Abstract
Cdx1, Cdx2, and Cdx4 comprise the caudal-like Cdx gene family in mammals, whose homologues regulate hematopoietic development in zebrafish. Previously, we reported that overexpression of Cdx4 enhances hematopoietic potential from murine embryonic stem cells (ESCs). Here we compare the effect of ectopic Cdx1, Cdx2, and Cdx4 on the differentiation of murine ESC-derived hematopoietic progenitors. The 3 Cdx genes differentially influence the formation and differentiation of hematopoietic progenitors within a CD41+c-kit+ population of embryoid body (EB)–derived cells. Cdx1 and Cdx4 enhance, whereas Cdx2 strongly inhibits, the hematopoietic potential of CD41+ckit+ EB-derived cells, changes that are reflected by effects on hematopoietic lineage-specific and Hox gene expression. When we subject stromal cell and colony assay cultures of EB-derived hematopoietic progenitors to ectopic expression of Cdx genes, Cdx4 dramatically enhances, whereas Cdx1 and Cdx2 both inhibit hematopoietic activity, probably by blocking progenitor differentiation. These data demonstrate distinct effects of Cdx genes on hematopoietic progenitor formation and differentiation, insights that we are using to facilitate efforts at in vitro culture of hematopoietic progenitors from ESC. The behavior of Cdx genes in vitro suggests how derangement of these developmental regulators might contribute to leukemogenesis.
Introduction
The caudal (Cdx) family of homeobox proteins, comprising Cdx1, Cdx2, and Cdx4 in mammals, is classically studied as regulators of axial elongation and anterior/posterior axis patterning via Hox gene modulation, although work in zebrafish suggests a novel role in hematopoietic development. Cdx4-deficient fish display diminished blood development, and further suppression of Cdx1 results in complete ablation of posterior embryonic blood.1,2 This behavior is reminiscent of the redundant regulation of anterior/posterior patterning because compound Cdx mutants show more severe homeotic transformations relative to individual mutant animals.3-5 Several master regulators of early hematopoietic development (eg, Scl/Tal-1 and Runx1/AML-1) were originally identified via association with leukemia, suggesting that embryonic pathways reactivate in adult disease.6 Similarly, Cdx genes have been implicated recently in leukemogenesis in mouse models (Cdx2 and Cdx4) and are shown to aberrantly express in t(14;18) lymphoma cell lines as well as many human acute myeloid leukemia (AML) patient samples.7-11 Elucidating the role of Cdx genes during normal development will inform studies of their transforming potential in leukemia.
Recently, our laboratory demonstrated that enforced Cdx4 expression could promote hematopoietic development in differentiating murine embryonic stem cells (ESCs), suggesting that Cdx-mediated regulation of hematopoietic development is a conserved pathway in mammals.12 However, the role of Cdx1 and Cdx2 in mammalian hematopoietic development has not yet been systematically explored. The study of early hematopoietic development is challenging in mammals, largely because the developing embryo is difficult to access. In contrast, ESC faithfully mimic early embryonic development when induced to differentiate in vitro as embryoid bodies (EBs). The 3 germ layers emerge in sequence, and gross hematopoietic differentiation is readily observed via the appearance of pigmented blood islands between day 6 and day 8 of differentiation. Thus, ESC have been elegantly exploited to elucidate the role of multiple genes and morphogens in hematopoietic development.13-22
To facilitate the appraisal of genes involved in mammalian development, our laboratory created a system in which any candidate gene can be placed under the control of a tetracycline-inducible promoter, allowing inducible and reversible expression during murine ESC in vitro differentiation23 (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Here, using this system, we systematically explore the effect of ectopic expression of each mammalian Cdx family member on both the generation of hematopoietic progenitors and their ability to expand and differentiate (Figure 1A). We demonstrate that each Cdx gene influences murine ESC-derived hematopoietic development in distinct ways by modulating the generation of CD41+c-kit+ hematopoietic progenitors.
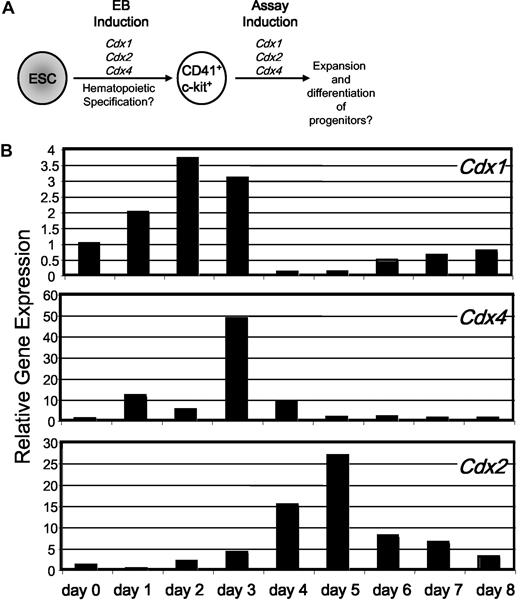
Endogenous Cdx gene expression in differentiating EBs. (A) A schematic representation of the questions under study: can Cdx genes affect either the specification of hematopoietic progenitors (ie, induction of Cdx gene expression during EB differentiation) and/or the expansion and differentiation of hematopoietic progenitors (ie, induction after plating purified progenitors in OP9 and m3434 assays)? (B) Quantitative RT-PCR analysis of endogenous Cdx1, Cdx2, and Cdx4 gene expression in differentiating AinV15 EBs from day 0 through day 8. Expression is presented relative to expression level of each gene in undifferentiated ESC, after normalization to the housekeeping gene β-actin. Shown is a representative example of several independent experiments in multiple distinct ESC lines (AinV15 and CCE).
Endogenous Cdx gene expression in differentiating EBs. (A) A schematic representation of the questions under study: can Cdx genes affect either the specification of hematopoietic progenitors (ie, induction of Cdx gene expression during EB differentiation) and/or the expansion and differentiation of hematopoietic progenitors (ie, induction after plating purified progenitors in OP9 and m3434 assays)? (B) Quantitative RT-PCR analysis of endogenous Cdx1, Cdx2, and Cdx4 gene expression in differentiating AinV15 EBs from day 0 through day 8. Expression is presented relative to expression level of each gene in undifferentiated ESC, after normalization to the housekeeping gene β-actin. Shown is a representative example of several independent experiments in multiple distinct ESC lines (AinV15 and CCE).
Methods
Cell culture and differentiation
Murine ESC were cultured as reported on irradiated mouse embryonic fibroblasts in Dulbecco modified Eagle medium with 15% fetal calf serum (HyClone Laboratories, Logan, UT), 1000 U/mL leukemia inhibitory factor (Chemicon International, Temecula, CA), 2 mM penicillin/streptomycin/glutamine (Invitrogen, Carlsbad, CA), 0.1 mM nonessential amino acids (Invitrogen), and 0.1 mM β-mercaptoethanol (Sigma-Aldrich, St Louis, MO) at 37°C/5% CO2.12,23 Media was refreshed daily, and cultures were passaged with trypsin (Invitrogen) every 2 to 3 days.
OP9 stromal cells (ATCC, Manassas, VA) were maintained in α-MEM supplemented with 2 mM penicillin/streptomycin/glutamine and 20% heat-inactivated fetal calf serum (Invitrogen). Cells were passaged every 4 to 6 days.
Murine ESC were differentiated as described previously.12,23 Confluent cultures were harvested and resuspended at a concentration of 100 cells/15 μL in EB differentiation media composed of Iscove modified Dulbecco medium (IMDM) plus 15% fetal calf serum (StemCell Technologies, Vancouver, BC), 2 mM penicillin/streptomycin/glutamine (Invitrogen), 4.5 mM monothioglycerol (Sigma-Aldrich), 200 μg/mL holo-transferrin (Sigma-Aldrich), and 50 μg/mL ascorbic acid (Sigma-Aldrich). EBs were cultured in 15 μL hanging drops for 48 hours and then transferred and cultured in 10 cm2 Petri dishes for an additional 4 days at 37°C/5% CO2 while shaking at 50 rpm. For ectopic gene expression, doxycycline (0.5-1.0 μg/mL; Sigma-Aldrich) was added as indicated.
Engineering of inducible Cdx1 and Cdx2 ES cells
Cdx1 cDNA was kindly provided by Christopher Wright (Vanderbilt University Medical Center, Nashville, TN), and Cdx2 cDNA was provided by Gary Gilliland (Brigham and Women's Hospital, Harvard Medical School, Boston, MA). Cdx1 and Cdx2 were each subcloned into the plox vector and electroporated with pSALK-Cre into 4 to 8 × 106 AinV15 ESC (ATCC).23 Cells were placed under selection with G418 (400 μg/mL; Sigma-Aldrich) for 10 to 14 days. Single colonies were collected, expanded, and screened via polymerase chain reaction (PCR) for correctly targeted hypoxanthine phosphoribosyltransferase loci (forward primer: ctagatctcgaaggatctggag, reverse primer: atactttctcggcaggagca) and inducible gene expression via immunoblot and real-time reverse transcriptase–polymerase chain reaction (RT-PCR).
Immunoblots
Cdx1, Cdx2, or Cdx4 transgene expression was induced with 1 μg/mL doxycycline (Sigma-Aldrich) at day 4 of EB differentiation. Protein extracts were prepared and immunoblot performed according to standard techniques. Briefly, 13 (iCdx1), 7 (iCdx2), or 16 μg (iCdx4) samples were loaded on a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel (Bio-Rad, Hercules, CA). After transfer of proteins onto polyvinylidene difluoride membrane (Bio-Rad), nonspecific binding was blocked by incubation with Tris-buffered saline/Tw (25 mM Tris, 25 mM glycine, 0.1% SDS, and 0.1% Tween-20) containing 5% milk powder and treated with rabbit Cdx1 antisera (kindly provided by P. Gruss, Max-Planck-Institute, Göttingen, Germany) diluted 1:1000 in blocking buffer, a monoclonal mouse Cdx2 antibody (Biogenex, San Ramon, CA) diluted 1:2000, or rabbit Cdx4 antisera (Aviva Biosystems, San Diego, CA) diluted 1:500. Secondary antibodies used were horseradish peroxidase–coupled rabbit or mouse antisera (GE Healthcare, Chalfont St Giles, United Kingdom) diluted 1:5000 in blocking buffer. Proteins were detected using the GE Healthcare enhanced chemiluminescence detection kit as described by the manufacturer. β-Actin was detected as a loading control using mouse β-actin antibody (BD Transduction Laboratories, San Jose, CA) diluted at 1:2000 and horseradish peroxidase coupled secondary antibody as described above.
Assays of hematopoietic colony formation
Hematopoietic progenitor colony assays were performed as described previously.12,23 EBs were collected at day 6 of differentiation and dissociated via treatment for 20 minutes at 37°C with an enzyme mixture diluted in phosphate-buffered saline to a final concentration of 2 mg/mL collagenase IV (Invitrogen), 10 mg/mL hyaluronidase (Sigma-Aldrich), and 160 U/mL DNase (Sigma-Aldrich), followed by trituration in 8 mL enzyme-free dissociation buffer (Invitrogen). Cells were counted manually via trypan-blue exclusion and plated at the indicated concentrations in methycellulose-based media (M3434; StemCell Technologies) in nonadherent 30 mm2 nontreated dishes (StemCell Technologies). For ectopic gene induction, 0.5 μg/mL doxycycline was used, unless otherwise indicated in the figure legend (Sigma-Aldrich). Ten days later, colonies were scored based on gross morphology.
OP9 colony activity was assessed via plating day 6 EB-derived cells at the indicated concentrations in 12-well tissue culture plates prepared 24 hours before with 12 500 OP9 stromal cells per well. Cells were cultured in IMDM supplemented with 10% heat-inactivated fetal calf serum (Invitrogen), 2 mM penicillin/streptomycin/glutamine (Invitrogen), 100 ng/mL hFlt3L, 100 ng/mL human stem cell factor, 40 ng/mL human thrombopoietin, and 40 ng/mL murine vascular endothelial growth factor. Cytokines were purchased from PeproTech (Rocky Hill, NJ). For ectopic gene expression, 0.5 μg/mL doxycycline was used unless otherwise indicated in figure legend (Sigma-Aldrich). Wells were scored 7 to 10 days later for colony number.
Cell fractionation
Day 6 EB-derived cells were stained with anti–CD41-phycoerythrin (PE; MWReg30; BD PharMingen, San Diego, CA) and/or anti–ckit-APC (2B8; BD PharMingen). Cells were fractionated via magnetic bead selection or fluorescence activated cell sorting (FACS). For FACS, a triple-LASER instrument (FACSAria; BD Biosciences, Franklin Lake, NJ) was used, and 7AAD (Sigma-Aldrich) was used to exclude dead cells. For magnetic bead selection, anti-PE microbeads (Miltenyi Biotec, Auburn, CA) and LS columns (Miltenyi Biotec) were used according to manufacturer instructions. The purity of fractionated populations was always assessed via postsort or postfractionation analysis and was routinely more than 90%.
Real-time RT-PCR
Cells were harvested in TRIZOL Reagent (Invitrogen), and total RNA was isolated according to manufacturer instructions. RNA samples were treated with DNaseI (Ambion, Austin, TX). cDNAs were prepared according to manufacturer instruction (Superscript II Reagent; Invitrogen). Real-time quantitative PCR was performed using SYBR Green reagent kits (Stratagene, La Jolla, CA) on an MX3000P Stratagene PCR instrument. Primer sequences were used as reported previously24 or listed in Table 1. The annealing temperature was 60°C for all reactions, and primers were used at 300 nM.
Sequences of primers employed in quantitative RT-PCR
| Gene . | Primer sequence . |
|---|---|
| Nkx2.5 | GACAAAGCCGAGACGGATGG |
| CTGTCGCTTGCACTTGTAGC | |
| Gata1 | CATTGGCCCCTTGTGAGGCCAG |
| CGCTCCAGCCAGATTCGACCC | |
| Gata2 | CTGCAACACACCACCCGATA |
| GGAGCGAGCCTTGCTTCTC | |
| Gata3 | CTGACGGAAGAGGTGGACGTACTTT |
| TAGCCCTGACGGAGTTTCCGTAGTA | |
| Pax6 | AGCCCGTCAGTGAATGGGCGGAGTTAT |
| TGGCCTCGATTACAGTAAAGAGAGAAGGAG | |
| LMO2 | GGA TCC TCG GCC ATC GAA AGG AAG AGC |
| ATC CCA TTG ATC TTG GTC CAC TC | |
| PU.1 | ATG GAA GGG TTT TCC CTC ACC GCC |
| GTC CAC GCT CTG CAG CTC TGT GAA | |
| HNF439 | ACA CGT CCC CAT CTG AAG GTG |
| CTT CCT TCT TCA TGC CAG CCC | |
| Cdx1 | GTA AGA CCC GAA CCA AGG AC |
| GGA ACC AGA TCT TTA CCT GC | |
| Cdx2 | CAG TCC CTA GGA AGC CAA GTG AAA |
| AAG TGA AAC TCC TTC TCC AGC TCC | |
| Cdx4 | GAG GAA GTC AGA GCT GGC AGT TA |
| GGC TCT GCG ATT CTG AAA CC | |
| HoxA2 | CGGCCACAAAGAATCCCTGGAAAT |
| AAAGCTGAGTGTTGGTGTACGCGGTT | |
| HoxA4 | TTATAACGGAGGCGAACCTAAGCG |
| GGT CAG GTA GCG GTT AAA GTG GAA | |
| HoxA613,40 | CCG AGC AGC AGT ACA AAC CTG |
| TGC CTT CCT CAT GGA GGG | |
| HoxA713,40 | TTT CCG CAT CTA CCC CTG G |
| TAG GTC TGG CGT CCC CG | |
| HoxA913,40 | CCG AAA ACA ATG CCG AGA AT |
| CCG GGT TAT TGG GAT CGA T | |
| HoxA1013,40 | TCG CCG GAG AAG GAC TCC |
| TTT GCT GTG AGC CAG TTG G | |
| HoxA13 | CTT CGC CGA CAA GTA CAT GGA CAC |
| CGT AGC CCT GAT GGT AGA AAG CAA | |
| HoxB1 | AAG GAA TCG CCT TGC TCG TCA GAA |
| ACC TTC GCT GTC TTA GGT GGG TTT | |
| HoxB4 | CCTGGATGCGCAAAGTTCA |
| CTTGGGCTCCCCGCC | |
| HoxB513,40 | TCC CCT GGA TGA GGA AGC TT |
| CGG GCC CTT TTT CCG T | |
| HoxB613,40 | CGTCTACCCGTGGATGCAG |
| TCGCCGACCGCTGG | |
| HoxB713,40 | TTG GCG GCC GAG AGT AAC |
| TCG GTC AGG CCC TGA GC | |
| HoxB8 | GACTCGCAAGCGGAGGATC |
| GAACCAGATTTTGACCTGTCTCTCT | |
| HoxB9 | ACC AAA TAC CAG ACG CTG GAG CTA |
| TCA GAT TGA GGA GTC TGG CCA CTT |
| Gene . | Primer sequence . |
|---|---|
| Nkx2.5 | GACAAAGCCGAGACGGATGG |
| CTGTCGCTTGCACTTGTAGC | |
| Gata1 | CATTGGCCCCTTGTGAGGCCAG |
| CGCTCCAGCCAGATTCGACCC | |
| Gata2 | CTGCAACACACCACCCGATA |
| GGAGCGAGCCTTGCTTCTC | |
| Gata3 | CTGACGGAAGAGGTGGACGTACTTT |
| TAGCCCTGACGGAGTTTCCGTAGTA | |
| Pax6 | AGCCCGTCAGTGAATGGGCGGAGTTAT |
| TGGCCTCGATTACAGTAAAGAGAGAAGGAG | |
| LMO2 | GGA TCC TCG GCC ATC GAA AGG AAG AGC |
| ATC CCA TTG ATC TTG GTC CAC TC | |
| PU.1 | ATG GAA GGG TTT TCC CTC ACC GCC |
| GTC CAC GCT CTG CAG CTC TGT GAA | |
| HNF439 | ACA CGT CCC CAT CTG AAG GTG |
| CTT CCT TCT TCA TGC CAG CCC | |
| Cdx1 | GTA AGA CCC GAA CCA AGG AC |
| GGA ACC AGA TCT TTA CCT GC | |
| Cdx2 | CAG TCC CTA GGA AGC CAA GTG AAA |
| AAG TGA AAC TCC TTC TCC AGC TCC | |
| Cdx4 | GAG GAA GTC AGA GCT GGC AGT TA |
| GGC TCT GCG ATT CTG AAA CC | |
| HoxA2 | CGGCCACAAAGAATCCCTGGAAAT |
| AAAGCTGAGTGTTGGTGTACGCGGTT | |
| HoxA4 | TTATAACGGAGGCGAACCTAAGCG |
| GGT CAG GTA GCG GTT AAA GTG GAA | |
| HoxA613,40 | CCG AGC AGC AGT ACA AAC CTG |
| TGC CTT CCT CAT GGA GGG | |
| HoxA713,40 | TTT CCG CAT CTA CCC CTG G |
| TAG GTC TGG CGT CCC CG | |
| HoxA913,40 | CCG AAA ACA ATG CCG AGA AT |
| CCG GGT TAT TGG GAT CGA T | |
| HoxA1013,40 | TCG CCG GAG AAG GAC TCC |
| TTT GCT GTG AGC CAG TTG G | |
| HoxA13 | CTT CGC CGA CAA GTA CAT GGA CAC |
| CGT AGC CCT GAT GGT AGA AAG CAA | |
| HoxB1 | AAG GAA TCG CCT TGC TCG TCA GAA |
| ACC TTC GCT GTC TTA GGT GGG TTT | |
| HoxB4 | CCTGGATGCGCAAAGTTCA |
| CTTGGGCTCCCCGCC | |
| HoxB513,40 | TCC CCT GGA TGA GGA AGC TT |
| CGG GCC CTT TTT CCG T | |
| HoxB613,40 | CGTCTACCCGTGGATGCAG |
| TCGCCGACCGCTGG | |
| HoxB713,40 | TTG GCG GCC GAG AGT AAC |
| TCG GTC AGG CCC TGA GC | |
| HoxB8 | GACTCGCAAGCGGAGGATC |
| GAACCAGATTTTGACCTGTCTCTCT | |
| HoxB9 | ACC AAA TAC CAG ACG CTG GAG CTA |
| TCA GAT TGA GGA GTC TGG CCA CTT |
Statistics
For all experiments, error bars represent the standard error, and P values are derived via the application of a 2-tailed, unpaired Student t test.
