Abstract
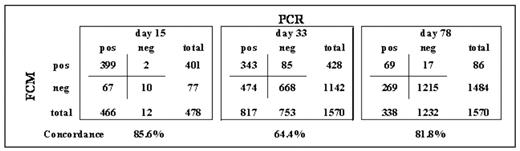
In the AIEOP-BFM ALL2000 trial, childhood acute lymphoblastic leukemia (ALL) patients were stratified mainly according to minimal residual disease (MRD) levels at day 33 (Time Point 1, TP1) and 78 (TP2) of treatment, as detected by PCR amplification of clonotypic immunoglobulin and T-cell receptor gene rearrangements. Overall, the results confirmed that PCR-based MRD detection is an independent prognostic indicator which overrides classical risk factors. In this context, MRD measurement by flow cytometry (FCM) on day 15 has been evaluated as an earlier predictor of relapse-free outcome in children with ALL. To understand how these two methods could be applied in a MRD-based clinical protocol, we analyzed the correlation of methodologies in data sets in which PCR and FCM were simultaneously applied. We studied 3,618 BM samples from 1,570 patients (32.5% of all patients on trial) derived from day15 (n=478, 30.4%), 33 (n=1,570) and 78 (n=1,570) of remission induction therapy of the AIEOP-BFM ALL 2000 protocol. Patients were enrolled from September 2000 to June 2006 in Italy, Austria and Germany, and selection was based only on available samples for both PCR and FCM. As in most MRD-based protocols, RQ-PCR was performed on BM mononuclear cells after Ficoll gradient separation, while 4-colors FCM was done on 300,000 nucleated cells (NC) from the whole BM sample after red blood cells lyse-wash procedure. MRD levels ≥10−4 were considered as ‘positive’, while levels below this threshold, not-quantifiable or undetectable were classified as ‘negative’. Overall, qualitative concordance was observed in 2,704/3,618 samples (74.7%) measured at d15, d33 and d78. Concordances at each TP are indicated in the Table.
Around 70% of discordant samples in all TPs were cases with low-positivity by PCR (10−4 log range) and negative by FCM, or viceversa. Concordance was also evaluated according to PCR-based MRD risk subgroups. Of note, 519/571 (90.9%) of Standard Risk by PCR-MRD (negative at d33 by 2 markers with sensitivity ≥10−4) were also FCM negative at d33; whereas 59/121 (48.8%) of cases PCR ≥10−3 at d78 (High Risk by PCR-MRD) were FCM negative. In order to evaluate the correlation of methodologies more exactly, we investigated similar cell preparations (same MNC divided for PCR and FCM), and we enhanced the resolution of FCM by using 7-colors and analyzing 500,000 MNC. Samples were collected at days 15, 33, 52, and 78 within a single center. Among a total of 266 samples, the concordance increased up to 87%. More specifically, 100 samples (37.6%) resulted undetectable by both methods; 56 samples were FCM-undetectable and PCR-positive <10−4, while only 13 samples were PCR-positive ≥10−4 but undetectable by FCM. In summary, several methodological issues limit the concordance of FCM and PCR as used by the AIEOP-BFM MRD study group. These include differences in the material analyzed by PCR and FCM, as well as in the amount of sample input (number of cells analyzed). The recent implementation of further technical developments in the FCM procedure, are designed to increase the concordance rate. In conclusion, within BFM-based protocols, FCM cannot simply substitute the current PCR-based MRD risk stratification at the same TPs. Instead, a tailored FCM-based risk definition may be independently reliable. Moreover, the two methodologies applied at different TPs might be complementary in more advanced MRD-based patient stratification protocols. Overall, the choice of the MRD methodology largely depends on the aims of the study protocol, resources available, and treatment strategy.
Disclosures: No relevant conflicts of interest to declare.
Author notes
Corresponding author

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal