Abstract
Erythropoiesis is a dynamic process regulated by oxygen in vertebrates. Recent evidence has indicated that erythropoietin (Epo) expression is regulated by hypoxia-inducible transcription factors (HIFs), HIF-2α in particular. In this study, we report that knockdown mutation of HIF-2α in mice (kd/kd) results in normocytic anemia, despite Epo induction in response to hypoxia not being severely affected. Transplantation analyses clearly demonstrated that the hematopoietic microenvironment, but not the hematopoietic cells, was altered in kd/kd. Furthermore, cell-type specific recovery of HIF-2α expression in endothelial cells (ECs) abrogated the anemic condition of the kd/kd mice, indicating that HIF-2α in EC plays an essential role in supporting erythropoiesis. In the absence of HIF-2α, the expression of vascular adhesion molecule-1 (VCAM-1) was reduced significantly and restoration of VCAM-1 expression in kd/kd ECs enhanced the development of erythroid progenitors. Finally, a chromatin immunoprecipitation assay and a reporter assay indicated that VCAM-1 gene transcription is directly regulated by HIF-2α. These data suggest that the hematopoietic microenvironment required for erythropoiesis is dynamically regulated by oxygen through the functions of HIF-2α in ECs.
Introduction
Adaptation to hypoxia is a process fundamental to vertebrates. Genes involved in glucose metabolism, vasculo/angiogenesis, vascular dilation, and erythropoiesis are required for this adaptation and are induced in response to hypoxia. The expression of these genes is controlled by hypoxia-inducible factors (HIFs), members of the basic-helix-loop-helix (bHLH)-PAS family of transcription factors. HIFs are heterodimeric factors consisting of α and β subunits that bind to the hypoxia-responsible element (HRE).1 To date, 3 α subunits (HIF-1α, -2α, and -3α) and one β subunit (HIF-1β, also called aryl hydrocarbon receptor nuclear translocator [Arnt]), have been identified.1-4 Their different expression profiles suggest that HIF-1α plays central roles in regulating hypoxia-inducible genes, whereas the roles of HIF-2α are more restricted and specific to certain biologic aspects. Peng et al reported that HIF-2α−/− embryos died by embryonic day 12.5 (E12.5) because of vascular defects, signifying that HIF-2α is essential for angiogenesis during embryonic development.5 However, other studies demonstrated that HIF-2α−/− mice generated with different mouse strains could survive for longer and showed different phenotypes.5-8 These studies suggest that the functions of HIF-2α are influenced by the genetic background.
There is no doubt that regulation of erythropoiesis is essential for maintaining oxygen homeostasis in vertebrates. Erythroid progenitors have the potential to proliferate rapidly in response to anemic and hypoxic stimuli, a process referred to as stress erythropoiesis.9 Erythropoietin (Epo) and its receptor (EpoR) play central roles in this process.9 Epo is a glycoprotein hormone produced mainly by kidney interstitial cells.10-14 The expression of Epo is significantly up-regulated by HIFs under hypoxic conditions.4 EpoR is expressed in erythroid progenitors, and Epo-EpoR signaling results in survival, proliferation, and differentiation of erythroid cells.15 Recent studies demonstrated that HIF-2α is a major regulator of Epo rather than HIF-1α in vivo.8,16,17 Deletion of the HIF-2α gene led to anemia in adult mice, which was rescued by exogenous Epo administration.16,17 Moreover, conditional inactivation of HIF-2α in the livers of anemic mice or infants severely affected the hypoxic induction of Epo mRNA expression.18 These data suggest that activation of Epo-EpoR signaling by HIF-2α is a central mechanism in the regulation of erythropoiesis in postnatal life.
It is noteworthy, however, that the Epo-EpoR system may not be able to act efficiently in the absence of an appropriate hematopoietic microenvironment. The hematopoietic microenvironment has been proposed as the area of hematopoietic tissue highly specialized for the proliferation and differentiation of hematopoietic cells.19 The hematopoietic microenvironment is supported by 5 distinct types of adherent cells: fibroblasts, adipocytes, osteoblasts, macrophages, and endothelial cells (ECs). There is increasing evidence that the hematopoietic microenvironment is required for the survival and self-renewal of hematopoietic stem cells (HSCs), as well as for the proliferation and differentiation of cells with a specific cell lineage.20-22 Remarkably, a recent study demonstrated that HIF-2α−/− mice generated by the F1 hybrids of 2 different mouse strains (C57BL/6J and 129S6/SvEvTac) exhibited pancytopenia because of a defective hematopoietic microenvironment.8 However, the precise roles of HIF-2α in the formation of the hematopoietic microenvironment have not been fully elucidated.
Here we show that knockdown mutation of HIF-2α in mice (HIF-2αkd/kd, hereafter referred to as kd/kd) generated with a C57BL/6J genetic background exhibited normocytic anemia, even though Epo expression was not severely affected. Transplantation analyses clearly demonstrated that reduction in HIF-2α affects the hematopoietic microenvironment, but not hematopoietic cells. Furthermore, cell-type specific recovery of HIF-2α expression in kd/kd EC abrogated anemia, indicating that HIF-2α plays an essential role in supporting erythropoiesis in EC. In kd/kd EC, reduced expression of the adhesion molecule VCAM-1, a target of the HIF-2α/Arnt heterodimer, is responsible for the impaired maintenance of erythropoiesis.
Methods
Animals were used for the study in accordance with the decision of the Animal Committee of the University of Tsukuba.
Mice
The HIF-2α knockdown allele was generated as reported previously.23 HIF-2α knockdown heterozygotes were back-crossed with C57BL/6J WT mice to obtain congenic mice (N8). Tie-1-Cre transgenic mice were purchased from The Jackson Laboratory (Bar Harbor, MA) and temperature-sensitive SV40 large T-antigen (SV40T) transgenic mice24 were a generous gift from Dr Masuo Obinata (Tohoku University, Sendai, Japan). Both transgenic mouse lines were maintained in a C57BL/6J genetic background. For hypoxic exposure, mice were subjected to 6% O2 for 48 hours in an isolated chamber (Natsume, Tokyo, Japan). In this chamber, a hypoxic atmosphere was obtained by mixing air with nitrogen, and oxygen levels were monitored using a polarographic oxygen sensor (Toray, Tokyo, Japan). To avoid the induction of HIF resulting from the collection of blood sample, 4 groups of mice (n = 3 per group) were prepared, and a blood sample was taken once at the indicated time point (0, 48, 96, or 144 hours) after the hypoxic treatment from each animal.
Analyses of blood cell counts and plasma Epo concentration
Peripheral blood samples were obtained from the orbital veins of anesthetized mice and collected into tubes containing ethylenediaminetetraacetic acid (EDTA). Automated blood cell counts were analyzed using Celltac (Nihon Kohden, Tokyo, Japan). Reticulocyte counts and the plasma Epo concentration were examined as previously described.25
In vitro colony assays and flow cytometric analysis
The erythroid burst-forming units (BFU-E)- and erythroid colony-forming unit (CFU-E)-derived colony assays were performed as previously described.26 Flow cytometric analysis was performed as previously,26 except that fluorescein isothiocyanate (FITC)-conjugated antibody was used for examining CD71 expression.
Isolation of ECs
Spleens were harvested from WT or kd/kd mice harboring SV40T transgene. The spleen cells were dispersed with 0.1% collagenase (Nitta Zelatin, Osaka, Japan)/phosphate-buffered saline (PBS)/20% fetal bovine serum (FBS; Hyclone, South Logan, UT), passed through a cell strainer (Falcon 2360, BD Biosciences, San Jose, CA), and centrifuged on Ficoll solution (1.077 g/mL; Nycomed, Oslo, Norway) to obtain mononuclear cell fraction. Cells were cultured on collagen type I–coated dishes (BD Biosciences) in HAVA medium in the presence of endothelial cell growth supplement (BD Biosciences) as described previously.27 After the cell growth reached confluence, cells were examined for CD31 and CD45 expression by flow cytometry (FACSVantage, BD Biosciences). The CD31+/CD45− cells were sorted and cultured on collagen type I–coated dishes in HAVA medium with endothelial cell growth supplement. Cloning of EC lines were done using cloning cylinder (Sigma-Aldrich, St Louis, MO). The established cells were maintained in HAVA medium at 33°C.
Coculture assay
A cell line established from spleen stromal cells (MSS31 cells) was maintained in RITC80-7 medium (Iwaki, Tokyo, Japan) as in an earlier report.28 MSS31 cells or isolated ECs derived from spleen were plated in a 24-well plates and incubated for 2 days before coculturing.29 These cells were cocultured with 2 × 104 fetal liver cells (E13.5) in methylcellulose medium (MethoCult M3234, StemCell Technologies, Vancouver, BC) supplemented with 30% FBS, 1% bovine serum albumin (BSA; Sigma-Aldrich), 100 μM 2-mercaptoethanol, and 1 U/mL Epo for 4 days as explained formerly.29 Erythroid colonies developed on MSS31 cells were examined and benzidine staining was performed.
Transplantation studies
To evaluate the functions of hematopoietic stem cells and the hematopoietic microenvironment, bone marrow (BM) cells were harvested from WT, kd/kd, Tie-1-Cre, or kd/kd::Tie-1-Cre mice and a BM reconstitution study was performed. BM cells (107 cells/mouse) were injected into lethally irradiated recipient mice (a total dose of 11 Gy) via tail veins. Mice with a C57BL/6J background were used for both donors and recipients. Peripheral blood samples from recipient mice 3 months after injection were analyzed. In addition, BM and spleens from recipient mice were analyzed for the expression of cell surface markers of the erythroid lineage by fluorescence-activated cell sorter (FACS).
Immunohistochemistry
Spleens and BM samples were dissected out and fixed with 4% paraformaldehyde overnight at 4°C. After washing with PBS, samples were embedded in polyester wax and sectioned at 5 to 7 μm. Sections were dewaxed in ethanol, treated with 0.3% hydrogen peroxide in methanol, and stained with anti–HIF-2α antibody,23 anti-CD31 antibody (BD Biosciences), or isotype-matched rat IgG overnight at 4°C as reported previously.30 After washing with PBS, slides were further incubated with biotinylated goat anti–rat IgG antibody (Vector Laboratories, Burlingame, CA), followed by streptavidin-conjugated ABC complex (Vector Laboratories). Specific staining was visualized by incubation in 3,3′-diaminobenzidine as described before.30 Counterstaining was carried out using Nuclear Fast Red.
Fluorescent immunohistochemistry
WT and kd/kd mice were injected with 200 μL of tetramethylrhodamine isothiocyanate (TRITC)-conjugated Bandeiraea simplicifolia Lectin (0.1 mg/mL, Sigma-Aldrich) from tail vein as reported previously.31 Mice were killed 10 minutes after the injection and spleens were fixed with 4% paraformaldehyde. Cryosections of the spleens were prepared by routine methods and fluorescent immunohistochemistry was performed using anti HIF-2α antibody23 and FITC-conjugated donkey anti–rabbit IgG antibody (Jackson ImmunoResearch Laboratories, West Grove, PA). Normal rabbit IgG was used as a negative control. Fluorescent intensity of TRITC was measured by National Institutes of Health images.
Microscopy analysis
Sample slides were viewed with a Olympus BX51 microscope system (Olympus, Tokyo, Japan) using UPlanSApo objective lenses at 4×/0.16PH, 10×/0.40PH and 20×/0.75PH (Olympus) and mounting reagent (Muto Chemical, Tokyo, Japan). Data acquisition was done with a DP70 digital camera attached to the microscope and DP controller software (Olympus). Images were processed using Adobe Photoshop version 8.0 software (Adobe Systems, San Jose, CA).
Quantitative reverse-transcribed polymerase chain reaction (RT-PCR)
Total RNA was prepared from BM, spleen, kidney, and liver and isolated EC using ISOGEN reagent (Wako, Osaka, Japan) and cDNAs were synthesized as previously described.31 The reaction mixtures for quantitative polymerase chain reaction (PCR) were prepared using POWER SYBR Green PCR master mix (Applied Biosystems, Foster City, CA) and analyzed by 7700 Sequence Detector (Applied Biosystems). Experiments were performed as triplicate and the data were calculated by ΔΔCt method. The sequence of primer sets used for the PCR reactions is shown in Table 1.
Primers used for PCR
| Genes and primers . | Sequence . |
|---|---|
| VCAM-1 | |
| 5′ | GAACACTTTTCCCAGACACTTTTCACGTGG |
| 3′ | TAAGGATCTGGGTTTTATAAATAATGTCTC |
| VECAD | |
| 5′ | GAACGAGGACAGCAACTTCACCCTCATA |
| 3′ | CTGACACATCATAGCTGGTGGTGTCCAT |
| Fibronectin | |
| 5′ | GGACCCCGCTAAACTCTTCCACCATTAT |
| 3′ | GGAGAGGGATGCTCTCATGTTGTTCGTA |
| UPAR | |
| 5′ | GCTTCGGGAATGGCAAGATGATAGAGAG |
| 3′ | CAGTGGGTGTAGTTGCAACACTTCAGGA |
| PECAM-1 | |
| 5′ | CACAGATAAGCCCACCAGAGACATGGA |
| 3′ | TCTCGCTGTTGGAGTTCAGAAGTGGAG |
| Flk-1 | |
| 5′ | AGGACCAAGGCGACTATGTTTGCTCTG |
| 3′ | ACGGTCCGTACGAGAATGACAAGAAGG |
| Tie-2 | |
| 5′ | CGTCAACAAGCATCCTTCCTACCTGCTA |
| 3′ | TGCATCCTTCTGGTCCACTACACCTTTC |
| HIF-1α | |
| 5′ | CAAGATCTCGGCGAACGAAAGAGTCTGA |
| 3′ | GAAGCACCTTCCACGTTGCTGACTTGAT |
| HIF-2α | |
| 5′ | CAGCTCAGAGCTGAGGAAGG |
| 3′ | GTTGTAGACTCACTTGCC |
| Epo | |
| 5′ | TGGCACCCTGCTGCTTTTACTCTCCTT |
| 3′ | CTGGAGGCGACATCAATTCCTTCTGAG |
| β-actin | |
| 5′ | GTCGTACCACAGGCATTGTGATGGACT |
| 3′ | CACCAGACAGCACTGTGTTGGCATAGA |
| Genes and primers . | Sequence . |
|---|---|
| VCAM-1 | |
| 5′ | GAACACTTTTCCCAGACACTTTTCACGTGG |
| 3′ | TAAGGATCTGGGTTTTATAAATAATGTCTC |
| VECAD | |
| 5′ | GAACGAGGACAGCAACTTCACCCTCATA |
| 3′ | CTGACACATCATAGCTGGTGGTGTCCAT |
| Fibronectin | |
| 5′ | GGACCCCGCTAAACTCTTCCACCATTAT |
| 3′ | GGAGAGGGATGCTCTCATGTTGTTCGTA |
| UPAR | |
| 5′ | GCTTCGGGAATGGCAAGATGATAGAGAG |
| 3′ | CAGTGGGTGTAGTTGCAACACTTCAGGA |
| PECAM-1 | |
| 5′ | CACAGATAAGCCCACCAGAGACATGGA |
| 3′ | TCTCGCTGTTGGAGTTCAGAAGTGGAG |
| Flk-1 | |
| 5′ | AGGACCAAGGCGACTATGTTTGCTCTG |
| 3′ | ACGGTCCGTACGAGAATGACAAGAAGG |
| Tie-2 | |
| 5′ | CGTCAACAAGCATCCTTCCTACCTGCTA |
| 3′ | TGCATCCTTCTGGTCCACTACACCTTTC |
| HIF-1α | |
| 5′ | CAAGATCTCGGCGAACGAAAGAGTCTGA |
| 3′ | GAAGCACCTTCCACGTTGCTGACTTGAT |
| HIF-2α | |
| 5′ | CAGCTCAGAGCTGAGGAAGG |
| 3′ | GTTGTAGACTCACTTGCC |
| Epo | |
| 5′ | TGGCACCCTGCTGCTTTTACTCTCCTT |
| 3′ | CTGGAGGCGACATCAATTCCTTCTGAG |
| β-actin | |
| 5′ | GTCGTACCACAGGCATTGTGATGGACT |
| 3′ | CACCAGACAGCACTGTGTTGGCATAGA |
Transfection
The kd/kd #10 EC stromal cells (designated as kd#10) were transfected with expression plasmids encoding wither full-length of VCAM-1 or HIF-2α cDNA using FuGENE 6 reagent (Roche, Mannheim, Germany). HIF-2α transfected kd#10 were analyzed for the expression of VCAM-1 mRNA by quantitative RT-PCR. To obtain stable transformant, pEF1-puromycin resistant gene was cotransfected and cells were screened by puromycin.