Results
Endogenous Cdx gene expression in differentiating murine ESC
ESC differentiation into EBs recapitulates the commitment events of early embryonic development and can be documented as temporal waves of lineage-specific gene expression.14 Brachyury-positive, primitive streak–like mesodermal cells emerge in the EB sequentially between day 2 and day 4 of EB development.14 Gene expression that indicates commitment to the hematopoietic and endothelial fate is first detectable between day 3 and day 4 of EB differentiation, with hematopoietic progenitor potential peaking around day 6.14 Focusing on day 0 to day 8 of differentiation, we assessed the endogenous expression of the 3 mammalian Cdx genes during EB differentiation via quantitative RT-PCR (Figure 1B).
Undifferentiated mouse ESC expressed Cdx1 but only low levels of Cdx4 and Cdx2 (Figure 1B; data not shown). When ESC were differentiated as EBs, Cdx1 expression increased, peaking between day 2 and day 3 of differentiation before extinguishing abruptly. Cdx4 was expressed in a relatively restricted interval of differentiation, peaking sharply at day 3 and then waning by day 5. Cdx2 expression was detectable at day 3 of differentiation, peaked at day 5, and dropped at day 6 (Figure 1B). Thus, the 3 known mammalian Cdx genes are expressed in overlapping temporal domains, with Cdx1 preceding Cdx4, which is then followed by Cdx2. All 3 Cdx genes are expressed during the reported window of hemangioblast and blood fate specification (day 3 to day 4 of EB differentiation),25 suggesting a potential role in hematopoiesis. Therefore, we sought to explore the individual effects of each Cdx gene on hematopoietic progenitors formed in EBs and on their expansion and differentiation in subsequent assays of hematopoietic colony formation on stromal cultures and in methylcellulose.
Cdx1, Cdx2, and Cdx4 distinctively regulate hematopoietic development in differentiating EBs
To test the effect of Cdx gene expression on blood formation, we generated mouse ESC with Cdx1 and Cdx2 under the control of a tetracycline-inducible promoter23 (Figure S1). ESC that conditionally express Cdx4 (iCdx4 ESC) were described previously.12 Exposure of iCdx1, iCdx2, or iCdx4 differentiating ESC to doxycycline resulted in a dramatic increase in Cdx transcripts and protein by 8 hours after induction, verifying the doxycycline-dependent nature of Cdx induction in these ESC lines (Figure 2A,B). No induction was observed when the unmodified parental ESC line (Ainv15) was exposed to doxycycline (Figure 2A,B).
Cdx genes distinctly affect hematopoietic progenitor formation during EB differentiation. (A) Quantitative RT-PCR analysis of inducible Cdx1, Cdx2, and Cdx4 gene expression in differentiating day 4 EBs assessed 8 hours after exposure to 1 μg/mL doxycycline (dox). Expression is presented relative to expression level of each gene in noninduced day 4 EBs harvested at the same time point. Normalization to the housekeeping gene β-actin has been performed for each individual sample. Data are representative of multiple independent experiments.(B) Immunoblot for Cdx1, Cdx2, and Cdx4 protein levels in day 4 iCdx1, iCdx2, or iCdx4 EBs collected 8 hours after exposure to 1 μg/mL doxycycline (+). The multiple bands detected by anti-Cdx2 represent distinct phosphorylation forms.41 Control samples (−) represent EBs of the same ESC line, harvested at the same time point, that have not been exposed to doxycycline. Data are representative of multiple independent experiments. iCdx1, iCdx4, and iCdx2 EBs were exposed to doxycycline during the indicated windows of differentiation, dissociated at day 6 of differentiation, and then plated into secondary assays with or without continued exposure to doxycycline. The effect of Cdx gene induction on OP9 colony forming potential (C) and hematopoietic progenitor potential (D; CFU formation) was assessed. To pool and average data from multiple experiments (each data point denotes between 3 and 8 replicates), the data are presented as fold change in colony potential relative to noninduced control. P values calculated using nonpaired, 2-tailed Student t test are depicted over bars.
Cdx genes distinctly affect hematopoietic progenitor formation during EB differentiation. (A) Quantitative RT-PCR analysis of inducible Cdx1, Cdx2, and Cdx4 gene expression in differentiating day 4 EBs assessed 8 hours after exposure to 1 μg/mL doxycycline (dox). Expression is presented relative to expression level of each gene in noninduced day 4 EBs harvested at the same time point. Normalization to the housekeeping gene β-actin has been performed for each individual sample. Data are representative of multiple independent experiments.(B) Immunoblot for Cdx1, Cdx2, and Cdx4 protein levels in day 4 iCdx1, iCdx2, or iCdx4 EBs collected 8 hours after exposure to 1 μg/mL doxycycline (+). The multiple bands detected by anti-Cdx2 represent distinct phosphorylation forms.41 Control samples (−) represent EBs of the same ESC line, harvested at the same time point, that have not been exposed to doxycycline. Data are representative of multiple independent experiments. iCdx1, iCdx4, and iCdx2 EBs were exposed to doxycycline during the indicated windows of differentiation, dissociated at day 6 of differentiation, and then plated into secondary assays with or without continued exposure to doxycycline. The effect of Cdx gene induction on OP9 colony forming potential (C) and hematopoietic progenitor potential (D; CFU formation) was assessed. To pool and average data from multiple experiments (each data point denotes between 3 and 8 replicates), the data are presented as fold change in colony potential relative to noninduced control. P values calculated using nonpaired, 2-tailed Student t test are depicted over bars.
iCdx1, iCdx2, and iCdx4 ESC were differentiated in the presence or absence of Cdx induction (doxycycline addition to culture medium between day 2 and day 4 or day 4 and day 6 of EB differentiation). At day 6, EBs were dissociated and assayed for hematopoietic potential as reflected by the ability to form colonies on OP9 stroma and colony-forming units (CFUs) in semisolid media supplemented with cytokines (Figure 2C,D; Tables S1,S2). EB-derived cells that expand on OP9 stroma are highly enriched in multipotent hematopoietic progenitor potential and, after genetic modification with HoxB4, display in vivo hematopoietic repopulating potential when injected into irradiated mice.12,23 Thus, OP9 coculture can support the expansion and/or maturation of cells with hematopoietic stem cell (HSC) activity. CFU potential reflects hematopoietic progenitor activity, including the downstream-committed progeny of HSC that gives rise to mixed colonies in these assays (CFU-GEMM, which designates a mixed colony composed of granulocytes, erythrocytes, macrophages, and megakaryocytes).
Induction of Cdx1 or Cdx4 either between day 2 and day 4 or day 4 and day 6 of EB differentiation significantly increased both OP9-colony number and CFUs, especially enhancing the formation of CFU-GEMM, the most multipotent progenitor detectable by this assay. The inductive effect of Cdx1 was most prominent when induced from day 4 to day 6, whereas Cdx4 had a stronger impact from day 2 to day 4 (Figure 2C,D EB induction). In contrast, overexpression of Cdx2 in either developmental window had the opposite effect of Cdx1 and Cdx4, nearly abolishing all EB-derived hematopoietic potential (Figure 2C,D EB induction). Cdx2 suppressed hematopoietic activity in a dose-dependent manner (Figure S2A; data were confirmed in 4 individually derived iCdx2 ESC clones). Importantly, no significant changes were observed in gross morphology or total cell numbers in EBs after doxycycline induction of Cdx2, suggesting that the suppressive effect on hematopoiesis was not attributable to unspecific inhibition of cell growth or induction of apoptosis (Figure S2B; data not shown).
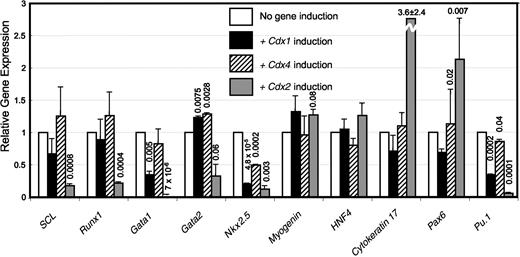
To further explore the effect of Cdx overexpression on hematopoiesis and lineage commitment in differentiating murine ESC, we examined the effect of Cdx induction on the expression levels of genes representative of the hematopoietic system (Scl, Runx1, Gata1, Gata2, PU.1) and nonhematopoietic tissues from the 3 germ layers (cardiac progenitors and endoderm: Nkx2.5; skeletal muscle: Myogenin; endoderm: HNF4, ectoderm: Pax6 and Cytokeratin 17). Ectopic Cdx2 induction dramatically suppressed all hematopoietic genes examined in these studies (Figure 3), mirroring the results of our functional assays (Figure 2C,D). Surprisingly, Cdx1 induction also resulted in the potent suppression of hematopoietic genes enriched in differentiated hematopoietic cells, such as Gata-1 (erythroid cells), and PU.1 (myeloid/B cells26-29 ; Figure 3), but had no effect on Scl or Runx1 expression. Both Cdx1 and Cdx4 slightly but consistently enhanced the expression of Gata-2, a marker of immature hematopoietic cells (Figure 3). This modest augmentation is probably attributable to the analysis of a mixed population in which only a small fraction of the cells are committed to the hematopoietic fate. These data are in agreement with our finding that both Cdx1 and Cdx4 promote the specification of multipotent hematopoietic progenitors but further suggest that Cdx1 may inhibit their terminal differentiation along hematopoietic lineages in the developing EB.
Cdx gene induction modulates lineage-specific gene expression in differentiating EBs. Quantitative real-time RT-PCR analysis of the effect of Cdx1, Cdx4, or Cdx2 induction on the expression of mesodermal (Scl, Runx1, Gata1, Gata2, PU.1, Nkx2.5, and Myogenin) nonmesodermal markers (Pax6, Cytokeratin 17, HNF4) in day 5 EB. Data represent averages of 2 independent biologic experiments and are presented as fold change in gene expression relative to noninduced control, after normalization to the housekeeping gene β-actin. Error bars show the standard error. P values calculated using nonpaired, 2-tailed Student t test are depicted over bars.
Cdx gene induction modulates lineage-specific gene expression in differentiating EBs. Quantitative real-time RT-PCR analysis of the effect of Cdx1, Cdx4, or Cdx2 induction on the expression of mesodermal (Scl, Runx1, Gata1, Gata2, PU.1, Nkx2.5, and Myogenin) nonmesodermal markers (Pax6, Cytokeratin 17, HNF4) in day 5 EB. Data represent averages of 2 independent biologic experiments and are presented as fold change in gene expression relative to noninduced control, after normalization to the housekeeping gene β-actin. Error bars show the standard error. P values calculated using nonpaired, 2-tailed Student t test are depicted over bars.
Cdx overexpression also affected the expression of ectodermal transcription factors (Figure 3), consistent with the reported role of Cdx in the development of ectodermal derivatives.30 Nkx2.5 was suppressed by ectopic expression of all Cdx genes, whereas Myogenin and HNF4 gene expression levels were unaffected (Figure 3). Thus, Cdx genes may affect hematopoietic commitment in differentiating EBs by skewing differentiation away from or toward hematopoietic lineages. Taken together, our data show that ectopic expression of Cdx1 and Cdx4 augments, whereas Cdx2 suppresses, the development of hematopoietic progenitors in developing EBs.
Cdx genes modulate Hox gene expression in differentiating EBs
Hox genes are crucial regulators of development that define positional identity during tissue formation.31,32 Because Cdx genes are known master regulators of Hox gene expression, we examined the effect of ectopic Cdx induction on Hox gene expression in differentiating EBs.33 Ectopic expression of each Cdx gene during EB differentiation significantly modulated Hox gene expression. Ectopic expression of Cdx1 in differentiating EBs up-regulated HoxA and HoxB posterior cluster genes (HoxA6, HoxB8, HoxB9), most notably, HoxA7, a known direct target of Cdx1 (Figure 4).3 Ectopic Cdx2 generally up-regulated the same Hox genes but effected greater fold changes in gene expression than Cdx1 (Figure 4) and modestly up-regulated HoxB4. Cdx4 induction induced only modest changes in Hox gene expression that generally paralleled those exacted by Cdx1 and Cdx2 (Figure 4), possibly echoing the modest functional effect observed after Cdx4 induction during this developmental window (Figure 2C,D). Thus, our gene expression data agree with the predicted role of the Cdx family as Hox regulators: all 3 Cdx genes induce posterior Hox genes that have been linked to blood development in the zebrafish,2 Cdx2 more prominently than Cdx1 or Cdx4. Moreover, in contrast to Cdx1 and Cdx4, Cdx2 also enhances the expression of anterior Hox genes.
Cdx gene induction modulates Hox gene expression in differentiating EBs. Quantitative real-time RT-PCR analysis of the effect of ectopic Cdx1, Cdx4, and Cdx2 gene induction on Hox A and B cluster genes in day 5 EBs. The data represent the average of 2 independent experiments and are presented as fold change relative to noninduction controls. Error bars represent the standard error. P values calculated using nonpaired, 2-tailed Student t test are depicted over bars.
Cdx gene induction modulates Hox gene expression in differentiating EBs. Quantitative real-time RT-PCR analysis of the effect of ectopic Cdx1, Cdx4, and Cdx2 gene induction on Hox A and B cluster genes in day 5 EBs. The data represent the average of 2 independent experiments and are presented as fold change relative to noninduction controls. Error bars represent the standard error. P values calculated using nonpaired, 2-tailed Student t test are depicted over bars.
Cdx overexpression modulates the differentiation and expansion of EB-derived hematopoietic progenitor cells
We next asked how ectopic Cdx expression influenced mouse ESC-derived hematopoietic progenitors. Cells isolated from EBs after 6 days of differentiation were cultured on OP9 stroma or in semisolid media supplemented with hematopoietic cytokines. Cdx induction dramatically affected the detection of hematopoietic progenitors isolated from day 6 EBs, as reflected by assays of colony formation on OP9 stroma or in methylcellulose (Figure 2C,D, OP9 and M3434 induction, respectively; Tables S1,S2). Cdx4 induction strongly enhanced OP9 colony number and CFUs (especially CFU-GEMM), particularly when Cdx4 had also been induced previously during EB development (day 4 to day 6; Figure 2C,D, OP9 and M3434 induction; Figure S3A,B). Cdx4 induction in these hematopoietic functional assays may further expand prepatterned cells, or alternatively, enhance their ability to respond to hematopoietic cytokines. Cdx2 induction after plating on OP9 stroma or in semisolid media strongly suppressed all hematopoietic colony formation, as did Cdx1, which was surprising given that Cdx1 was the most potent inducer of hematopoiesis during EB differentiation (Figure 2C,D OP9 and M3434 induction; Figure S3A,B). This suppression is apparently not attributable to the induction of cell death because colonies re-emerge after doxycycline removal even after 1 week of Cdx1 induction (Figure S3C,D). Thus, the Cdx1-mediated squelching of hematopoietic progenitor activity may represent a block of differentiation. In sum, Cdx gene overexpression modulates the colony forming ability of EB-derived hematopoietic progenitors. Cdx4 promotes OP9-colony formation and CFU development, whereas both Cdx1 and Cdx2 abolish colony formation.
Hematopoietic potential is enriched in CD41+ckit+ cells
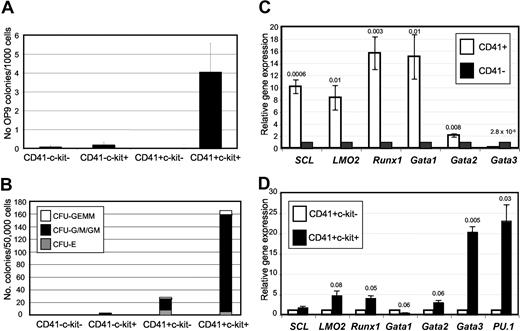
Previous reports suggested that in the mouse, most EB-derived hematopoietic activity falls in the CD41+c-kit+ population.34,35 To confirm this observation in our system, and to determine whether the OP9-colony initiating cell is likewise contained in this cell compartment, we fractionated day 6 EBs with respect to the expression of CD41 and c-kit via FACS (Figure S4A) and performed assays of OP9 and hematopoietic CFU activity. OP9–colony activity was enriched between 5- and 37-fold in CD41+c-kit+ cells, and all OP9–colony forming potential was restricted to CD41+c-kit+ EB-derived cells (Figure 5A; Figure S4B). Likewise, EB-derived CFU potential was strongly enriched in the CD41+c-kit+ compartment, with low but detectable CFU activity also present in the CD41+c-kit− compartment, as reported previously (Figure 5B).32,33
CD41+c-kit+ EB-derived cells contain all hematopoietic colony forming potential and are enriched for hematopoietic gene expression. OP9–colony-forming potential (A) and hematopoietic progenitor activity (B) of day 6 EB-derived cells fractionated with respect to c-kit and CD41 expression by FACS. OP9–colony-forming potential is presented as the average colony number/1000 cells plated for 9 independent experiments. Error bars represent the standard error. The data presented in panel B represent the pooled total number of colonies detected for each compartment in 6 independent experiments per 50 000 cells plated. Not all compartments were assessed in each independent experiment, but each were assessed a minimum of 4 (A) or 2 (B; CD41−c-kit− cells) times. (C) Quantitative real-time RT-PCR of hematopoietic gene expression in CD41+ and CD41− day 6 EB-derived cells. (D) CD41+ cells were significantly enriched in Scl, LMO2, Runx1, and Gata-2 gene expression relative to CD41− cells. Normalization to expression levels of the housekeeping gene β-actin was performed for each sample. Results represent the average of 3 independent biologic experiments. CD41+ day 6 EB cells were further fractionated with respect to c-kit expression, and analyzed by quantitative real-time RT-PCR for the expression of hematopoietic genes. Data are presented relative to expression level of each gene in CD41+c-kit− cells, after individual normalization of each sample to β-actin. CD41+c-kit+ cells were significantly enriched for Runx1, Gata-3, and PU.1 gene expression. For panels C and D, P values calculated using nonpaired, 2-tailed Student t test are depicted over bars.
CD41+c-kit+ EB-derived cells contain all hematopoietic colony forming potential and are enriched for hematopoietic gene expression. OP9–colony-forming potential (A) and hematopoietic progenitor activity (B) of day 6 EB-derived cells fractionated with respect to c-kit and CD41 expression by FACS. OP9–colony-forming potential is presented as the average colony number/1000 cells plated for 9 independent experiments. Error bars represent the standard error. The data presented in panel B represent the pooled total number of colonies detected for each compartment in 6 independent experiments per 50 000 cells plated. Not all compartments were assessed in each independent experiment, but each were assessed a minimum of 4 (A) or 2 (B; CD41−c-kit− cells) times. (C) Quantitative real-time RT-PCR of hematopoietic gene expression in CD41+ and CD41− day 6 EB-derived cells. (D) CD41+ cells were significantly enriched in Scl, LMO2, Runx1, and Gata-2 gene expression relative to CD41− cells. Normalization to expression levels of the housekeeping gene β-actin was performed for each sample. Results represent the average of 3 independent biologic experiments. CD41+ day 6 EB cells were further fractionated with respect to c-kit expression, and analyzed by quantitative real-time RT-PCR for the expression of hematopoietic genes. Data are presented relative to expression level of each gene in CD41+c-kit− cells, after individual normalization of each sample to β-actin. CD41+c-kit+ cells were significantly enriched for Runx1, Gata-3, and PU.1 gene expression. For panels C and D, P values calculated using nonpaired, 2-tailed Student t test are depicted over bars.
We examined the CD41+c-kit+ compartment for the expression of genes characteristic of hematopoietic stem and progenitor cells. In agreement with the functional data presented above, CD41+ cells were highly enriched for Scl, Runx1, LMO2, Gata-1, and Gata-2 (Figure 5C), whereas CD41+ckit+ cells were only modestly further enriched for genes such as LMO2, Runx1, and Gata-2, but showed a dramatic enrichment of Gata-3 and PU.1 expression (Figure 5D). Gata-3 has been described recently as a marker of multipotent hematopoietic repopulating cells in the AGM (aorta-gonad-mesonephros).36 Thus, CD41+ckit+ EB-derived cells may selectively display a gene expression profile reminiscent of immature hematopoietic populations in the developing embryo. PU.1 is characteristic of myeloid and B cells26-29 but has also been described in AGM cells displaying an HSC phenotype.37
Cdx genes selectively modulate the hematopoietic potential of CD41+c-kit+ EB-derived cells
We next examined whether induction of Cdx genes acts in a cell autonomous manner on hematopoietic progenitors or indirectly by altering tissues within EBs that augment or suppress blood formation. We found that the numbers of CD41+c-kit+ cells in day 6 EBs were not consistently enhanced or suppressed by exposure to ectopic Cdx1 or Cdx4, whereas ectopic Cdx2 expression abolished the generation of CD41+c-kit+ cells (data not shown), consistent with the suppressive effect of Cdx2 on hematopoietic colony forming assays. Although the numbers of CD41+c-kit+ cells were not affected by Cdx1 or Cdx4 gene induction, their capacity to form hematopoietic colonies on OP9 stroma or in methylcellulose was dramatically enhanced, mirroring the effects seen in unfractionated EB-derived cells (Figure 6A,B EB induction; Figure S4C,D; Tables S3,S4). These data suggest that Cdx gene induction acts on a subpopulation of CD41+c-kit+ cells.
Cdx genes selectively effect the hematopoietic potential and hematopoietic-specific gene expression of CD41+c-kit+ EB-derived cells. The effect of ectopic Cdx1 and Cdx4 expression on the OP9–colony-forming potential (A) and hematopoietic progenitor activity (B) of CD41+c-kit+ EB-derived cells was assessed. Data are presented as fold change in colony activity relative to noninduced controls. Error bars represent the standard error. The data represent the average of 4 and 7 independent experiments for iCdx1 and iCdx4, respectively. Ectopic Cdx1 and Cdx4 both enhanced all hematopoietic colony activity when induced during EB differentiation. When induced during secondary assay, Cdx1 suppressed all colony potential. Hematopoietic gene expression in CD41+c-kit− and CD41+c-kit+ cells subjected to ectopic Cdx1 (C) or Cdx4 (D) gene expression was also examined by quantitative real-time RT-PCR. Although Cdx1 induction significantly enhanced the expression of several hematopoietic genes in CD41+c-kit+ cells, relative to CD41+c-kit− cells, Cdx4 induction had only modest effects. Each data point represents the average of between 4 and 6 independent experiments. Error bars represent standard error. P values calculated using nonpaired, 2-tailed Student t test are depicted over bars. Purities were typically more than 90%.
Cdx genes selectively effect the hematopoietic potential and hematopoietic-specific gene expression of CD41+c-kit+ EB-derived cells. The effect of ectopic Cdx1 and Cdx4 expression on the OP9–colony-forming potential (A) and hematopoietic progenitor activity (B) of CD41+c-kit+ EB-derived cells was assessed. Data are presented as fold change in colony activity relative to noninduced controls. Error bars represent the standard error. The data represent the average of 4 and 7 independent experiments for iCdx1 and iCdx4, respectively. Ectopic Cdx1 and Cdx4 both enhanced all hematopoietic colony activity when induced during EB differentiation. When induced during secondary assay, Cdx1 suppressed all colony potential. Hematopoietic gene expression in CD41+c-kit− and CD41+c-kit+ cells subjected to ectopic Cdx1 (C) or Cdx4 (D) gene expression was also examined by quantitative real-time RT-PCR. Although Cdx1 induction significantly enhanced the expression of several hematopoietic genes in CD41+c-kit+ cells, relative to CD41+c-kit− cells, Cdx4 induction had only modest effects. Each data point represents the average of between 4 and 6 independent experiments. Error bars represent standard error. P values calculated using nonpaired, 2-tailed Student t test are depicted over bars. Purities were typically more than 90%.
We next plated iCdx1 and iCdx4 day 6 EB-derived cells fractionated based on CD41 and c-kit expression in hematopoietic colony assays with or without doxycycline to evaluate whether ectopic Cdx expression acted directly on hematopoietic target cells or was capable of respecifying nonhematopoietic cells toward the hematopoietic fate. Only CD41+c-kit+ EB-derived cells were significantly affected by ectopic Cdx1 or Cdx4 (Figure 6A,B OP9 and m3434 induction; Figure S4C,D; Tables S3,S4). In particular, Cdx4 induction yielded its most dramatic effect on CD41+c-kit+ cells when induced in both EB and colony assays (Figure 6A,B). These data demonstrate that Cdx1 and Cdx4 activation selectively enhance the hematopoietic activity of CD41+c-kit+ cells without conferring blood potential on other cellular compartments.
In agreement with our functional results, Cdx1 induction during EB differentiation (day 4 to day 6) selectively enhanced the expression of hematopoietic genes (eg, Scl and Gata-2) in CD41+ckit+ cells (Figure 6C). Induction of Cdx4 during this window only modestly affected gene expression in CD41+ckit+ cells (Figure 6D), a finding perhaps reminiscent of the relatively modest functional outcome affected by Cdx4 when ectopically expressed in day 4–differentiating EBs (Figure 2C,D). Although analysis of Hox expression profiles in CD41−c-kit+ and CD41+c-kit+ EB-derived cells after Cdx1 or Cdx4 induction during EB differentiation revealed the up-regulation of multiple Hox genes in both compartments, this up-regulation was often more dramatic in the CD41+c-kit+ compartment, especially in response to Cdx1 (Figures S5,S6).
Discussion
Studies in zebrafish have suggested redundant roles for Cdx1 and Cdx4 in hematopoietic development.1,2 In this study, we evaluated how the 3 mammalian Cdx genes influence hematopoietic development, using murine ESC differentiation as a model of embryonic blood specification. We show that EB-derived CD41+ckit+ cells comprise not only the majority of hematopoietic CFU-initiating cells but also selectively give rise to OP9 colonies with reported HSC properties.23 We demonstrate that Cdx1 and Cdx4 both promote, whereas Cdx2 suppresses, hematopoietic specification when ectopically expressed during mouse EB differentiation, chiefly by specifically modulating the activity of the CD41+c-kit+ compartment and via the modulation of distinct downstream genetic pathways (Figure 7). Importantly, Cdx expression did not confer hematopoietic potential on any other EB-derived cell compartment (ie, CD41−), revealing that Cdx genes cannot respecify nonhematopoietic populations to the hematopoietic fate. Ectopic expression of Cdx1 and Cdx4 during EB development resulted in the gross up-regulation of posterior Hox expression concomitant with a modest suppression of multiple anterior Hox genes, a pattern previously associated with enhanced blood production.2,22 In contrast, Cdx2 induction up-regulated rather than suppressed multiple anterior Hox genes. These data are consistent with the interpretation that Cdx genes modulate hematopoietic fate by effects on Hox genes but do not allow us to define a specific Cdx-driven Hox gene signature for blood formation.
Summary model of the differential effects of Cdx genes on ESC-derived hematopoiesis. Our data support a model in which Cdx genes differentially affect both the formation (ie, induction of Cdx gene expression during EB differentiation) and the hematopoietic activity (ie, induction of Cdx genes expression in purified CD41+c-kit+ hematopoietic progenitors) of already specified hematopoietic progenitor cells. Cdx1 and Cdx4 both promote the specification of hematopoietic progenitors, whereas Cdx2 inhibits this process. However, only Cdx4 can enhance the expansion/differentiation of hematopoietic progenitors; Cdx1 and Cdx2 both suppress these functions.
Summary model of the differential effects of Cdx genes on ESC-derived hematopoiesis. Our data support a model in which Cdx genes differentially affect both the formation (ie, induction of Cdx gene expression during EB differentiation) and the hematopoietic activity (ie, induction of Cdx genes expression in purified CD41+c-kit+ hematopoietic progenitors) of already specified hematopoietic progenitor cells. Cdx1 and Cdx4 both promote the specification of hematopoietic progenitors, whereas Cdx2 inhibits this process. However, only Cdx4 can enhance the expansion/differentiation of hematopoietic progenitors; Cdx1 and Cdx2 both suppress these functions.
Despite sequence conservation among the Cdx genes, we observed dramatic functional differences when Cdx genes were ectopically expressed during murine EB differentiation and subsequently during culture on OP9 stroma and in methylcellulose assays (Figure 7). Cdx4 further enhanced the formation of hematopoietic colonies on OP9 stroma and in CFU assays, whereas Cdx1 and Cdx2 suppressed both these activities. Cdx4 expression consistently enhanced hematopoietic potential when induced either during EB formation or in subsequent assays of hematopoietic colony formation, whereas Cdx2 consistently suppressed blood formation. Cdx1 showed distinct effects, enhancing blood formation within EBs but suppressing hematopoietic colony formation in OP9 stromal culture. Although Cdx1 overexpression suppressed hematopoietic colony formation in OP9 culture, the progenitors nevertheless persist because withdrawal of doxycycline allows the colonies to reappear (Figure S3C,D). These data suggest that Cdx1 may act by inhibiting the maturation of hematopoietic progenitors (Figure 7). Indeed, Cdx1 can inhibit the differentiation of adult bone marrow hematopoietic progenitors, a role that contributes to leukemogenesis (D. Bansal and G. Gilliland, personal communication, March 2007). Cdx2 has even greater suppressive effects than Cdx1, an effect also observed in adult bone marrow progenitors that probably contributes to the association of Cdx2 with AML (Figure 7).11
Many genes involved in early hematopoietic development (such as Scl) were originally identified by their roles in leukemogenesis.38 Understanding the role these genes play during development can yield insights as to how their irregular expression in adult cells results in malignancy. A series of papers have recently focused a spotlight on Cdx genes and their apparent capacity to contribute to leukemogenesis when expressed aberrantly: ectopic Cdx4 expression in whole bone marrow (WBM) induces AML in mouse models and has also been detected in the blast cells of AML patients.7 Cdx2 is expressed in a high percentage of AML patients and causes AML when overexpressed in murine WBM.8,10 Although Cdx2 aberrant expression appears to play a causal role in AML,10 Cdx2 expression in differentiating EB and colony assays suppresses hematopoiesis, most likely by inhibiting hematopoietic differentiation, a property apparently mimicked by overexpression of Cdx1. Interestingly, Cdx1 and Cdx2 are reportedly more potent inducers of leukemia than Cdx4,7,10,11 and Cdx4 failed to suppress hematopoietic differentiation in our assays. Molecularly, Cdx1 and Cdx2 strongly up-regulate the expression of multiple posterior Hox genes like HoxA9, which may provide a molecular mechanism for their leukemogenic effect.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Grigoriy Losyev for invaluable FACS support. We also thank M. William Lensch and members of the Daley laboratory for comments on this manuscript.
This work was supported by the American Cancer Society (S.L.M.-F.), the Eleanor and Miles Shore Scholarship Program (S.L.M.-F.), the Dr Mildred Scheel Foundation for Cancer Research (C.L.), the Human Frontier Science Program (S.S.), an NIH hematology training grant (Y.W.), additional NIH grants, the NIH Director's Pioneer Award, and the Burroughs Wellcome Fund Clinical Scientist Award in Translational Research (G.Q.D.).
National Institutes of Health
Authorship
Contribution: S.L.M.-F. and C.L. designed the research, performed experiments, analyzed results, and composed this manuscript. M.P., I.-H.J., S.S., and J.S. performed experiments. Y.W. contributed critical reagents. G.Q.D. designed the research, analyzed results, and edited this manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: George Daley, Children's Hospital, 300 Longwood Ave, Karp-7, Boston, MA 02115; e-mail: george.daley@childrens.harvard.edu.
References
Author notes
*S.L.M.-F. and C.L. contributed equally to this work.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal