80 | 5 | 141 | tgc-tac | Cys-Tyr | PM | Ts | p.Cys141Tyr | c.422G>A | 105 | 9,84 | |
| 12 | 12 | 87 | 4 | 98 | ccttcc-cc**** | Frameshift | DEL | — | p.Pro98fs* | c.294-297delTTCC | NA | 0 | |
| 61 | 0 | 87 | 6 | 209 | aga-**a | Frameshift | DEL | — | p.Arg209fsX5 | c.626_627delGA | 14 | 0 | |
| 31 | 9 | 89 | 5 | 179 | cat-cgt | His - Arg | PM | Ts | p.His179Arg | c.536A>G | 150 | 13,02 | |
| 9 | 39 | 97 | 5 | 167 | cag-tag | Stopcodon | PM | Ts | p.Gln167X | c.499C>T | 42 | 0 | |
| 150 | 75 | 98 | 9 | 317 | cag-tag | Stopcodon | PM | Ts | p.Gln317X | c.949C>T | 25 | 0 | |
| 3 | 79 | — | 7 | 232 | atc-ttc | Ile-Phe | PM | Tv | p.Ile232Phe | c.694A>T | 11 | 2,46 | |
| 22 | 17 | — | 6 | 214 | cat-cgt | His-Arg | PM | Ts | p.His214Arg | c.641A>G | 69 | 3,12 | |
| 35 | 54 | — | 4 | 62 | gaa-taa | Stopcodon | PM | Tv | p.Glu62X | c.184G>T | 9 | 0 | |
| 54 | 0 | — | 5 | 163 | tac-tgc | Tyr-Cys | PM | Ts | p.Tyr163Cys | c.488A>G | 145 | 18,30 | |
| 149 | 89 | — | 5 | 139 | aag-aat | Lys-Asp | PM | Tv | p.Lys139Asn | c.417G>T | 6 | 10,40 | |
| 149 | 89 | — | 5 | 176 | tgc-ttc | Cys-Phe | PM | Tv | p.Cys176Phe | c.527G>T | 192 | 22,88 | |
| 7 | 122 | Yes | 5 | 181 | cgc-cac | Arg-His | PM | Ts | p.Arg181His | c.542G>A | 35 | 34,07 | |
| 26 | 8 | Yes | Intron5 | 187-1 | tag-tac | pot. Splice site | PM | Tv | c.560-1G>C | NA | 0 |
PM indicates point mutation; DEL, deletion; CpG, missense mutation at CpG; and —, not applicable.
Frequency and predicted p53 residual activity were retrieved from the 2007_R1 release of the UMD_TP53 Mutation database (http://p53.free.fr).30 The frequency refers to the number of reported mutations in all cancers.
Profile of TP53 mutations
Eighteen of 19 mutations were located in the DNA binding domain of exons 4 to 9. We detected 2 small deletions and one splice site mutation. Of the most common hot spot mutations in cancer, we found only one mutation (p.Arg273His, c.818G > A), suggesting that the common hot spot mutations may not be as prevalent in CLL (Table 2). Although transitions were common (12 of 19), transitions at CpG were rare (1 of 19) (Table 2). The mutation pattern of cases without 17p deletion showed a trend toward more transversions in the patients with mutation of TP53 in the absence of 17p deletion (4 of 6 transversions vs 1 of 11 in cases with deletion and mutation) (P = .03). The residual (predicted) p53 activity in these groups was similar (mean ± SD: 9.527 ± 9.357, TP53 mutation vs 7.886 ± 10.17, deletion and TP53 mutation).30
Genetic complexity and 17 deletion/TP53 mutation
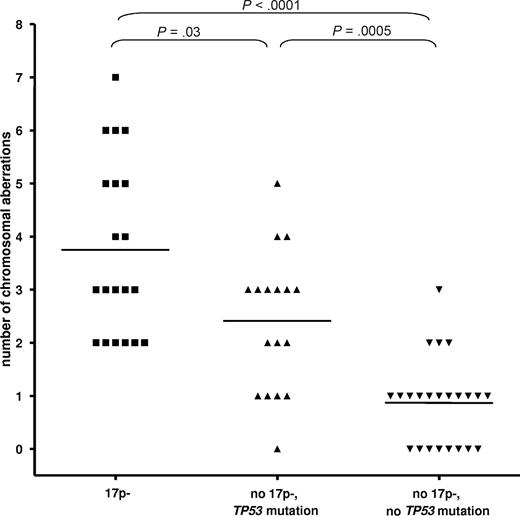
We assessed the genetic complexity in relation to 17p- and TP53 mutation status by the use of array-CGH and SNP-array analysis (500k) in a cohort of 60 CLL patients. The different techniques showed similar results, so we combined the data for the analysis. To arrive at meaningful numbers, we included patients from other cohorts. The analysis of the number of chromosomal gains or losses showed a significant increase of the number of cytogenetic aberrations in the cases with 17p deletion (3.75 ± 1.65; P < .001) and TP53 mutations (no 17p deletion, 2.4 ± 1.3; P = .001) vs cases without either abnormality (0.9 ± 0.8; Figure 1). CLL cases with 17p deletion showed a higher complexity compared with cases with the mutation of TP53 only (P = .03).
CLL with 17p deletion and TP53 mutation show an increased but differential degree of genetic complexity. The analysis of the number of chromosomal gains or losses showed a significant increase of the number of cytogenetic aberrations in the cases with 17p deletion (mean ± SD, 3.75 ± 1.65) and TP53 mutations (no 17p deletion, 2.4 ± 1.3) versus cases without either abnormality (0.9 ± 0.8; P < .001 and P = .0005, respectively). CLL cases with 17p deletion showed a higher complexity compared with cases with the mutation of TP53 only (P = .03).
CLL with 17p deletion and TP53 mutation show an increased but differential degree of genetic complexity. The analysis of the number of chromosomal gains or losses showed a significant increase of the number of cytogenetic aberrations in the cases with 17p deletion (mean ± SD, 3.75 ± 1.65) and TP53 mutations (no 17p deletion, 2.4 ± 1.3) versus cases without either abnormality (0.9 ± 0.8; P < .001 and P = .0005, respectively). CLL cases with 17p deletion showed a higher complexity compared with cases with the mutation of TP53 only (P = .03).
Acquisition of mutations over time
Because there remain questions about the acquisition of TP53 mutations, their occurrence in relation to therapy, and their impact on response to treatment, we were particularly interested to assess the mutations in serial samples over many years. In addition, the demonstration of clonal evolution in cases with monoallelic inactivation can be considered as evidence of biologic significance. In cases with TP53 mutation and serial samples, we tracked the first time point with the mutation by going back to previous samples by sequencing and sensitive DHPLC. In cases without the detection of the mutation with these techniques, we sequenced 40 to 50 different clones of the respective exon, thus reaching a sensitivity of 2% to 2.5% mutant allele. A total of 6 patients with mutations were assessed serially. Two patients (31, 116) showed a high proportion of cells with 17p deletion with constant TP53 mutation (80%) over 20 to 26 months. One patient (150) showed an acquired 17p deletion and TP53 mutation after multiple lines of chemotherapy (eg, cyclophosphamide, Oncovin vincristine, prednisone; fludarabine), but we could not analyze the timing of acquisition resulting from lack of DNA.
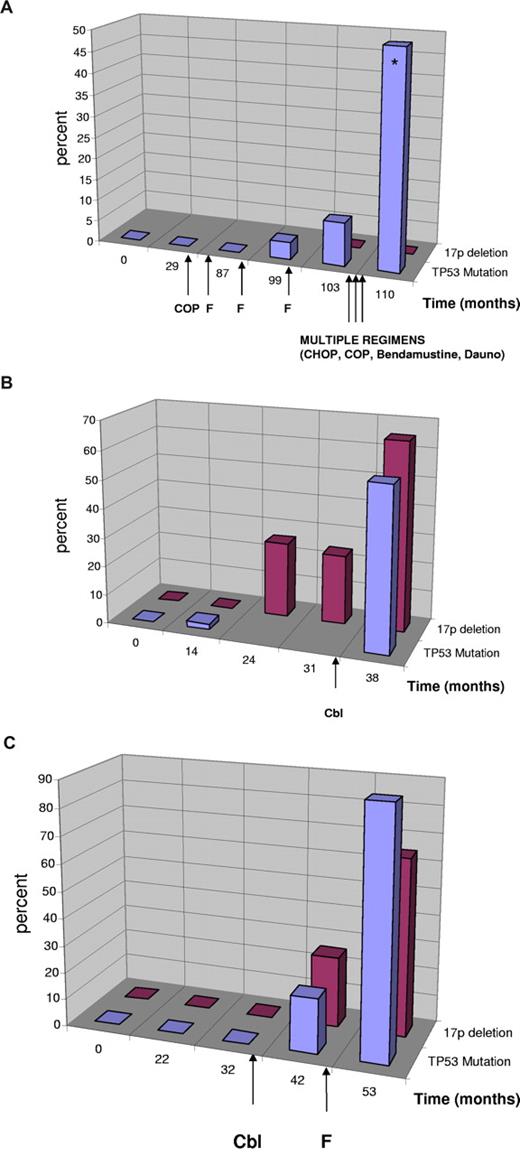
In 3 patients, we were able to investigate the time point of mutation occurrence in detail with DHLPC and cloning in serial samples collected over several years (38-110 months; Figure 2). In the first patient (Figure 2A, UPN 149), the mutation occurred in the absence of a 17p deletion but showed definitive evidence of clonal selection resulting from in vivo treatment resistance with increasing allele frequency from 4% to 50% over a period of 23 months. At the last analysis, the patient had acquired a second p53 mutation (p.Lys139Asn, c.417G > T), which was shown to have occurred on the other allele by cloning (20 clones) and sequencing. During this time the patient was treated with multiple chemotherapeutic agents, suggesting that the clone with the monoallelic TP53 mutation was refractory to treatment and selected. In the last negative sample before mutation detection, we could not detect the mutation when sequencing 40 clones, suggesting that the mutation was either present in less than 2.5% of the alleles or was acquired later.
Clonal evolution of TP53 mutation and 17p deletion in patients with CLL. Sequential analyses of TP53 mutation status and 17p deletion in patients with CLL. Treatment is indicated by arrows. Cbl indicates chlorambucil; F, fludarabine; Dauno, daunorubicin. (Blue) TP53 mutation in percent of total DNA as quantified by sequencing including the sequencing of clones. (Red) Percentage of cells with single signal for TP53 (cases with no evidence of 17p deletion were quantified as 0%). (A) Evolution of monoallelic TP53 mutation in the absence of 17p deletion. Clinically, the patient had chemorefractory disease with the emergence of the TP53 mutation and poor survival despite multiple different substances supporting the relevance of the monoallelic TP53 mutation. At the last mutation analysis, the patient had 2 TP53 mutations (*) on different alleles (p.Cys176Phe, p.Lys139Asn). (B) Evolution of biallelic TP53 inactivation. The mutation preceded the detection of a 17p deletion and was only detectable by cloning (month 14). The deletion was constant at 2 time points and increased (was selected for) after treatment with chlorambucil. (C) Evolution of 17p deletion and TP53 mutation in a patient with CLL after therapy with chlorambucil and fludarabine.
Clonal evolution of TP53 mutation and 17p deletion in patients with CLL. Sequential analyses of TP53 mutation status and 17p deletion in patients with CLL. Treatment is indicated by arrows. Cbl indicates chlorambucil; F, fludarabine; Dauno, daunorubicin. (Blue) TP53 mutation in percent of total DNA as quantified by sequencing including the sequencing of clones. (Red) Percentage of cells with single signal for TP53 (cases with no evidence of 17p deletion were quantified as 0%). (A) Evolution of monoallelic TP53 mutation in the absence of 17p deletion. Clinically, the patient had chemorefractory disease with the emergence of the TP53 mutation and poor survival despite multiple different substances supporting the relevance of the monoallelic TP53 mutation. At the last mutation analysis, the patient had 2 TP53 mutations (*) on different alleles (p.Cys176Phe, p.Lys139Asn). (B) Evolution of biallelic TP53 inactivation. The mutation preceded the detection of a 17p deletion and was only detectable by cloning (month 14). The deletion was constant at 2 time points and increased (was selected for) after treatment with chlorambucil. (C) Evolution of 17p deletion and TP53 mutation in a patient with CLL after therapy with chlorambucil and fludarabine.
In a second patient (Figure 2B, UPN 122), we found the mutation (p.Met246Val, c.736A > G) by cloning before a detectable 17p deletion (1 of 44 clones, month 14), which may be attributed to the different sensitivity of the techniques. In this case, the acquisition of the mutation preceded therapy. The deletion was constant at 2 time points and increased (was selected for) after treatment with chlorambucil.
The third cases (Figure 2C, UPN 152) showed both deletion and mutation (p.His193Arg, c.578A > G) occurring after the initiation of therapy (chlorambucil). We could not detect the mutation in the last sample with disomy of 17p before the 17p deletion occurred (months 32; 0 of 42 clones).
Effect on survival
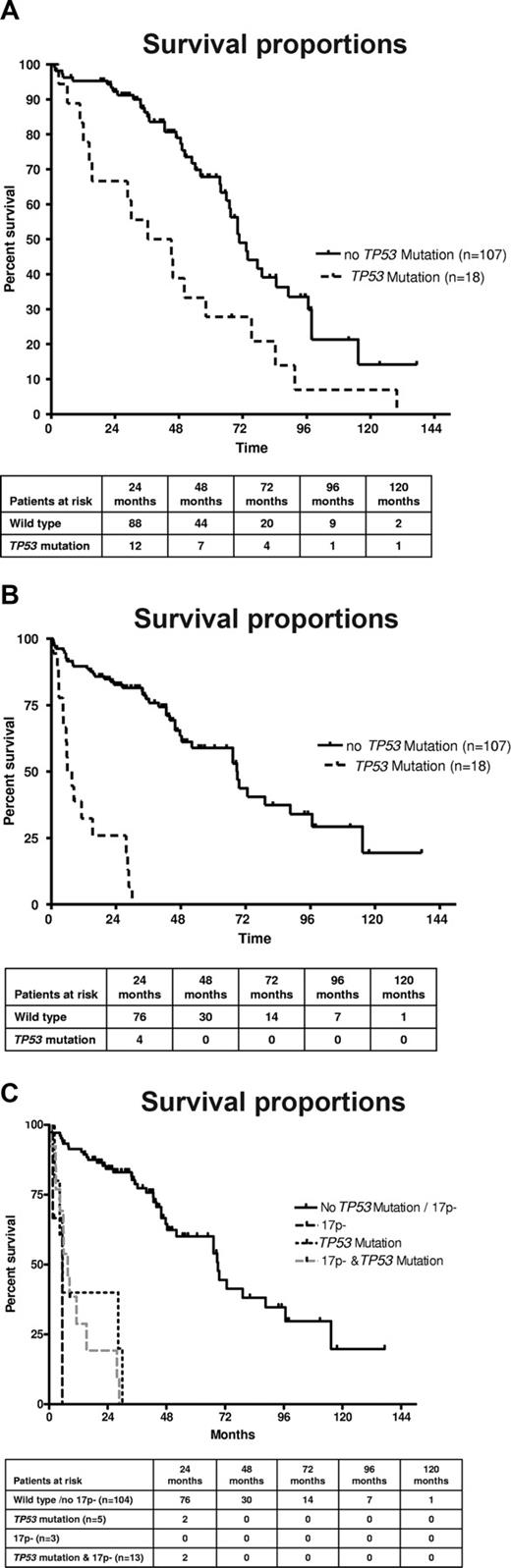
The presence of TP53 mutations was associated with a poorer survival from the time point of CLL diagnosis (Figure 3A). Patients with a TP53 mutation had a median survival of 40.8 months versus 70.7 months for patients with wild-type TP53 (P = .002). Similar results were observed when restricting the analysis to patients whose sample was analyzed within 12 months of CLL diagnosis. A significant number of patients, and particularly patients with TP53 mutations, were analyzed at a longer time interval from diagnosis (median, 14.5 vs 0.52 months; P = .009). We therefore reanalyzed the data from the time of occurrence of TP53 mutations and observed that the prognosis was very poor once TP53 mutations occurred (6.8 months) compared with patients without mutations (68.8 months; P < .001; Figure 3B). To assess the differential effect of TP53 mutation or 17p deletion, we investigated the subgroups separately according to 17p / TP53 mutational status. Again, to avoid lead time bias, we analyzed the data from the time of study (date the sample was obtained). The presence of any “p53 abnormality” (ie, deletion with mutation, deletion without mutation, mutation without deletion) was associated with inferior survival (Figure 3C). The median overall survival (from the time of study) was inferior in the groups with deletion 17p and mutation (7.6 months), sole TP53 mutation (5.5 months), and 17p deletion without TP53 mutation (5.4 months) compared with patients without TP53 loss or mutation (69 months; P < .001), although groups with monoallelic inactivation were small. The comparison of the different categories of p53 inactivation (mutation vs deletion vs both) suggests that the prognosis of these patients may be similar. To test the independent prognostic impact of TP53 mutations in the absence of 17p deletion, we performed Cox regression analysis, including stage at time of study, FISH results (13q-, 11q-, and 17p-), and VH mutation status as variables. We found an independent prognostic impact of the TP53 mutation (without 17p deletion; hazard ratio = 3.71; 95% confidence interval, 1.27-10.8; P = .02; Table 3). Similarly, the full model, including FISH results separately (13q-, 11q-, 17p-) and the presence or absence of chemotherapy before mutation analysis, showed a significantly increased risk of death for the TP53 mutation only cohort (hazard ratio = 3.19; 95% confidence interval, 1.07-9.5; P = .04).
(A) TP53 mutation is associated with poorer survival in CLL. Overall survival from the time point of diagnosis of the cohort of CLL patients according to absence or presence of TP53 mutation (n = 125; P < .002). (B) TP53 mutation is associated with poorer survival in CLL. Overall survival of the cohort of CLL patients according to absence or presence of TP53 mutation from the time of mutation analysis (n = 125; P < .001). (C) TP53 mutation without 17p deletion, 17p deletion without TP53 mutation, and the combination of TP53 mutation plus 17p deletion are all associated with poor survival in CLL. Overall survival of the cohort of CLL patients according to absence or presence of TP53 mutation/17p deletion from the time of mutation analysis (n = 125; P < .001).
(A) TP53 mutation is associated with poorer survival in CLL. Overall survival from the time point of diagnosis of the cohort of CLL patients according to absence or presence of TP53 mutation (n = 125; P < .002). (B) TP53 mutation is associated with poorer survival in CLL. Overall survival of the cohort of CLL patients according to absence or presence of TP53 mutation from the time of mutation analysis (n = 125; P < .001). (C) TP53 mutation without 17p deletion, 17p deletion without TP53 mutation, and the combination of TP53 mutation plus 17p deletion are all associated with poor survival in CLL. Overall survival of the cohort of CLL patients according to absence or presence of TP53 mutation/17p deletion from the time of mutation analysis (n = 125; P < .001).
Cox regression on overall survival (from the time of study)
| Variable . | Cox regression model (n = 113, d = 55) . | P . | |
|---|---|---|---|
| Effect . | Hazard ratio for death (95% CI) . | ||
| Binet stage at time of genetic study | C vs A,B | 3.54 (1.96, 6.37) | < .001 |
| Unmutated VH status | Yes vs no | 1.95 (0.93, 4.09) | .08 |
| 11q- | Yes vs no | 1.75 (0.94, 3.24) | .08 |
| 17p- | Yes vs no | 11.2 (5.00, 24.9) | < .001 |
| TP53 mutation w/o 17p- | Yes vs no | 3.71 (1.27, 10.8) | .02 |
| Variable . | Cox regression model (n = 113, d = 55) . | P . | |
|---|---|---|---|
| Effect . | Hazard ratio for death (95% CI) . | ||
| Binet stage at time of genetic study | C vs A,B | 3.54 (1.96, 6.37) | < .001 |
| Unmutated VH status | Yes vs no | 1.95 (0.93, 4.09) | .08 |
| 11q- | Yes vs no | 1.75 (0.94, 3.24) | .08 |
| 17p- | Yes vs no | 11.2 (5.00, 24.9) | < .001 |
| TP53 mutation w/o 17p- | Yes vs no | 3.71 (1.27, 10.8) | .02 |
n indicates the number of patients analyzed; and d, number of deaths.
Discussion
The prognostic role and clinical impact of 17p deletion in patients with CLL who need treatment are undisputed and confirmed in randomized trials.9-11 In contrast, the role of TP53 mutation is less clear. The findings of the current study underline the tight correlation of 17p deletion and TP53 mutation, suggesting that most if not all cases with 17p deletion will have biallelic loss of functional p53 resulting from mutation of the remaining allele. Because of lacking RNA, we could not test the hypothesis that the cases with 17p deletion where we could not identify a mutation in the exons or intron-exon boundary, still carry “mutations” as, for example, splicing variants with exon skipping or skewed expression of rare splicing variants. The striking similarity of clinical course and the presence of mutations in over 80% of cases with 17p deletion suggest that TP53 is indeed the culprit in cases with 17p deletion.
For cases with monoallelic defects (mutation without deletion), the proof of clinical consequence is much more difficult because of the low overall incidence (4.5%). Most studies show incidences of mutations of p53 without 17p deletion less than 5%, suggesting that, in light of the other known prognostic factors, larger studies are needed from prospective trials to confirm the prognostic impact of TP53 mutation alone.9,15 Although there have been numerous small studies looking at TP53 mutations, no study has so far documented the prognostic and biologic significance of mutations in the absence of 17p deletion.15-19,21 A recent analysis from a prospective trial failed to show a prognostic significance of monoallelic TP53 mutations, but the lumping of unconfirmed mutations and polymorphisms in the mutation group could explain these findings as this group made up for more than 50% of the cases in the “mutation group.”9 Showing the biologic effect in a CLL population is further complicated by the potentially different biologic consequences of different mutations. In addition, different selective pressures based on different therapies may pose a challenge to the detailed prediction of clinical consequence outside homogeneously treated cohorts. In this respect, the current cohort is informative as the treatment was primarily based on single-agent chemotherapy (chlorambucil, fludarabine) with very long follow-up.
In the current study, we have used the predicted residual p53 activity as derived from in vitro screens, and the majority of mutations had low residual activity, suggesting that these mutations will lead to reduced p53 function.31 The overall numbers did not allow a more detailed analysis of clinical consequences of different TP53 mutations. Similarly to previous studies, there was an association of TP53 mutations with prior therapy.12,18,19 To dissect the relation to therapy, we have analyzed serial samples with high resolution using DHPLC and cloning approaches. In a very informative patient with monoallelic defect (mutation) and follow-up samples over many years, we show definite evidence of clonal evolution and tracked the mutation by DHPLC and cloning even in samples that were negative by direct sequencing or DHPLC. Although cloning of each exon will not be a practical way of mutation detection in clinical practice, the case documents clear evidence of clonal selection in cases with TP53 mutations (in the absence of 17p deletion). This finding argues strongly for the biologic significance of TP53 mutations irrespective of 17p deletions. Further evidence comes from the analysis of cases with F-refractory CLL where we found a very high incidence of TP53 mutations, even in the absence of 17p deletion, suggesting that there is a biologic link between resistance to therapy and monoallelic TP53 mutations.22
Despite the low overall incidence of mutations without deletion, we found a strikingly similar clinical course of the patients with a mutation measured from the time of the initial mutation detection, although the subgroups with TP53 mutation only and 17p deletion without TP53 mutation were small. Nonetheless, the multivariate analysis confirmed the prognostic impact of TP53 mutations (without 17p deletion) and showed its independence of stage and the known prognostic subgroups with unmutated VH status, 11q deletion, and 17p deletion. This finding argues for the addition of TP53 mutation analysis to the prognostic assessment before treatment. To confirm these findings and to get a more precise estimate of the prognosis of monoallelic TP53 inactivation, we will need the analysis of samples from large prospective trials with adequate patient numbers and detailed genetic analysis. Although we show that the clinical consequence of TP53 mutation was similar to the deletion 17p in our cohort, the size of the deleted region on chromosome 17 is very large, suggesting that phenotypic differences of cases with 17p deletion and TP53 mutation might exist. Although the 17p deletion has been associated with a “complex karyotype” in recent metaphase studies, an open question remains if TP53 mutations without 17p deletion are associated with complex aberrations.32 We assessed the genetic complexity in relation to 17p- and TP53 mutation status by the use of array-CGH and SNP-array analysis (500k) and showed a significant increase of the number of cytogenetic aberrations in the cases with 17p deletion but also in cases with TP53 mutations in the absence of 17p deletion.
Although the recent fludarabine-based combination therapies have improved the response rate and progression-free survival, overall survival has not been shown to be improved in randomized trials. It is possible that patients with TP53 mutations will not benefit unless treated with agents for which activity independent of p53 function can be documented. Indeed, cases as described in the current report with (1) clonal evolution (ie, de novo emergence of TP53 mutated subclones) and (2) increasing clone size during chemotherapy suggest that there may be patients where this type of treatment may be potentially harmful. There is also evidence from other reports that chemotherapy may select p53 dysfunctional cells, which are probably to change the biology of the disease.33
Arguably the proportion of patients with TP53 mutations in the absence of 17p deletion is quite small, but with the advent of first line treatments that act independently of the DNA damage machinery, the early stratification of patients is gaining clinical significance.34 In addition, with the availability of array based diagnostic TP53 mutation platforms, routine testing for this mutation is possible and has successfully been used in CLL and other diseases.35,36
On the other hand, there may be a subgroup of cases with 17p deletion (and TP53 mutation) that follow a relatively benign course. Some patients with Binet A disease and a mutated VH status have been shown to survive for years without treatment.37 In our study, we found no clear evidence of such cases, but the differences in cohort size and patient profile might explain these findings. In our cohort, only one patient with p53 inactivation of any kind did not require therapy, but follow-up was short in this patient. Definite results on the prognosis of patients with 17p deletion and early-stage disease will emerge from prospective trials of these patient groups.
One obvious approach to improve overall survival in CLL patients will remain the more precise definition of patients who will not benefit from (intensive) chemotherapy similarly to patients with 17p deletion. In this respect, the group of CLL patients with evidence of TP53 mutation are an obvious contender.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Elsbeth Brückle for excellent technical assistance.
This work was supported by the Jose Carreras Foundation (R06/28v, R08/26), Else Kröner Fresenius Stiftung (P20/07//A11/07), and Global CLL Research Foundation.
Authorship
Contribution: T.Z., A.K., K.S., S.H., A. Bühler, T.D., J.E., and C.S. performed experiments; and T.Z., A.K., K.S., S.H., A. Benner, J.E., C.S., H.D., and S.S. designed the research, analyzed results, and wrote the paper.
Conflict-of-interest disclosure: T.Z. has had the costs of participating in scientific meetings reimbursed by and has received honoraria from the pharmaceutical industry. S.S. has had the costs of participating in scientific meetings reimbursed by, and has received research support honoraria from the pharmaceutical industry. The other authors declare no competing financial interests.
Correspondence: Stephan Stilgenbauer, Department of Internal Medicine III, University of Ulm, Robert-Koch-Strasse 8, 89081 Ulm, Germany; e-mail: stephan.stilgenbauer@uniklinik-ulm.de.