Western blot analysis
kd#10 were transfected into 60-mm dish with pEF-HIF2α or pEF-BOS empty vector using FuGENE 6 reagent (Roche). At 48 hours after the transfection, cells were cultured under 1% O2 for 6 hours, Western blot analysis was performed as described previously using nuclear extracts (20 μg) prepared from these cells.32
Luciferase reporter assay
A reporter assay was performed by cotransfecting pGL3 0.7-kb-VCAM-1-Luciferase (Luc) or pGL3 0.7-kb-VCAM-1-mut-Luc (10 ng), pBOS-HIF-2α (10 ng), and pBOS-Arnt (10 ng) into 293T cells. A plasmid of pEF-renilla-Luc (1 ng) was cotransfected as a control in both reporter assays.
Chromatin immunoprecipitation assay
ECs (5 × 106 cells) were fixed with 1% formaldehyde for 10 minutes at room temperature, washed with PBS containing 1 mM protease inhibitor cocktail (PIC; Roche), harvested and treated with hypotonic solution (5 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 85 mM KCl, 0.5% NP-40, and 1 mM PIC). Nuclei were collected from cells centrifuged at 14 000g for 5 minutes, then lysed with lysis buffer (50 mM Tris-HCl, pH 8.1, 10 mM EDTA, 1% sodium dodecyl sulfate [SDS], and 1 mM PIC). After fragmenting DNA by sonication, immunoprecipitation reactions were performed using a rotating mixer at 4°C with 1 μg/mL anti-HIF-1α antibody (Novus Biologicals, Littleton, CO) or anti–HIF-2α antibody.23 Normal rabbit IgG was used as a negative control to verify the specificity of the reaction. After incubation with antibody, reaction mixtures were incubated with preblocked protein A-agarose beads (Calbiochem, La Jolla, CA) at 4°C for 1 hour and precipitated complexes were collected by centrifugation at 3000g for 5 minutes. These complexes were washed 3 times with wash buffer (0.25 mM LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl, pH 8.1) and eluted from the protein A-agarose beads with elution buffer (1% SDS and 0.1 M NaHCO3), then denatured at 65°C for 4 hours. DNA fragments were extracted with phenol/chloro-form, precipitated with ethanol, and used as template for PCR analysis. The VCAM-1 HRE sequence was amplified with forward (5′-GCCCTTTCGGAGCTGAAGGTCAGG-3′) and reverse (5′-CTCTGCTTCAAAGCCTTCTTTGTGCC-3′) primers.
Statistical analysis
Statistical evaluations of data were conducted using the Student t test for per-comparison analysis. Data are presented as means plus or minus SD.
Results
HIF-2α knockdown mice exhibit normocytic anemia
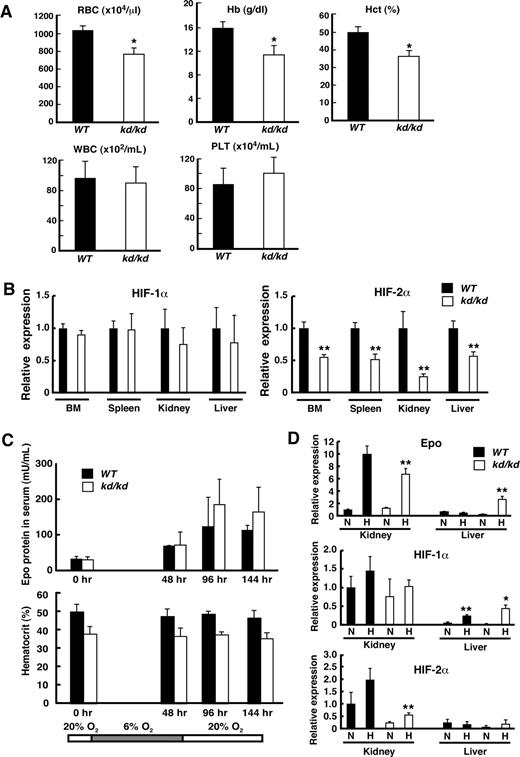
Because the functions of HIF-2α are influenced by the genetic background, wild-type (WT) and kd/kd mice with a C57BL/6J background were examined in this study. Peripheral blood indices showed that hematocrit levels were significantly lower in kd/kd mice compared with WT (Figure 1A). The red blood cell (RBC) count for kd/kd mice reflected normocytic anemia, with no increase in the reticulocyte count (Figure 1A and data not shown). Anemia became prominent 4 weeks after birth, similar to that observed in HIF-2α−/− mice in a previous study.8 Whereas HIF-2α−/− mice showed pancytopenia, white blood cell (WBC) and platelet counts were within the normal range in kd/kd peripheral blood. Quantitative RT-PCR analysis revealed that HIF-2α mRNA levels were significantly lower in kd/kd mice compared with WT in BM, spleen, kidney, and liver, whereas HIF-1α mRNA levels were comparable between kd/kd and WT mice in these organs (Figure 1B).
Analyses of anemia observed in kd/kd mice. (A) Peripheral blood from 3-month-old WT and kd/kd mice was analyzed. Erythrocytes (RBC), hemoglobin (Hb), hematocrit (Hct), leukocytes (WBC), and platelets (PLT) were determined (*P < .05, compared with WT). (B) The mRNA expression levels of HIF-1α (left panel) and HIF-2α (right panel) were examined in BM, spleen, kidney, and liver by quantitative RT-PCR. The expression levels seen in WT were normalized to a value of 1 as the standard in each tissue. ■ represents WT; □ represents kd/kd (**P < .01, compared with WT). (C) Epo protein levels in sera and hematocrit were analyzed after exposure to a low oxygen concentration (6%) for 48 hours. Peripheral blood was harvested 0, 48, 96, and 144 hours after starting the experiment. ■ represents WT; □ represents kd/kd. (D) The mRNA expression levels of Epo (top), HIF-1α (middle), and HIF-2α (bottom) were examined in kidney and liver by quantitative RT-PCR. RNA samples were collected from WT and kd/kd mice under normoxic (N) or hypoxic (H, 48-hour exposure to 6% O2) conditions. The expression levels in WT kidneys at normoxia were normalized to a value of 1 as the standard. ■ represents WT; □ represents kd/kd (*P < .05 and **P < .01, compared with normoxic conditions). Error bars represent SD.
Analyses of anemia observed in kd/kd mice. (A) Peripheral blood from 3-month-old WT and kd/kd mice was analyzed. Erythrocytes (RBC), hemoglobin (Hb), hematocrit (Hct), leukocytes (WBC), and platelets (PLT) were determined (*P < .05, compared with WT). (B) The mRNA expression levels of HIF-1α (left panel) and HIF-2α (right panel) were examined in BM, spleen, kidney, and liver by quantitative RT-PCR. The expression levels seen in WT were normalized to a value of 1 as the standard in each tissue. ■ represents WT; □ represents kd/kd (**P < .01, compared with WT). (C) Epo protein levels in sera and hematocrit were analyzed after exposure to a low oxygen concentration (6%) for 48 hours. Peripheral blood was harvested 0, 48, 96, and 144 hours after starting the experiment. ■ represents WT; □ represents kd/kd. (D) The mRNA expression levels of Epo (top), HIF-1α (middle), and HIF-2α (bottom) were examined in kidney and liver by quantitative RT-PCR. RNA samples were collected from WT and kd/kd mice under normoxic (N) or hypoxic (H, 48-hour exposure to 6% O2) conditions. The expression levels in WT kidneys at normoxia were normalized to a value of 1 as the standard. ■ represents WT; □ represents kd/kd (*P < .05 and **P < .01, compared with normoxic conditions). Error bars represent SD.
The hypoxic induction of Epo is conserved in kd/kd mice
Although HIF-2α has been suggested as a major regulator of Epo gene expression in vivo,8,16,17 we found that the reduced expression of HIF-2α did not severely affect serum Epo levels in kd/kd mice exposed to hypoxia. Epo levels in the sera of both kd/kd and WT mice increased and reached a maximum level at 96 hours after the hypoxic treatment. Hematocrit values were not affected by the hypoxic exposure in both kd/kd and WT mice (Figure 1C). To elucidate molecular basis of this observation, we collected mRNAs from kidneys and liver at normoxia (0 hour) and 48 hours after the hypoxic treatment and examined mRNA levels of Epo, HIF-1α, and HIF-2α by quantitative RT-PCR (Figure 1D). Consistent with the observation of serum Epo levels, Epo mRNA expression was significantly increased on hypoxia in kidneys of both kd/kd and WT mice (Figure 1D). Of note is that HIF-2α mRNA expression was increased on hypoxia in the kd/kd kidneys, albeit at lower levels compared with that of WT. These results suggest that the induction of HIF-2α by hypoxia might be sufficient to activate Epo mRNA expression in the kd/kd kidneys. Interestingly, Epo mRNA level in liver was increased on hypoxia in kd/kd, but not WT, mice (Figure 1D). Moreover, the exposure to hypoxia induced HIF-1α, but not HIF-2α, mRNA expression in the liver of both kd/kd and WT mice, and the magnitude of the induction was greater in the kd/kd liver compared with WT (Figure 1D). These results suggest the compensatory role of HIF-1α for activating Epo mRNA expression on hypoxia in kd/kd mice. Collectively, these results indicate that the induction of Epo on hypoxia was not impaired in the kd/kd mice, despite that mild, yet significant, anemia was observed in these mice.
The maturation of erythroblasts is attenuated in kd/kd mice
To evaluate maturation status of erythroid cells, we examined the expressions of CD71 and TER119 by FACS. A previous study showed that TER119+/CD71+ cells and TER119+/c-Kit+ cells consist of immature erythroblasts, whereas TER119+/CD71− cells correspond to mature erythroblasts.33 In the BM of kd/kd mice, the frequencies of both TER119+/CD71+ and TER119+/c-Kit+ cells were significantly greater than those of WT (Figure 2A). In contrast, the frequency of TER119+/CD71− mature erythroblasts was lower in kd/kd compared with WT in both BM and spleen (Figure 2A,B). Furthermore, absolute number of BM cells (2.46 ± 0.32 and 2.47 ± 0.36 × 107 cells/femur for WT and kd/kd, respectively, P = .98) as well as weight of spleen (52 ± 4.0 and 50.7 ± 5.1 mg/spleen for WT and kd/kd, respectively, P = .74) were comparable between WT and kd/kd. These data indicate that maturation of early to late erythroblast is attenuated in kd/kd mice, which presumably leads to normocytic anemia, as observed in peripheral blood (Figure 1A).
Analyses of erythroid progenitors in WT and kd/kd mice. (A) Left: Flow cytometric analyses of the expression of TER119, CD71, and c-Kit in total BM cells prepared from femur of WT (top panels) and kd/kd (bottom panels) mice. Frequencies (%) of cells in gated fractions (squared) are shown. In right panels, frequencies (%) of CD71+/TER119− (top left region) and CD71−/TER119+ (bottom right region) cells are also indicated. Right: Frequencies (%) of CD71+/TER119+ cells (gated fractions of the left panels). ■ represents WT; □ represents kd/kd. n = 3 per group; *P < .05. (B) Left: Flow cytometric analyses of the expression of TER119 and CD71 in spleen cells prepared from WT (left panel) and kd/kd (right panel) mice. Frequencies (%) of CD71−/TER119+ cells in gated fractions (squared), CD71+/TER119− (top left region) and CD71+/TER119+ (top right region) cells are shown. Right: Frequencies (%) of CD71−/TER119+ cells (gated fractions of the left panels). ■ represents WT; □ represents kd/kd. n = 3 per group (*P < .05). (C) In vitro colony assay using BM cells from WT (■) and kd/kd (□) mice. A total of 50 000 BM mononuclear cells were plated and cultured with methylcellulose media. Note that the numbers of BFU-E– and CFU-E–derived colonies were significantly increased in kd/kd BM compared with WT (*P < .05, **P < .01). Error bars represent SD.
Analyses of erythroid progenitors in WT and kd/kd mice. (A) Left: Flow cytometric analyses of the expression of TER119, CD71, and c-Kit in total BM cells prepared from femur of WT (top panels) and kd/kd (bottom panels) mice. Frequencies (%) of cells in gated fractions (squared) are shown. In right panels, frequencies (%) of CD71+/TER119− (top left region) and CD71−/TER119+ (bottom right region) cells are also indicated. Right: Frequencies (%) of CD71+/TER119+ cells (gated fractions of the left panels). ■ represents WT; □ represents kd/kd. n = 3 per group; *P < .05. (B) Left: Flow cytometric analyses of the expression of TER119 and CD71 in spleen cells prepared from WT (left panel) and kd/kd (right panel) mice. Frequencies (%) of CD71−/TER119+ cells in gated fractions (squared), CD71+/TER119− (top left region) and CD71+/TER119+ (top right region) cells are shown. Right: Frequencies (%) of CD71−/TER119+ cells (gated fractions of the left panels). ■ represents WT; □ represents kd/kd. n = 3 per group (*P < .05). (C) In vitro colony assay using BM cells from WT (■) and kd/kd (□) mice. A total of 50 000 BM mononuclear cells were plated and cultured with methylcellulose media. Note that the numbers of BFU-E– and CFU-E–derived colonies were significantly increased in kd/kd BM compared with WT (*P < .05, **P < .01). Error bars represent SD.
To ascertain whether erythroid cell differentiation is impaired at the progenitor stage in kd/kd BM, a colony assay was performed. Notably, both BFU-E– and CFU-E–derived colonies were significantly increased in kd/kd compared with WT (Figure 2C). No difference was noted between kd/kd and WT in the morphology of colonies or benzidine staining intensity (data not shown). These results suggest that the frequency of erythroid progenitors was rather increased in the kd/kd BM compared with WT. Taken together, these findings indicate that a reduced HIF-2α expression leads to impaired maturation at the erythroblast stage, whereas the ability of erythroid progenitors to form erythroid colonies in vitro remains unaffected.
Decreased number of endothelial cells and formation of microcapillaries in the red pulp of kd/kd spleen
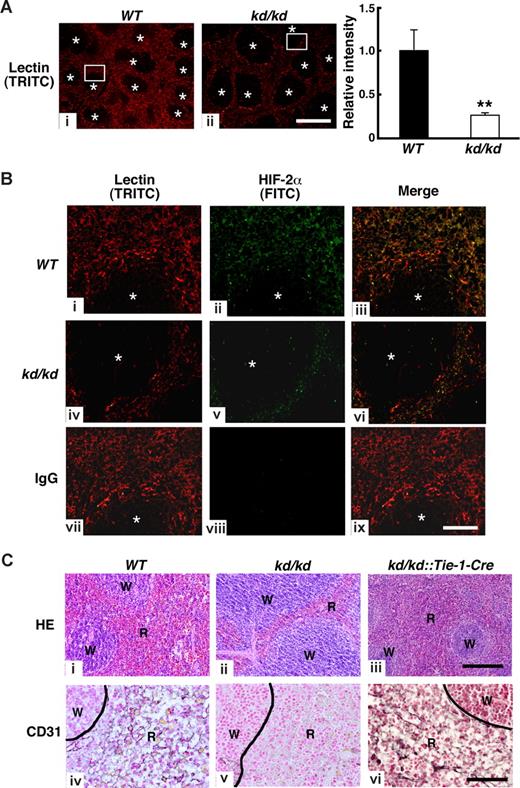
We next examined whether impaired erythroid cell maturation is accompanied by morphologic abnormality in the kd/kd spleen. We prepared spleen sections of mice treated with the TRITC-conjugated lectin that specifically binds to ECs.31 Fluorescent micrographs revealed that lectin-bound cells were assembled in red pulp area of spleen where microcapillary formation was enriched (Figure 3A,B). We found the area of red pulp relative to white pulp was reduced in the kd/kd spleen compared with WT. Indeed, the fluorescent intensity was significantly reduced in kd/kd compared with WT, indicating lectin-bound cells were decreased in the kd/kd spleen (Figure 3A). Moreover, fluorescent immunohistochemistry of HIF-2α using FITC-conjugated secondary antibody revealed that HIF-2α protein resides in the lectin-bound cells (appeared in yellow, Figure 3Biii-vi). Consistent with these observations, CD31, a marker for ECs, was barely detectable in the red pulp of kd/kd spleen by immunohistochemistry (Figure 3Cv). To prove that HIF-2α expression in EC contributes microcapillary formation in red pulps, kd/kd mice were intercrossed with Tie-1-Cre mice, EC-specific Cre transgenic mice.34 In kd/kd mice, HIF-2α expression is reduced by the insertion of a floxed-PGK promoter-neomycin gene cassette in the first exon, and excision of the neo-cassette by Cre recombinase results in recovery of HIF-2α mRNA expression.23 Thus HIF-2α expression is expected to recover specifically in ECs in the kd/kd::Tie-1-Cre mice. As expected, CD-31–positive cells were clearly observed in the red pulp of kd/kd::Tie-1-Cre mice (Figure 3Cvi). Moreover, hematoxylin and eosin (HE) staining revealed that the reduced red pulp of kd/kd mice was recovered by the expression of Tie-1-Cre (Figure 3Cii,iii). Collectively, these results indicate that reduced HIF-2α expression in EC causes a reduction of microcapillary formation in the red pulp of kd/kd spleen.
Comparison of HIF-2α and CD31 expressions among WT, kd/kd, and kd/kd::Tie-1-Cre mouse spleens. (A) Left and center panels: Structures of microcapillary in the spleen of WT (i) and kd/kd (ii) were examined by the injection of TRITC-conjugated lection. Note that lection-bound cells form capillary-like structures in red pulps. *Area of white pulps in spleens. Bar represents 500 μm. Right panel: Fluorescent intensity of TRITC was measured in 9 separate fields of view in the red pulp area. The fluorescent frequency measured in a field of WT was normalized to a value of 1 and the average of relative fluorescent intensity was indicated (**P < .01). Error bars represent SD. (B) Left columns (TRITC; i, iv, and vii) represent magnified view of rectangle area shown in panel A. Middle columns (FITC; ii, v, and viii) represent fluorescent immunohistochemistry of HIF-2α in spleen. Right columns (iii, vi, and ix) represent merged pictures to examine HIF-2α-positive microcapillaries (yellow color). Sections from WT (i-iii) and kd/kd (iv-vi) mice are shown. (vii-ix) Normal rabbit IgG was used in WT sections as a negative control. *Area of white pulps. Bar represents 100 μm. (C) Histochemical staining was undertaken in spleens using hematoxylin and eosin (HE) (i-iii) and anti-CD31 antibody (iv-vi). Left panels, middle panels, and right panels represent WT, kd/kd, and kd/kd::Tie-1-Cre mice, respectively. W and R represent white pulp and red pulp, respectively. Bar represents 100 μm.
Comparison of HIF-2α and CD31 expressions among WT, kd/kd, and kd/kd::Tie-1-Cre mouse spleens. (A) Left and center panels: Structures of microcapillary in the spleen of WT (i) and kd/kd (ii) were examined by the injection of TRITC-conjugated lection. Note that lection-bound cells form capillary-like structures in red pulps. *Area of white pulps in spleens. Bar represents 500 μm. Right panel: Fluorescent intensity of TRITC was measured in 9 separate fields of view in the red pulp area. The fluorescent frequency measured in a field of WT was normalized to a value of 1 and the average of relative fluorescent intensity was indicated (**P < .01). Error bars represent SD. (B) Left columns (TRITC; i, iv, and vii) represent magnified view of rectangle area shown in panel A. Middle columns (FITC; ii, v, and viii) represent fluorescent immunohistochemistry of HIF-2α in spleen. Right columns (iii, vi, and ix) represent merged pictures to examine HIF-2α-positive microcapillaries (yellow color). Sections from WT (i-iii) and kd/kd (iv-vi) mice are shown. (vii-ix) Normal rabbit IgG was used in WT sections as a negative control. *Area of white pulps. Bar represents 100 μm. (C) Histochemical staining was undertaken in spleens using hematoxylin and eosin (HE) (i-iii) and anti-CD31 antibody (iv-vi). Left panels, middle panels, and right panels represent WT, kd/kd, and kd/kd::Tie-1-Cre mice, respectively. W and R represent white pulp and red pulp, respectively. Bar represents 100 μm.
HIF-2α in ECs is required for normal erythropoiesis in a non–cell-autonomous manner
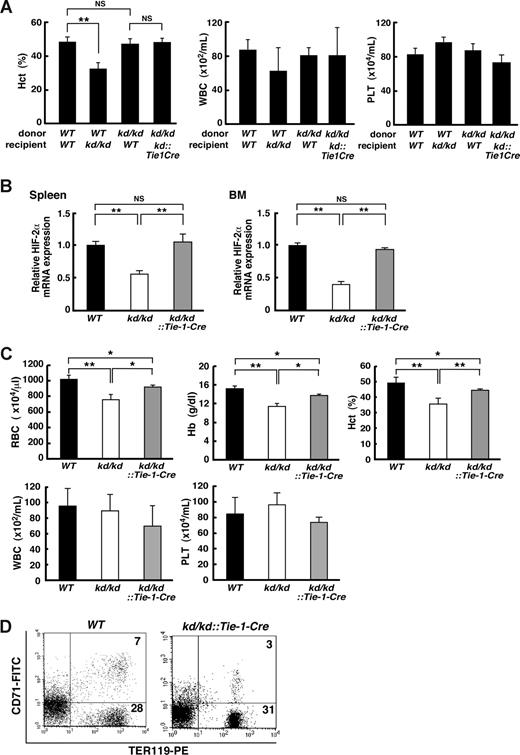
Overall, these observations suggest a possibility that a reduction in HIF-2α in EC may affect the hematopoietic microenvironment in a non–cell-autonomous manner. To test this possibility, we performed reciprocal transplantation assays using BM cells prepared from kd/kd, WT, and kd/kd::Tie-1-Cre mice (Figure 4A). When WT BM cells were transplanted, hematocrit levels were significantly lower in kd/kd recipient mice compared with WT recipients (49.0% ± 2.3% and 32.9% ± 3.3% for WT and kd/kd recipients, respectively, P < .01). On the other hand, BM cells from kd/kd mice successfully reconstituted hematopoiesis in lethally irradiated WT recipients. Furthermore, the hematocrit level of kd/kd::Tie-1-Cre recipient mice transplanted with kd/kd BM cells was akin to that of WT recipient mice (45.1% ± 1.5% and 47.6% ± 2.9% for kd/kd::Tie-1-Cre and WT recipients, respectively, P = .26). Interestingly, even in the kd/kd recipient mice, neither WBC nor platelet counts were significantly affected (Figure 4A middle and right panels, respectively). Together, these outcomes unequivocally indicate that HIF-2α in EC plays an essential role in maintaining a hematopoietic microenvironment that supports erythropoiesis.
Analysis of the ability of the hematopoietic microenvironment to support erythropoiesis. (A) A transplantation assay. Hematopoietic cells (107 total BM cells/mouse) derived from donor mice (WT or kd/kd) were transplanted into lethally irradiated recipient mice (WT, kd/kd, or kd/kd::Tie-1-Cre, n = 3 for each group). Peripheral blood cells were measured for hematocrit levels (Hct), leukocyte numbers (WBC), and platelet numbers (PLT). **P < .01. NS indicates not significant. (B) The mRNA expression levels of HIF-2α in spleen (left panel) and BM (right panel) were examined in WT, kd/kd, and kd/kd::Tie-1-Cre mice by quantitative RT-PCR. The expression levels seen in WT were normalized to a value of 1 as the standard in each tissue (P < .01). NS indicates not significant. (C) Peripheral blood analysis (RBC, Hb, Hct, WBC, and PLT) was performed in WT, kd/kd, and kd/kd::Tie-1-Cre mice (*P < .05, **P < .01). Error bars represent SD. (D) Flow cytometric analysis of spleen cells prepared from WT and kd/kd::Tie-1-Cre mice. The numbers shown depict the frequency (%) of CD71+/TER119+ and CD71−/TER119+ cells.
Analysis of the ability of the hematopoietic microenvironment to support erythropoiesis. (A) A transplantation assay. Hematopoietic cells (107 total BM cells/mouse) derived from donor mice (WT or kd/kd) were transplanted into lethally irradiated recipient mice (WT, kd/kd, or kd/kd::Tie-1-Cre, n = 3 for each group). Peripheral blood cells were measured for hematocrit levels (Hct), leukocyte numbers (WBC), and platelet numbers (PLT). **P < .01. NS indicates not significant. (B) The mRNA expression levels of HIF-2α in spleen (left panel) and BM (right panel) were examined in WT, kd/kd, and kd/kd::Tie-1-Cre mice by quantitative RT-PCR. The expression levels seen in WT were normalized to a value of 1 as the standard in each tissue (P < .01). NS indicates not significant. (C) Peripheral blood analysis (RBC, Hb, Hct, WBC, and PLT) was performed in WT, kd/kd, and kd/kd::Tie-1-Cre mice (*P < .05, **P < .01). Error bars represent SD. (D) Flow cytometric analysis of spleen cells prepared from WT and kd/kd::Tie-1-Cre mice. The numbers shown depict the frequency (%) of CD71+/TER119+ and CD71−/TER119+ cells.
To confirm that recovered HIF-2α expression in EC abrogates the impaired erythropoiesis in kd/kd mice, hematologic indices and differentiation of erythroid cells were assessed in kd/kd::Tie-1-Cre mice (Figure 4B-D). Quantitative RT-PCR analysis demonstrated that HIF-2α mRNA levels were completely recovered in kd/kd::Tie-1-Cre mice in spleen and BM (Figure 4B). These results indicate that EC is a major site of HIF-2α expression in these tissues. Hematologic analysis revealed that the decreased RBC count, hemoglobin, and hematocrit levels in kd/kd peripheral blood were increased to normal range in kd/kd::Tie-1-Cre peripheral blood, whereas neither WBC nor platelet counts were affected (Figure 4C). Furthermore, the frequency of TER119+/CD71− mature erythroblasts was comparable between WT and kd/kd::Tie-1-Cre spleens (Figure 4D). Collectively, these results signify that cell type–specific recovery of HIF-2α expression in kd/kd EC is sufficient to correct the defective erythroid cell differentiation.
VCAM-1 expression was reduced in kd/kd EC
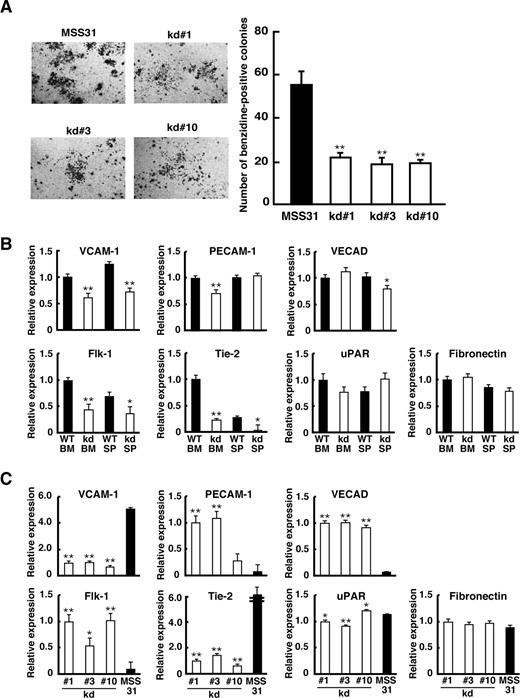
To examine directly the role of HIF-2α in erythropoiesis in EC, we established 3 independent cell lines, kd#1, kd#3, and kd#10, derived from ECs of the kd/kd spleen. As a control, we used an EC-derived cell line, MSS31 derived from WT spleen. The ability to support erythropoiesis was assessed by coculturing these cells with erythroid progenitors isolated from fetal livers (Figure 5A). After 4 days of coculturing, erythroid colonies were visualized by benzidine staining. As shown in Figure 5A, the number of erythroid colonies sustained by each kd/kd cell line was less than half that of MSS31 cells. This suggests that the ability to support erythroid cells is attenuated in kd/kd EC compared with WT.
Analyses of kd/kd-derived EC in vitro. (A) A coculture assay was performed using MSS31 (positive control) and kd/kd spleen-derived stromal cells (kd#1, kd#3, kd#10). The photographs represent benzidine-stained erythroid colonies developed on stromal cells. The number of benzidine-positive colonies consisting of more than 200 cells was scored in each well, and the average of colony number in triplicate wells for each stromal cell line is shown (**P < .01). (B) ECs were harvested from WT and kd/kd (kd) BM and spleens (SP). The mRNA expressions of adhesion molecules and endothelial receptors were examined by quantitative RT-PCR. The expression levels seen in WT BM were normalized to a value of 1 as the standard. ■ represents WT; □ represents kd/kd (*P < .05, **P < .01, compared with WT). (C) kd/kd spleen-derived stromal cells (□ represent kd#1, kd#3, kd#10) and MSS31 cells (■) were analyzed for the mRNA expression of adhesion molecules and endothelial receptors by quantitative RT-PCR. The expression levels seen in kd#1 cells were normalized to a value of 1 as the standard (*P < .05, **P < .01, compared with values from MSS31). Error bars represent SD.
Analyses of kd/kd-derived EC in vitro. (A) A coculture assay was performed using MSS31 (positive control) and kd/kd spleen-derived stromal cells (kd#1, kd#3, kd#10). The photographs represent benzidine-stained erythroid colonies developed on stromal cells. The number of benzidine-positive colonies consisting of more than 200 cells was scored in each well, and the average of colony number in triplicate wells for each stromal cell line is shown (**P < .01). (B) ECs were harvested from WT and kd/kd (kd) BM and spleens (SP). The mRNA expressions of adhesion molecules and endothelial receptors were examined by quantitative RT-PCR. The expression levels seen in WT BM were normalized to a value of 1 as the standard. ■ represents WT; □ represents kd/kd (*P < .05, **P < .01, compared with WT). (C) kd/kd spleen-derived stromal cells (□ represent kd#1, kd#3, kd#10) and MSS31 cells (■) were analyzed for the mRNA expression of adhesion molecules and endothelial receptors by quantitative RT-PCR. The expression levels seen in kd#1 cells were normalized to a value of 1 as the standard (*P < .05, **P < .01, compared with values from MSS31). Error bars represent SD.
Prior studies demonstrated that several cell adhesion molecules expressed on EC induce the proliferation and differentiation of hematopoietic cells.35-37 We therefore examined the mRNA levels of several adhesion molecules in ECs isolated from the BM and spleens of kd/kd and WT mice by quantitative RT-PCR (Figure 5B). Remarkably, the expression of vascular cell adhesion molecule (VCAM-1) mRNA was significantly lower in kd/kd than WT in both BM and spleen. Levels of fibronectin and urokinase-type plasminogen activator receptor mRNAs were comparable between kd/kd and WT ECs. For platelet endothelial cell adhesion molecule and vascular endothelial cadherin, the mRNA level was lower in kd/kd compared with WT in either BM or spleen, but not in the other tissue. Previous studies have shown that 2 receptor molecules for endothelial growth factors, Flk-1 and Tie-2, are regulated by HIF-2α6,38 and that Tie-2 protein expression was significantly reduced in HIF-2α−/− embryos.39 Consistent with these observations, Flk-1 and Tie-2 mRNA levels were lower in kd/kd EC compared with WT (Figure 5B).
We next examined the mRNA levels of these factors in the 3 kd/kd-derived EC lines by quantitative RT-PCR (Figure 5C). Consistent with the results for the tissue mRNAs, the expressions of VCAM-1 and Tie-2 were consistently lower in kd#1, kd#3, and kd#10 than in MSS31, whereas no clear difference in the expression of fibronectin and a significant but not large difference in urokinase-type plasminogen activator receptor were observed between kd/kd and MSS31 cells. Interestingly, vascular endothelial cadherin mRNA was undetectable and the levels of platelet endothelial cell adhesion molecule and Flk-1 mRNA were extremely low in MSS31 cells, yet moderate levels of these molecules were observed in kd/kd cells. Given that these molecules were clearly detectable in freshly isolated ECs from WT mice (Figure 5B), we assumed that the expression of these molecules was lost during the clonal expansion of MSS31 cells.
The ability to support erythropoiesis was increased by the forced expression of VCAM-1 mRNA in kd/kd EC
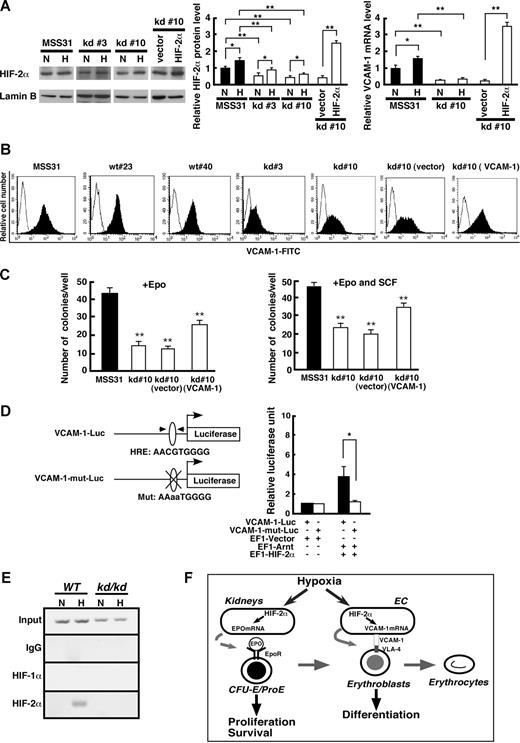
We next examined HIF-2α protein levels of kd/kd EC and MSS31 cells cultured under normoxic or hypoxic conditions (Figure 6A). HIF-2α protein levels of kd#3 and kd#10 cells were lower than that of MSS31 cells under both normoxic and hypoxic conditions, although the levels were increased on hypoxia. To test whether reduced VCAM-1 mRNA expression is rescued by the overexpression of HIF-2α in kd/kd cells, HIF-2α cDNA was transfected into kd#10 cells. The level of HIF-2α protein in transfected cells was nearly 2-fold increased compared with that of MSS31 cells in hypoxic condition (Figure 6A middle panel). As expected, the overexpression of HIF-2α significantly increased VCAM-1 mRNA level in kd#10 cells (Figure 6A right panel). The expression of VCAM-1 was further assessed by a flow cytometry. The EC lines derived from WT (wt#23 and #40) and kd/kd (kd#3 and #10) mice were incubated with FITC-conjugated VCAM-1-antibody and the intensity of FITC was examined (Figure 6B). The peak intensity of FITC was comparable with that of MSS31 cells in wt#23 and #40 cells. In contrast, the intensity was lower in kd#3 and #10 cells. Overall, these findings suggest that VCAM-1 expression in spleen-derived EC is likely to be regulated by HIF-2α.
VCAM-1 expression in EC is regulated by HIF-2α. (A) The HIF-2α protein levels in kd/kd stromal cells (kd#3, kd#10) and MSS31 cells were examined by Western blot (left panel). Nuclear extracts were prepared from the cells cultured under normoxic (N, 20% O2) or hypoxic (H, 1% O2 for 6 hours) conditions. Right 2 lanes represent samples from kd#10 transfected with either HIF-2α cDNA (HIF-2α) or empty vector (vector). Quantification of the band intensities from 3 independent assays is shown in the middle column. The HIF-2α expression levels seen in MSS31 cells under normoxic conditions were normalized to a value of 1 as the standard. Right column represents quantitative RT-PCR analysis of VCAM-1mRNA. The VCAM-1 expression levels seen in MSS31 cells under normoxic conditions were normalized to a value of 1 as the standard (**P < .01, *P < .05). (B) The expression of VCAM-1 was examined by flow cytometry. Stromal cell lines derived from WT (wt#34 and wt#40) and kd/kd (kd#3 and kd#10) spleens were examined. Right 2 panels represent data from kd#10 cells transfected with either full-length VCAM-1 cDNA (VCAM-1) or vector alone (vector). (C) A coculture assay was performed using VCAM-1–transfected kd#10 cells. E13.5 fetal liver cells (2 × 104 cells/well) were cultured on stromal cells in the presence of Epo (left) or Epo and SCF (right) for 4 days. The number of benzidine-positive colonies consisting of more than 200 cells was scored in each well and the average of colony number in triplicate wells for each stromal cell line is shown (**P < .01, compared with MSS31). (D) A luciferase reporter assay was performed using a WT VCAM-1-0.7 kb promoter (VCAM-1-Luc, top) and that with a mutation in HRE (VCAM-1-mut-Luc, bottom) as reporters. The pEF1–HIF-2α and pEF1-Arnt expression vectors were cotransfected with the reporter plasmid into 293T cells. Luciferase activities were measured for the VCAM-1-Luc (■) and VCAM-1-mut-Luc (□) reporters. The values shown are the averages of 3 independent experiments (means ± SD). The luciferase activity seen in 293T cells transfected with the reporter plasmid and empty vector was normalized to a value of 1 as the standard (*P < .05). (E) A chromatin immunoprecipitation assay was performed using anti-HIF-1α and anti–HIF-2α antibodies with WT and kd/kd EC under normoxic (N, 20% O2) or hypoxic (H, 1% O2 for 6 hours) conditions. Rabbit IgG was used as a negative control. Note that VCAM-1 expression was seen in WT EC using anti–HIF-2α antibody after treatment with 1% O2. (F) Schematic models for roles of HIF-2α on erythropoiesis.
VCAM-1 expression in EC is regulated by HIF-2α. (A) The HIF-2α protein levels in kd/kd stromal cells (kd#3, kd#10) and MSS31 cells were examined by Western blot (left panel). Nuclear extracts were prepared from the cells cultured under normoxic (N, 20% O2) or hypoxic (H, 1% O2 for 6 hours) conditions. Right 2 lanes represent samples from kd#10 transfected with either HIF-2α cDNA (HIF-2α) or empty vector (vector). Quantification of the band intensities from 3 independent assays is shown in the middle column. The HIF-2α expression levels seen in MSS31 cells under normoxic conditions were normalized to a value of 1 as the standard. Right column represents quantitative RT-PCR analysis of VCAM-1mRNA. The VCAM-1 expression levels seen in MSS31 cells under normoxic conditions were normalized to a value of 1 as the standard (**P < .01, *P < .05). (B) The expression of VCAM-1 was examined by flow cytometry. Stromal cell lines derived from WT (wt#34 and wt#40) and kd/kd (kd#3 and kd#10) spleens were examined. Right 2 panels represent data from kd#10 cells transfected with either full-length VCAM-1 cDNA (VCAM-1) or vector alone (vector). (C) A coculture assay was performed using VCAM-1–transfected kd#10 cells. E13.5 fetal liver cells (2 × 104 cells/well) were cultured on stromal cells in the presence of Epo (left) or Epo and SCF (right) for 4 days. The number of benzidine-positive colonies consisting of more than 200 cells was scored in each well and the average of colony number in triplicate wells for each stromal cell line is shown (**P < .01, compared with MSS31). (D) A luciferase reporter assay was performed using a WT VCAM-1-0.7 kb promoter (VCAM-1-Luc, top) and that with a mutation in HRE (VCAM-1-mut-Luc, bottom) as reporters. The pEF1–HIF-2α and pEF1-Arnt expression vectors were cotransfected with the reporter plasmid into 293T cells. Luciferase activities were measured for the VCAM-1-Luc (■) and VCAM-1-mut-Luc (□) reporters. The values shown are the averages of 3 independent experiments (means ± SD). The luciferase activity seen in 293T cells transfected with the reporter plasmid and empty vector was normalized to a value of 1 as the standard (*P < .05). (E) A chromatin immunoprecipitation assay was performed using anti-HIF-1α and anti–HIF-2α antibodies with WT and kd/kd EC under normoxic (N, 20% O2) or hypoxic (H, 1% O2 for 6 hours) conditions. Rabbit IgG was used as a negative control. Note that VCAM-1 expression was seen in WT EC using anti–HIF-2α antibody after treatment with 1% O2. (F) Schematic models for roles of HIF-2α on erythropoiesis.
A previous study demonstrated that erythroid progenitor cell development was inhibited by an antibody blocking VCAM-1.29 We therefore investigated whether VCAM-1 is a key adhesion molecule supporting erythropoiesis using a coculture assay (Figure 6C). An expression plasmid containing full-length of VCAM-1 cDNA was transfected into kd#10 cells (kd#10 [VCAM-1]). FACS analysis verified that VCAM-1 expression was greater in cells transfected with VCAM-1 cDNA than in control cells (Figure 6B). The kd#10 (VCAM-1) cells supported a 2-fold greater erythroid colony number than the parental kd#10 cells, and these cells transfected with an empty vector when cultured in the presence of Epo (Figure 6C left panel). However, even though the transfected cells expressed a level of VCAM-1 comparable with MSS31 cells, they sustained less erythroid colonies compared with MSS31 cells (Figure 6B,C). This suggests that additional factors might be required to support erythropoiesis in the coculture assay. Because stem cell factor (SCF)–c-Kit signaling plays an important role in supporting erythropoiesis in fetal liver stromal cells,40 SCF was added to the coculture with EPO (Figure 6C right panel). The number of erythroid colonies cultured on kd#10 (VCAM-1) cells was increased but was still lower than that of MSS31, suggesting that additional factors required for erythroid colony formation might be reduced in the kd#10 cells.
VCAM-1 mRNA expression is directly regulated by HIF-2α
To test whether the expression of VCAM-1 is directly regulated by HIF-2α, we performed a luciferase reporter assay using a plasmid containing a 0.7-kb region upstream of the predicted start codon of the VCAM-1 gene (VCAM-1-Luc) as a reporter (Figure 6D). The luciferase activity was significantly increased when the reporter plasmid was cotransfected with expression plasmids for HIF-2α and Arnt. In contrast, when a putative HRE sequence resides within this region was mutated (VCAM-1-mut-Luc), activation of the reporter was blunted. These results indicate that HIF-2α and Arnt activate VCAM-1 gene transcription via the putative HRE sequence.
To investigate whether HIF-2α binds to the VCAM-1 promoter region containing the HRE sequence in ECs, we performed a chromatin immunoprecipitation (Figure 6E). CD31+CD45− ECs were isolated from WT and kd/kd BM and cultured under normoxic or hypoxic conditions. HIF-1α failed to bind to the VCAM-1 promoter in either condition or cell genotype. HIF-2α only associated with the VCAM-1 promoter in WT EC under hypoxic conditions. Taken together with the results from the reporter assays, these data suggest that the HIF-2α/Arnt heterodimer binds to the HRE sequence within the VCAM-1 promoter to activate VCAM-1 gene transcription when ECs are exposed to hypoxia.
Discussion
There is increasing evidence that HIF-2α is a key regulator for Epo, a central player in the regulation of erythropoiesis in vivo.16-18 In the present study, however, we found a novel, Epo-independent role of HIF-2α for erythropoiesis, maintaining erythropoietic microenvironment. HIF-2α/Arnt heterodimer regulates the gene transcription of several factors in ECs that control erythroid cell differentiation in adult hematopoietic tissue. Especially, our results indicated that the adhesion molecule VCAM-1 is directly regulated by HIF-2α/Arnt and plays an essential role in promoting a hematopoietic microenvironment appropriate for erythropoiesis. On the basis of previous observations and the data presented here, we propose that HIF-2α activates erythropoiesis on hypoxia via at least 2 distinct mechanisms (Figure 6F). In kidneys, HIF-2α activates Epo gene transcription in response to hypoxia.8,16,17 Epo has an endocrine effect on the proliferation of erythroid progenitors through EpoR signaling. On the other hand, in EC of hematopoietic tissues, HIF-2α activates VCAM-1, which exerts a paracrine effect on the differentiation of erythroblasts via its partner VLA-4.29,41,42 Interestingly, whereas Epo-EpoR signaling promotes cell survival and proliferation at the CFU-E/proerythroblast stage,43 VCAM-1 expression in EC affects the maturation of erythroblasts, later stages of erythroid cell differentiation. Thus, the 2 distinct mechanisms may act cooperatively to control the proliferation and differentiation of erythroid cells in response to hypoxia.
Using kd/kd::Tie-1-Cre compound mice, we found that recovered HIF-2α expression in ECs abrogated anemia and abnormal spleen architecture (Figure 3). Notably, the impaired hematopoietic microenvironment for erythropoiesis was accompanied by the reduction of red pulp area, microcapillary formation, and CD31-positive cells in the kd/kd spleen. Moreover, we found that the mRNA levels of Tie-2, a receptor for a growth factor angiopoietin, were consistently decreased in cell lines derived from kd/kd ECs (Figure 5C). These observations suggest that HIF-2α may play broad, fundamental roles on EC biology, such as cell survival, differentiation, proliferation, and migration in spleen. Further studies are required to fully understand roles of HIF-2α in EC.
As for EC function of supporting erythropoiesis, our data provided evidence that HIF-2α/Arnt heterodimer directly activates VCAM-1 gene transcription in EC. Contrary to our observation, a previous study demonstrated that VCAM-1 mRNA expression level was greater in the BM of HIF-2α−/− mice compared with WT, suggesting HIF-2α as a negative regulator of VCAM-1. In that study, mRNA levels from whole BM tissues containing both hematopoietic and stromal cells were examined. VCAM-1 is expressed in vascular smooth muscle cells and other stromal cells in addition to ECs in BM.44,45 Collectively, roles of HIF-2α in the regulation of VCAM-1 gene expression might be dependent on the cellular context of hematopoietic microenvironment.
EC express several adhesion molecules on their surfaces that mediate the interactions between hematopoietic cells and ECs.46,47 Our data here show that VCAM-1 expressed in EC plays a major role in supporting erythropoiesis. Importantly, however, we found evidence suggesting that VCAM-1 may not be the only target gene of HIF-2α involved in the regulation of erythropoiesis in ECs. VCAM-1 transfected-kd#10 cells increased the erythroid colonies compared with parental cells, but still less than that of MSS31 cells even in the addition of SCF (Figure 6C). Thus, it is conceivable that HIF-2α regulates expression of multiple molecules in ECs and these factors may act cooperatively to support erythropoiesis. Further studies are needed to determine the subset of genes regulated by HIF-2α required for supporting erythropoiesis.
In conclusion, our results show the first evidence that the hematopoietic microenvironment required for erythropoiesis is dynamically regulated by oxygen through the functions of HIF-2α in EC. Together with the fact that Epo is mainly regulated by HIF-2α, our findings demonstrate that HIF-2α plays central roles in the oxygen-dependent regulation of erythropoiesis.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Tania O'Connor for critical reading of the manuscript and Ms N. Kaneko for histologic analyses.
This work was supported by grants from Solution Oriented Research for Science and Technology (SORST), the Japan Science and Technology Agency.
Authorship
Contribution: T.Y., A.S., and F.I. performed research; O.O. and K.O. designed research and wrote the paper; Y.F.-K. designed the research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Osamu Ohneda, Department of Regenerative Medicine, University of Tsukuba, 1-1-1 Tennoudai, Tsukuba 305-8575, Japan; e-mail: oohneda@md.tsukuba.ac.jp.
References
Author notes
T.Y. and O.O. contributed equally to this work.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal