Oncogenic Notch1 mutations are found in most T-lineage acute lymphoblastic leukemias in humans and T-cell lymphomas in mice. However, the mechanism by which Notch1 promotes transformation or maintains malignant cell survival has not been determined fully. Here, we report that expression of the transcription factor lymphoid enhancer factor 1 (Lef1) is Notch dependent in murine T-cell lymphomas in vitro and in vivo, and that the intracellular domain of Notch1 (ICN1) is present at the Lef1 promoter. Lef1 expression is not Notch dependent in primary T-cell progenitors, but Lef1 mRNA is increased by ectopic expression of ICN1 in these cells. We show that Lef1 is required for survival of T-cell lymphoma lines, and that ectopic expression of Lef1 delays lymphoma cell death in the absence of Notch signaling, indicating that Lef1 is an important Notch target in these cells. Therefore, Notch1 co-opts Lef1 during the process of transformation to maintain survival of T-cell lymphomas.
Introduction
Notch1 encodes a transmembrane receptor protein that is essential for T-lymphocyte fate specification from multipotent hematopoietic progenitors.1 Notch1 is also required for differentiation, proliferation, survival, and proper metabolism of immature T-lymphocytes.2,–4 Notch1 signaling is tightly controlled during T-cell development since deregulation leads to malignant transformation. An oncogenic role for Notch1 was first suggested by the identification of chromosome rearrangements in approximately 2% of human T-lineage acute lymphoblastic leukemias (T-ALLs) that translocate the intracellular domain of Notch1 (ICN1) to the T-cell receptor β (TCRB) gene.5 This rearrangement leads to constitutive expression of ICN1 in T-lymphocytes, and in mice, ectopic expression of ICN1 in hematopoietic progenitors leads to T-lymphocyte transformation.6 Recently, mutations in Notch1 were identified in more than 50% of patients with T-ALL, indicating that Notch1 plays a widespread role in T-lymphocyte progenitor transformation.7,8 Activation of Notch1 is associated with the development of T-cell lymphoma in numerous mutant mouse strains.9,,–12 Therefore, Notch1 is a major contributor to T-lymphocyte development and malignancy
Notch1 is a heterodimeric transmembrane receptor in which extracellular and intracellular domains are stably associated through the heterodimerization domain.13 Notch1 becomes activated after interaction with its ligands Delta-like 1 (DL1), DL3, DL4, Jagged-1, and Jagged-2 on neighboring cells.13 Ligand binding produces a conformational change in Notch1, allowing cleavage of the extracellular domain by a metalloprotease, followed by cleavage of the intracellular domain by enzymes with γ-secretase (GS) activity. These cleavage events release ICN1 from the membrane, allowing translocation of ICN1 to the nucleus, where it interacts with a DNA-bound transcription factor (CSL) and coactivators of the MAML family.13 ICN1 is composed of multiple domains, including the RAM domain, and a series of ankyrin repeats that function in protein-protein interactions, a nuclear localization signal, a PEST (proline, glutamine, serine, and threonine-rich) domain involved in protein degradation, and a C-terminal activation domain. Interestingly, mutations in human T-ALL cluster within the heterodimerization domain, resulting in ligand independent activation, and/or the PEST domain, leading to stabilization of ICN1.8,14,15
In T-lymphocytes, the ICN1/CSL/MAML complex promotes transcription of Notch1, Notch3, Notch-regulated ankyrin repeat protein (Nrarp), Deltex1, Hes1, and Ptcra (pre-Tα).16,,,–20 However, the targets of Notch1 that promote and maintain T-cell transformation, proliferation, and survival remain to be determined fully. Transformation of T-lymphocytes by Notch1 is facilitated by Notch1-mediated induction of pre-Tα, a component of the pre-TCR.20,21 Previous studies indicate that an essential function of pre-TCR signaling is inhibition of E2A transcription factor activity, loss of which leads to T-lymphocyte progenitor transformation.22,23 Nonetheless, we recently demonstrated that Notch1 signaling is essential for survival of E2A−/− T-cell lymphomas, indicating that Notch1 has important targets in addition to E2A.9 Interestingly, while pre-TCR signaling is not essential for transformation in the absence of E2A, it can contribute to lymphoma cell survival, a finding that is consistent with other studies.9,24 Therefore, Notch1 promotes transformation through both pre-TCR–dependent and – independent pathways. These findings highlight the adaptability of the signaling pathways that contribute to T-lymphocyte progenitor malignancies.
A number of recent studies have demonstrated that c-myc is regulated by Notch signaling and the ICN1/MAML/CSL complex in T-ALL.25,–27 c-myc is a well-characterized oncogene that collaborates with multiple proteins to transform T-lineage cells.28,–30 Importantly, lymphomas arising in E2A−/− mice frequently have amplification of chromosome 15 in a region containing the c-myc gene, and have increased levels of c-myc mRNA compared with nontransformed thymocytes.31 Thus, c-myc deregulation in E2A−/− lymphomas may result from both oncogenic Notch signaling and gene amplification.
To gain further insight into the mechanisms by which Notch1 promotes transformation of T-lymphocyte progenitors, we identified Notch-dependent genes in E2A−/− lymphomas. Here, we report that the transcription factor lymphoid enhancer factor 1 (Lef1) is a Notch1 target gene in lymphomas arising in E2A−/−, p53−/−, and ICN1 transgenic mice. Lef1 is not essential for T-lymphocyte development because it shares redundant functions with the related transcription factor Tcf1.32,33 In contrast, Lef1 is required for survival of T-cell lymphomas and is able to delay cell death induced by inhibition of Notch signaling. Therefore, Notch1-directed Lef1 expression contributes to survival and expansion of T-cell lymphomas, and indicates that Lef1 may be a rational therapeutic target in T-cell malignancies.
Materials and methods
Northern blot analysis
RNA was extracted using Trizol reagent and analyzed as described previously.9 The Lef1 and Tcf1 probes were created by polymerase chain reaction (PCR) using the following primers: Lef1-forward, 5′-CCCTTTCTCCACCCATCC-3′ and Lef1-reverse, 5′-GTGCTCGTCGCTGTAGGT-3′; or Tcf1 (Tcf7)–forward, 5′-TCTCACTCTTCCAGCCCCTAAGTC-3′ and Tcf1(Tcf7)–reverse, 5′-CAACCCATCTGACCTACAGCAAAC-3′. Deltex1, Notch3, c-myc, and actin probes were described previously.9 The cyclin D1 probe was digestion from pCMV–cyclin D1 with BamHI (gift of Dr Charles Sherr, St Jude Children's Hospital, Memphis TN).
Quantitative real-time PCR
RNA was isolated from lymphomas using Trizol and reverse transcribed using Superscript III (Invitrogen, Carlsbad, CA). PCR reactions were set up with first-strand cDNA, gene-specific primers, passive reference dye, and SYBR Green QPCR Master Mix (Bio-Rad, Hercules, CA) according to the manufacturer's instructions. Real-time PCR was performed in triplicate, and fluorometric data were collected at the annealing step of each cycle using an iCycler (Bio-Rad). A dissociation curve was performed at the end of 40 cycles to confirm specificity of amplification. The Lef1-specific primers were as follows: Lef1 RT5′, 5′-CCCTTTCTCCACCCATCCC-3′ and Lef1 RT3′, 5′-GTGCTGGTCGCTGTAGGTG-3′.
Western blot analysis
Total and nuclear protein extracts were prepared and analyzed by Western blot analysis as described previously.9 Primary antibodies used include anti- Lef1 (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-Notch1 antibody (V1744) reactive with the cleaved cytoplasmic domain (Cell Signaling Technology, Danvers, MA).
ChIP assays
Chromatin immunoprecipitation (ChIP) was performed using ChIP assay kits (Upstate Biotechnology, Lake Placid, NY). The 0531 lymphoma line was treated with DMSO or GS inhibitor (GSI; 10 μM DAPT) for 18 hours prior to fixation. Cells were fixed with 1% paraformaldehyde at room temperature for 10 minutes, washed, and lysed with SDS lysis buffer (50 mM Tris-HCl, 1% SDS, 10 mM EDTA, and protease inhibitor cocktail [Sigma, St Louis, MO]). The lysates were sonicated to produce DNA between 200 and 600 bp. The extracts were diluted, precleared with salmon sperm DNA/protein A–agarose, then divided into 2 tubes and incubated with 5 mL antiserum specific for Notch1 TAD domain or normal rabbit IgG.14 The immune complexes were then precipitated with protein A–agarose and eluted with elution buffer (0.1 M NaHCO2 containing 1% SDS). The eluted material was reverse–cross-linked and treated with proteinase K (20 μg/mL). DNA was purified using a PCR purification kit (Qiagen, Valencia, CA) and eluted with H2O. Real-time quantitative PCR (QPCR) was performed as described using the following primers: +11 forward, 5′-TCTTTGCTTTGACCGAGGGG-3′ and +11 reverse, 5′-CACACACACACACACACACACACTC-3′; −2 kb forward, 5′-TAAAGACAGCACACTGCGGAGACC-3′ and −2 kb reverse, 5′-GCCAGAGGGTATTTTTTGAAAGCC-3′; and intron forward, 5′-TCGGTTGAGATGATGTGTGGG C-3′ and intron reverse, 5′-CAGGTTTGACTTACAGGTCCTTTGG-3′. The Hes1 promoter primers were described previously.25
Cell culture and retroviral infection
Lymphomas were cultured in RPMI 1640 supplemented with 10% FBS, 50 μM 2-mercaptoethanol, 100 μg/mL penicillin, 100 μg/mL streptomycin, and 300 μg/mL glutamine in a humidified incubator with 5% CO2. DAPT was used at a concentration of 10 μM (Calbiochem, San Diego, CA). Viral supernatants were made in Phoenix cells, and transductions were as described previously.9 The Lef1 siRNA was created in the Banshee retroviral vector using the sequence 5′-CCCAAGCTTAAAAAAGTTACTCTGGCTACATAATGATGCGCTACGAAGCATCATTATGTAGCCAGAGTAACGGTGTTTCGTCCTTTCC-3′. MigR1-Lef1 was created by PCR of Lef1 from 0531 cDNA using the primers 5′-GAAGATCTCCGCCGGCATGCCCAAACTTTCCG-3′ and 5′-CGGAATTCTTACCACCATGTTTCAGATG-3′, cloned into pBluescript (Stratagene, La Jolla, CA), and sequenced prior to cloning into MigR1.
In vivo analysis of tTA-Nic mice
Primary cell culture
Mice were housed in accordance with the guidelines of the University of Chicago Institutional Animal Care and Use Committee (IACUC). Ter119−Gr1−CD117+CD27+ hematopoietic progenitor cells were isolated from the liver of embryonic day (E) 13.5 embryos by cell sorting and cultured in vitro on a confluent layer of OP9-DL1 in OPTI-MEM supplemented with 10% FBS (Invitrogen), 50 μM 2-mercaptoethanol, 100 μg/mL penicillin, 100 μg/mL streptomycin, 300 μg/mL glutamine, IL-7 (1:100 dilution of J558-IL7 culture media), and 5 ng/mL Flt3 ligand (Peprotech, Rocky Hill, NJ). On day 8 of culture, the cells were treated with DMSO or 10 μM DAPT, or infected with MigR1 or MigR1-ICN1. The cells were harvested after 24 hours, and RNA was extracted for QPCR.
Flow cytometry
Cells were analyzed on a LSRII using FacsDiva Software (Becton Dickinson, San Jose, CA), and data were analyzed using FlowJo (Tree Star, Ashland, OR). Cells were fixed in 1% formaldehyde/PBS prior to analysis. For cell-cycle analysis, cells were labeled for 30 minutes by addition of 20 μM BrdU followed by fixation in 70% ethanol-PBS and treated with freshly prepared 2.5 M HCl with 0.5% Triton-X100 for 25 minutes at room temperature. The cells were washed extensively and stained with anti-BrdU–FITC (Pharmingen, San Diego, CA) prior to analysis as described previously.35
RACE
The 5′ end of the mouse Lef1 gene was cloned using the 5′–-rapid amplification of cDNA ends (RACE) system from Invitrogen following the manufacturer's protocol using mLef gsp1, 5′-TGATTTCGGTGATTTG-3′ followed by Lef1 gsp2, 5′-AGTCCACTTCCTTCAGAGTAAAC-3′ in conjunction with the anchor primer, 5′-GGCCACGCGTCGACTAGTACGGGIIGGGIIGGG IIG-3′. The resulting PCR products were cloned into pBSK and sequenced.
EMSA
Electrophoretic mobility shift assay (EMSA) was performed as described previously using the following sequences: Jκ3, 5′-GAAGTCAAATTTCCCACGAAGTC-3′; Lef1p+11 CSL, 5′-GAAGCGGTGGGAAGGCGATC-3′; Lef1p-2179 CSL, 5′-CCAAAAATTCCCATCTCGATC-3′; Lef1p-4138 CSL, 5′-CTTGGCCTTCTCAGCTGGGAG-3′; and Oct, 5′-TCTTAATATTTGCATACCCTCAC-3′.36,37 Probes were labeled with γP32-ATP using polynucleotide kinase. Approximately 0.1 ng of labeled probe was added to 10 μg of nuclear extract in binding buffer containing 1 μg BSA/0.8 μg poly-d(I)d(C). Competitor oligos were added to protein for 10 minutes prior to labeled probe, and the mixture was incubated for a further 20 minutes at room temperature. The binding reaction was separated on a 6% nondenaturing 0.5 × TBE acrylamide gel and dried on Whatman paper (Whatman, Florham Park, NJ) prior to exposure to autoradio-graphic film.
Results
Identification of Notch-dependent genes in E2A−/− T-cell lymphomas
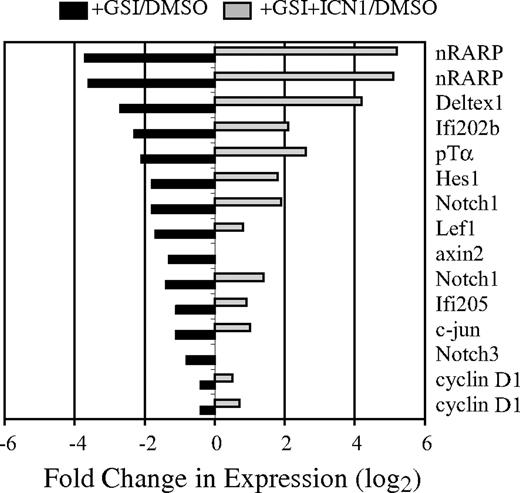
Notch signaling is essential for the survival of T-cell lymphomas arising in E2A−/− mice, suggesting that oncogenic Notch1 regulates genes distinct from those functioning downstream of E2A.9 To identify Notch-dependent genes that are independent of E2A, we examined gene expression in an E2A−/− lymphoma line (0531) with or without active Notch signaling. 0531 cells were treated with GSI to block endogenous Notch signaling, or with DMSO for 24 hours prior to harvesting RNA for screening of Affymetrix 430A microarrays (Affymetrix, Santa Clara, CA). To confirm that changes in gene expression were the consequence of reduced Notch signaling, 0531 cells were infected with a retrovirus producing ICN1 (MigR1-ICN1), which does not require GS processing, prior to treatment with GSI. Using this approach, we identified approximately 50 mRNA decreased by GSIs, compared with DMSO, and restored by ICN1 (Figure 1; data not shown). Among these mRNAs were known Notch1 targets, including Nrarp, Deltex1, Ifi202b, Ptcra, Hes1, Notch3, Ifi205, and Notch1. In addition, Lef1 mRNA and the potential Lef1 targets c-jun, cyclin D1, and axin 2 also required Notch signaling in this lymphoma (Figure 1).38 c-myc mRNA was only modestly affected by GSI (< 2-fold; data not shown).
Identification of Notch1-dependent genes in an E2A−/− lymphoma. The E2A−/− lymphoma line 0531 was transduced with the MigR1-ICN retrovirus; 24 hours later, transduced and nontransduced cultures were treated with GSI or DMSO. RNA was extracted 24 hours after addition of GSI or DMSO and used to screen Affymetrix 430A microarrays. The fold change in mRNA expression is shown for nontransduced (■) or MigR1-ICN1 transduced (▩) cells treated with GSI relative to nontransduced cells treated with DMSO. Gene expression was determined by microarray analysis; Nrarp, Notch1, and cyclin D1 were represented by 2 unique identifiers on the microarray.
Identification of Notch1-dependent genes in an E2A−/− lymphoma. The E2A−/− lymphoma line 0531 was transduced with the MigR1-ICN retrovirus; 24 hours later, transduced and nontransduced cultures were treated with GSI or DMSO. RNA was extracted 24 hours after addition of GSI or DMSO and used to screen Affymetrix 430A microarrays. The fold change in mRNA expression is shown for nontransduced (■) or MigR1-ICN1 transduced (▩) cells treated with GSI relative to nontransduced cells treated with DMSO. Gene expression was determined by microarray analysis; Nrarp, Notch1, and cyclin D1 were represented by 2 unique identifiers on the microarray.
Lef1 is a Notch-dependent gene in E2A−/− T-cell lymphomas
We next examined the dependence of Lef1 mRNA on Notch signaling in multiple E2A−/− lymphoma lines (0531, 1.F9, and 0714), a p53−/− lymphoma that does not express Notch1 (16610D9), and a lymphoma line derived from ICN3 transgenic mice (N3T).39 By Northern blot analysis, we detected a 3- to 5-fold decrease in Lef1 mRNA after culture of the E2A−/− lymphoma lines in the presence of GSI for 24 hours compared with that of cells cultured in DMSO (Figure 2A). Similarly, mRNA for the Notch1 target genes Deltex1 and Notch3 were decreased in GSI-treated cells (Figure 2A). The N3T cells were derived from a T-cell lymphoma initiated using a Notch3 transgene containing only intracellular sequences.39 Thus, these cells should not be sensitive to GSI, and as expected, GSI treatment had little effect on Lef1, Deltex1, or endogenous Notch3 expression (Figure 2A). We also tested the requirement for CSL-dependent transcription for Lef1, Deltex1, and Notch3 mRNA expression by infecting lymphomas with retrovirus producing a dominant-negative (DN) form of the coactivator MAML.40 Expression of DN-MAML resulted in decreased Lef1, Deltex1, and endogenous Notch3 mRNA in all lymphoma lines (Figure 2A). Lef1 protein expression is also decreased after inhibition of Notch signaling by GSI (Figure 2B). The requirement for Notch signaling to maintain Lef1 mRNA was not restricted to these lymphomas since the E2A−/−Rag2−/− lymphoma line 531026 and the p53−/− lymphoma line K052FA2C1, both of which require Notch1 signaling for their growth and survival,9 also had decreased Lef1 mRNA after treatment with GSI (Figure 2C). GSI also reduced Deltex1 expression in these lymphomas, demonstrating that Notch signaling was efficiently antagonized (Figure 2D). Therefore, Lef1 mRNA and protein are dependent on Notch signaling and the ICN1/CSL/MAML complex in T-cell lymphoma lines derived from E2A−/−, p53−/−, and ICN3 transgenic mice.
Lef1 expression is dependent on Notch signaling. (A) Northern blot of RNA isolated from the p53−/− lymphoma line 16610D9 (which does not have active Notch signaling), N3T, and E2A−/− lymphoma lines 0531, 1.F9, and 0714 (which have active Notch signaling) 24 hours after treatment with DMSO or GSI or 48 hours after infection with MigR1-DN-MAML (DN). (B) Western blot of 16610D9, 0531, or 1.F9 cells 20 or 40 hours after treatment with DMSO (−) or GSI (+) using Lef1-specific antibody. Relative expression of Lef1 (C) or Deltex1 (D) in the E2A−/− lymphoma 1.F9, the E2A−/−Rag2−/− lymphoma 531026, and the p53−/− lymphoma K052FA2C1 (FA2C1) cultured in DMSO or GSI for 24 hours. Expression was determined by QPCR and standardized to HPRT. Error bars represent the standard error of triplicate measurements.
Lef1 expression is dependent on Notch signaling. (A) Northern blot of RNA isolated from the p53−/− lymphoma line 16610D9 (which does not have active Notch signaling), N3T, and E2A−/− lymphoma lines 0531, 1.F9, and 0714 (which have active Notch signaling) 24 hours after treatment with DMSO or GSI or 48 hours after infection with MigR1-DN-MAML (DN). (B) Western blot of 16610D9, 0531, or 1.F9 cells 20 or 40 hours after treatment with DMSO (−) or GSI (+) using Lef1-specific antibody. Relative expression of Lef1 (C) or Deltex1 (D) in the E2A−/− lymphoma 1.F9, the E2A−/−Rag2−/− lymphoma 531026, and the p53−/− lymphoma K052FA2C1 (FA2C1) cultured in DMSO or GSI for 24 hours. Expression was determined by QPCR and standardized to HPRT. Error bars represent the standard error of triplicate measurements.
Lef1 is highly expressed in primary Notch-dependent T-cell lymphomas
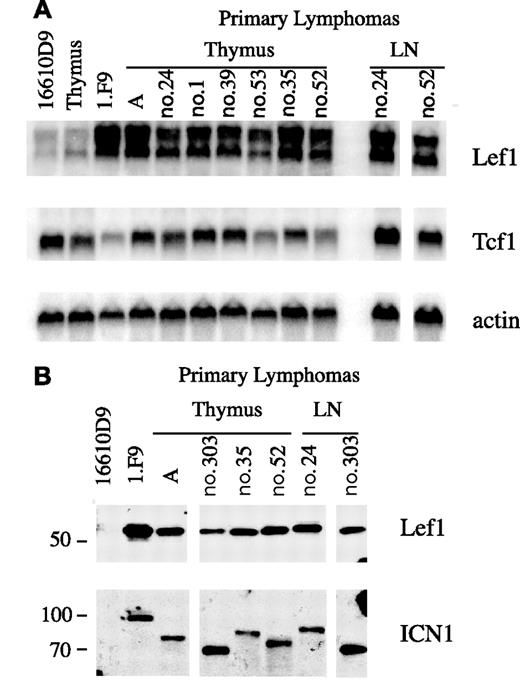
We next analyzed Lef1 mRNA expression in a panel of primary lymphomas isolated directly from diseased E2A−/− or E2A−/−Rag2−/− mice. In all cases, Lef1 mRNA was highly expressed compared with thymocytes or the 16610D9 lymphoma, which lacks Notch1 (Figure 3A). In contrast, Tcf1 mRNA was expressed at similar levels in primary lymphomas and thymocytes or 16610D9 cells (Figure 3A). Consistent with the high level of Lef1 mRNA, Lef1 protein is also detected in Notch-dependent primary lymphomas (Figure 3B). Each of these primary lymphomas has detectable levels of ICN1 with a unique size that was predicted based on mutations identified near the PEST domain (Figure 3B; data not shown). Therefore, Lef1 mRNA and protein are highly expressed in primary lymphomas with activated Notch1.
Expression of Lef1 in primary T-cell lymphomas. (A) Northern blot of RNA from primary lymphomas isolated from thymus or lymph nodes (LNs) of E2A−/− (A, and nos. 24, 1, 39) or E2A−/−Rag2−/− (nos. 53, 35, 52) mice compared with the E2A−/− lymphoma line 1.F9 or the p53−/− lymphoma line 16610D9. Probes specific for Lef1, Tcf1, and actin mRNA were used as indicated. Vertical lines have been inserted to indicate where a gel was cut. All lanes are from the same gel. (B) Western blot of primary lymphoma lines using Lef1 specific antibody (top panel). The same blot was subsequently probed with anti-Notch1 reactive with ICN1. ICN1 is a unique size in each lymphoma due to mutations resulting in truncation within the PEST domain. Vertical lines have been inserted to indicate where a gel was cut. All lanes are from the same gel.
Expression of Lef1 in primary T-cell lymphomas. (A) Northern blot of RNA from primary lymphomas isolated from thymus or lymph nodes (LNs) of E2A−/− (A, and nos. 24, 1, 39) or E2A−/−Rag2−/− (nos. 53, 35, 52) mice compared with the E2A−/− lymphoma line 1.F9 or the p53−/− lymphoma line 16610D9. Probes specific for Lef1, Tcf1, and actin mRNA were used as indicated. Vertical lines have been inserted to indicate where a gel was cut. All lanes are from the same gel. (B) Western blot of primary lymphoma lines using Lef1 specific antibody (top panel). The same blot was subsequently probed with anti-Notch1 reactive with ICN1. ICN1 is a unique size in each lymphoma due to mutations resulting in truncation within the PEST domain. Vertical lines have been inserted to indicate where a gel was cut. All lanes are from the same gel.
Notch1 maintains Lef1 expression in T-cell lymphomas in vivo
To determine whether Notch1 is required for Lef1 expression in T-cell lymphomas in vivo, we took advantage of the ability to control ICN1 expression in tTA-Nic transgenic mice.34 tTA-Nic transgenic mice are compound hemizygous for the Top-Nic transgene, in which ICN1 is under the control of the tetracycline operator and the EμSrα-tTAOFF transgene, which produces a doxycycline-repressible tetracycline transactivator protein in T-lymphocytes.41 These mice were used previously to investigate the requirements for ICN1 in initiation and maintenance of T-cell lymphoma in vivo.34 As in previous experiments, tTA-Nic mice with overt signs of lymphoma were killed, and splenocytes were injected into multiple wild-type FVB/NJ recipients (Figure 4A). Within 2 to 4 weeks, the recipients began to show signs of disease, including lethargy, cachexia, and labored breathing, and were given doxycycline to repress ICN1 (Figure 4A). Under these conditions, ICN1 mRNA and protein are extinguished within 6 hours, and significant apoptosis of lymphoma cells is observed by 36 hours.34 Northern blot analysis of splenocyte RNA at intervals after initiation of doxycycline treatment revealed that Lef1 mRNA begins to decline between 6 and 24 hours, shortly after the decline in ICN1 (Figure 4B). Importantly, the decline in Lef1 mRNA was more rapid than the decline in Tcf1 mRNA, indicating that the loss of Lef1 is not due simply to loss of lymphoma cells (Figure 4B). Lef1 protein also declines within 12 to 24 hours, consistent with the loss of Lef1 mRNA (Figure 4C). Therefore, ICN1 is required to maintain Lef1 mRNA and protein in primary ICN1-dependent lymphomas in vivo.
In vivo regulation of Lef1 by Notch1 in T-cell lymphomas. (A) Schematic of experimental design. (B) Northern blot of mRNA isolated from the spleens of tTA-Nic lymphoma–carrying mice with disease at the indicated time point (hours) after initiation of doxycycline treatment. Two independent experiments are shown. The blots were probed sequentially with cDNA probes specific for Lef1, Tcf1, or actin as indicated. (C) Western blot of protein extracts prepared from splenocytes of diseased mice at the indicated time (hours) after initiation of doxycycline treatment. The blot was probed sequentially with anti-Lef1 and anti-actin antibodies as indicated.
In vivo regulation of Lef1 by Notch1 in T-cell lymphomas. (A) Schematic of experimental design. (B) Northern blot of mRNA isolated from the spleens of tTA-Nic lymphoma–carrying mice with disease at the indicated time point (hours) after initiation of doxycycline treatment. Two independent experiments are shown. The blots were probed sequentially with cDNA probes specific for Lef1, Tcf1, or actin as indicated. (C) Western blot of protein extracts prepared from splenocytes of diseased mice at the indicated time (hours) after initiation of doxycycline treatment. The blot was probed sequentially with anti-Lef1 and anti-actin antibodies as indicated.
Lef1 is not Notch-dependent in primary T-cell progenitors in vitro but is co-opted by ICN1
To determine whether Lef1 mRNA is Notch-dependent in primary T-lymphocyte progenitors, we examined Lef1 mRNA by QPCR in in vitro–cultured T-lymphocyte progenitors after treatment with DMSO or GSI. T-lymphocyte progenitors were generated by culture of Lin− E13.5 fetal liver cells on OP9-DL1 stromal cells for 8 days prior to addition of DMSO or GSI for 12 hours. At this time point, approximately 50% of cells express CD25, and a subset of these have down-regulated c-kit, indicating that the cells are at the DN2 and DN3 stages of development. Under these conditions, Deltex1 mRNA was reduced more than 8-fold by GSI, indicating that Notch signaling was inhibited effectively (Figure 5B). Notably however, Lef1 mRNA was expressed at similar levels in GSI- and DMSO-treated T-lymphocyte progenitors (Figure 5A). In contrast, transduction of the same progenitor population with the ICN1 retrovirus resulted in a 7-fold increase in Lef1 mRNA, and a 25-fold increase in Deltex1 mRNA relative to cells transduced with retrovirus producing green fluorescent protein (GFP) only (Figure 5A,B). Therefore, Lef1 mRNA does not appear to be regulated by Notch signaling during T-lymphocyte development in vitro, but can be co-opted by ectopic ICN1.
Regulation of Lef1 by ICN1 in primary T-cell progenitors and T-cell lymphoma. (A) QPCR analysis of Lef1 and Deltex1 expression in primary T-cell progenitors cultured in vitro on OP9-DL1 in the presence of DMSO or GSI for 12 hours or after infection with MigR1 or MigR1-ICN1 retrovirus. mRNA expression was standardized to HPRT. Comparison of the mouse and human Lef1 gene sequence surrounding the mouse Lef1p + 11-bp CSL site (B) and the Lef1p −2179-bp CSL site (C). The CSL-binding sites are shaded. (D) EMSA using the Lef1p +11-bp and Lef1p −2179-bp CSL sites. Complexes were competed with self-competitor, a CSL consensus sequence (Jκ3), or nonspecific competitor (Oct). The position of the free oligonucleotides and shifted CSL complex are shown. Vertical lines have been inserted to indicate where a gel was cut. All lanes are from the same gel. (E) ChIP analysis. DNA precipitated with IgG or anti-Notch1 from 0531 cells treated with GSI (squlf]) or DMSO (▩) for 15 hours was analyzed by QPCR using primers specific for sequences near the Lef1p −2-kb and Lef1p +11-bp CSL-binding sites, in Lef1 intron 11, and the Hes1 promoter. Results are representative of 4 precipitations. Hes1 promoter P < .001; Lef1 intron P < .25; and Lef1p −2 kb P < .05 using a paired Student t test. The Lef1p +11-bp site was precipitated specifically with anti-Notch1 in 3 of 4 experiments, resulting in P < .25. (F) QPCR analysis of DNA precipitated with IgG or antiacetylated histone H4 (AcH4) from the same samples as in panel E. Histograms represent the means (± SE) of triplicate QPCR amplifications.
Regulation of Lef1 by ICN1 in primary T-cell progenitors and T-cell lymphoma. (A) QPCR analysis of Lef1 and Deltex1 expression in primary T-cell progenitors cultured in vitro on OP9-DL1 in the presence of DMSO or GSI for 12 hours or after infection with MigR1 or MigR1-ICN1 retrovirus. mRNA expression was standardized to HPRT. Comparison of the mouse and human Lef1 gene sequence surrounding the mouse Lef1p + 11-bp CSL site (B) and the Lef1p −2179-bp CSL site (C). The CSL-binding sites are shaded. (D) EMSA using the Lef1p +11-bp and Lef1p −2179-bp CSL sites. Complexes were competed with self-competitor, a CSL consensus sequence (Jκ3), or nonspecific competitor (Oct). The position of the free oligonucleotides and shifted CSL complex are shown. Vertical lines have been inserted to indicate where a gel was cut. All lanes are from the same gel. (E) ChIP analysis. DNA precipitated with IgG or anti-Notch1 from 0531 cells treated with GSI (squlf]) or DMSO (▩) for 15 hours was analyzed by QPCR using primers specific for sequences near the Lef1p −2-kb and Lef1p +11-bp CSL-binding sites, in Lef1 intron 11, and the Hes1 promoter. Results are representative of 4 precipitations. Hes1 promoter P < .001; Lef1 intron P < .25; and Lef1p −2 kb P < .05 using a paired Student t test. The Lef1p +11-bp site was precipitated specifically with anti-Notch1 in 3 of 4 experiments, resulting in P < .25. (F) QPCR analysis of DNA precipitated with IgG or antiacetylated histone H4 (AcH4) from the same samples as in panel E. Histograms represent the means (± SE) of triplicate QPCR amplifications.
The Lef1 gene contains CSL-binding sites
Using 5′ RACE, we determined the start site of transcription for the Lef1 mRNA to be 146 bp upstream of the reported transcription start site for Lef1 mRNA (NCBI accession number NM_010703) and 20 bp downstream of the major start site identified for human Lef1.42 A consensus CSL-binding site was identified 11 bp downstream of this start site that falls within a region of Lef1 that is highly conserved between mouse and human and has a positive effect on promoter activity in transient reporter-luciferase assays (Figure 5B).42 EMSA demonstrated that an oligonucleotide spanning the Lef1 promoter (Lef1p) +11-bp sequence is bound by proteins in lymphoma cell extracts that comigrate with proteins binding to a CSL consensus sequence (Figure 5D; data not shown). This protein complex was competed by specific and nonspecific sequences in a manner consistent with binding of CSL (Figure 5D). Interestingly, by titrating the Lef1p +11-bp oligonucleotide as a competitor against the Jκ3 oligonucleotide, we determined that the Lef1p +11-bp sequence has an approximately 25-fold lower affinity for CSL than Jκ3, even though these oligonucleotides share the consensus CSL-binding sequence (GTGGGAA; data not shown). Two additional conserved CSL-binding sites were identified at −2179 bp and −4381 bp, and these were also tested by EMSA (Figure 5C; data not shown). A probe spanning the Lef1p −4381-bp site failed to bind CSL, and the Lef1p −2179-bp site containing probe bound CSL with 2-fold higher affinity than the Lef1p +11-bp site (Figure 5D; data not shown). Taken together, our data indicate that CSL is able to interact with sequences in the Lef1 gene that may function in the regulation of Lef1 expression.
To determine whether ICN1/CSL interacts with the Lef1 gene, we performed ChIP using an anti-Notch1 antibody followed by QPCR for sequences near the Lef1p +11-bp (+11-bp) and Lef1p −2179-bp (−2-kb) sites. As a negative control, we examined sequences in intron 11 that are nearly 6 kb away from the nearest potential CSL-binding site. Sequences near the −2-kb CSL site were precipitated with the anti-Notch antibody more efficiently than with control IgG, and in 3 of 4 experiments, sequences near the +11-bp site were also precipitated specifically with anti-Notch1 (Figure 5E). Importantly, the ability of the anti-Notch1 antibody to precipitate the +11-bp and −2-kb CSL sites was reduced by treating the lymphomas with GSI but not DMSO. The fold enrichment with anti-Notch1 over IgG at both of these sites was less than that observed at the Hes-1 promoter, a known Notch1 target gene. However, the Lef1p −2-kb sequence was precipitated more efficiently than the intron 11 sequence, which was not affected by GSI treatment (Figure 5E). Therefore, ICN1 interacts with proteins bound near the Lef1p −2-kb CSL site and possibly near the Lef1p +11-bp CSL site. Interestingly, all of the Lef1 gene sequences could be precipitated with an antibody against acetylated histone H4; however, treatment of the cells with GSI decreased association of the Lef1p −2-kb site with this histone modification (Figure 5F). Our experiments indicate that Lef1 is a direct target of ICN1.
Lef1 is essential for survival and expansion of Notch-dependent T-cell lymphomas
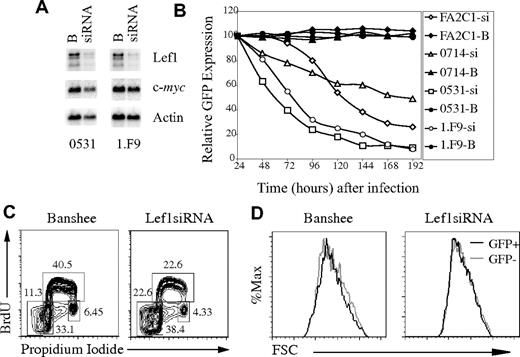
To determine the consequences of loss of Lef1 after inhibition of Notch signaling, we created siRNA that inhibits Lef1 expression. Transduction of the 0531 or 1.F9 lymphoma cell lines with Lef1 siRNA virus resulted in an approximate 4-fold decrease in Lef1 mRNA, and Lef1 protein was also decreased (Figure 6A; data not shown). Remarkably, multiple Notch-dependent lymphomas infected with Lef1 siRNA retrovirus initially expressed GFP, but the GFP+ cells were rapidly lost from culture (Figure 6B). In contrast, lymphomas infected with retrovirus producing GFP only, or a scrambled siRNA, stably expressed GFP for more than 8 days (Figure 6B; data not shown). Therefore, Lef1 is required for survival or proliferation of these T-cell lymphomas.
Lef1 is required for survival of T-cell lymphomas. (A) Northern blot of mRNA extracted from 0531 and 1.F9 cells infected with Banshee (“B”) or LEF1 siRNA (siRNA) virus 48 hours after infection. The blot was probed sequentially with cDNA probes complementary to Lef1, c-myc, and actin. (B) Lymphoma lines were infected with Banshee control (“B”; closed symbols) or Lef1 siRNA (si; open symbols) retrovirus, and the percentage of cells expressing GFP was determined every 24 hours by flow cytometry. The relative percentage of GFP+ compared with 24 hours is shown. (C) Flow cytometric cell-cycle analysis of 0531 cells infected with Banshee (left) or Lef1 siRNA virus (right) after a 30-minute pulse with BrdU and staining with anti-BrdU and propidium iodide. The percentage of cells in each gate is indicated. (D) Forward light scatter for GFP+ (■) and GFP− (▩) cells from Banshee- or Lef1 siRNA-infected lymphomas 48 hours after infection.
Lef1 is required for survival of T-cell lymphomas. (A) Northern blot of mRNA extracted from 0531 and 1.F9 cells infected with Banshee (“B”) or LEF1 siRNA (siRNA) virus 48 hours after infection. The blot was probed sequentially with cDNA probes complementary to Lef1, c-myc, and actin. (B) Lymphoma lines were infected with Banshee control (“B”; closed symbols) or Lef1 siRNA (si; open symbols) retrovirus, and the percentage of cells expressing GFP was determined every 24 hours by flow cytometry. The relative percentage of GFP+ compared with 24 hours is shown. (C) Flow cytometric cell-cycle analysis of 0531 cells infected with Banshee (left) or Lef1 siRNA virus (right) after a 30-minute pulse with BrdU and staining with anti-BrdU and propidium iodide. The percentage of cells in each gate is indicated. (D) Forward light scatter for GFP+ (■) and GFP− (▩) cells from Banshee- or Lef1 siRNA-infected lymphomas 48 hours after infection.
To further examine the consequences of Lef1 siRNA expression in T-cell lymphomas, we analyzed the cell-cycle profile of Lef1 siRNA or control virus–expressing cells. Lymphomas were pulsed for 20 minutes with BrdU starting 48 hours after infection, and GFP+ cells were isolated by flow cytometry. Intracellular staining with anti-BrdU antibody and propidium iodide allowed us to resolve the proportion of cells in each phase of the cell cycle (G1, S, and G2/M) as well as apoptotic cells (sub-G1 DNA content). Interestingly, there was a greater than 5-fold increase in the proportion of Lef1 siRNA–expressing cells with sub-G1 DNA content when compared with control virus–infected cells, indicating that cells with decreased Lef1 were undergoing apoptosis (Figure 6C). Lef1 siRNA–expressing cells also showed a decreased proportion of cells in the S-phase of the cell cycle, which may be the result of reduced cell-cycle progression or selective death of cells entering S-phase (Figure 6C). We note, however, that Lef1 siRNA–expressing cells maintain expression of c-myc and are indistinguishable from control virus–infected cells in their forward light scatter, leading us to conclude that these cells are not undergoing a G1-phase arrest (Figure 6A,D). Taken together, these data indicate that Lef1 promotes survival of T-cell lymphomas.
Ectopic Lef1 expression promotes survival of T-cell lymphomas in the absence of Notch signaling
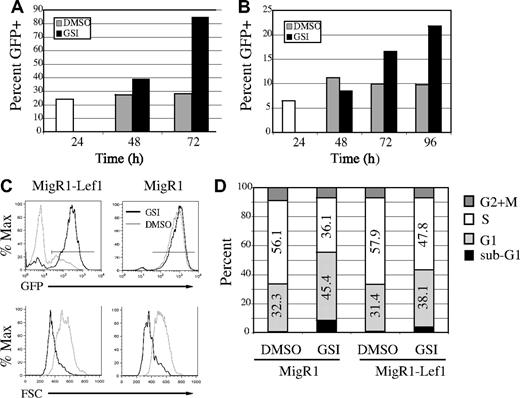
Notch regulates expression of multiple genes that are critical for lymphoma cell growth and survival. Therefore, we questioned whether Lef1 could rescue lymphomas from the effects of GSI. To address this question, we ectopically expressed Lef1 in lymphomas and examined the ability of these cells to withstand GSI treatment. 0531 and 1.F9 cells were transduced with MigR1 or MigR1-Lef1 retrovirus; 24 hours later, the cells were divided and treated with GSI or DMSO. Analysis of GFP expression in MigR1-Lef1–transduced cultures revealed a progressive increase in the percentage of GFP+ cells after GSI treatment (Figure 7A-C). Therefore, Lef1-expressing/GFP+ cells had a selective advantage over GFP− cells in the presence of GSI. As expected, Lef1-expressing/GFP+ cells were not enriched in cultures treated with DMSO (Figure 7A-C). Similarly, no enrichment of GFP+ cells was observed in lymphomas infected with MigR1 whether treated with DMSO or GSI (Figure 7C; data not shown). Although there was a selection for cells with high levels of GFP, there was very little expansion of Lef1-expressing cells in the presence of GSI, and these cells became smaller than DMSO-treated cells (Figure 7C). In this respect, Lef1-expressing cells were indistinguishable from MigR1-infected cells (Figure 7C). To investigate whether Lef1 was affecting proliferation and/or survival of lymphomas, we treated sorted GFP+ cells with GSI or DMSO for 18 hours, followed by labeling with BrdU and cell-cycle analysis. Our analysis revealed that GSIs caused a significant reduction of cells in S-phase and an increase of cells in G1-phase independent of whether the cells expressed Lef1 (Figure 7D). However, Lef1 had some protective effect on the cells since the relative change in S- and G1-phase cells induced by GSI was reduced in cells expressing Lef1. Similarly, fewer Lef1-expressing cells had sub-G1 DNA content when compared with control virus–expressing cells after treatment with GSI (Figure 7D-E). Importantly, a proportion of Lef1-expressing lymphomas remained viable 4 to 5 days after addition of GSI, whereas all MigR1 virus–infected lymphomas had died by this time, further indicating that Lef1 prolonged the survival of GSI-treated lymphomas. However, all Lef1-expressing lymphomas eventually died in the presence of GSI. Therefore, ectopic expression of Lef1 reduced the rate of apoptosis after inhibition of Notch signaling. Taken together, our data indicate that Lef1 is a Notch1 target gene that promotes survival of T-cell lymphomas.
Lef1 delays apoptosis after inhibition of Notch1 signaling. E2A−/− lymphoma lines 0531 (A) and 1.F9 (B) were infected with MigR1-Lef1 for 24 hours (□), divided in 2, and treated with DMSO (▩) or GSI (■). The percentage of cells expressing GFP was measured at the indicated time by flow cytometry. One of 2 experiments for 0531 and 1 of 3 experiments for 1.F9 are shown. P < .03, paired Student t test, for the percentage of GFP in DMSO versus GSI-treated Lef1-expressing cultures at 72 (0531) or 96 (1.F9) hours. (C) MigR1 and MigR1-Lef1–infected cultures were divided in 2 and treated with DMSO (light gray histogram) or GSI (black histogram); 48 hours later, expression of GFP (top row) or forward light scatter on GFP+ cells (bottom row) was measured by flow cytometry. (D) GFP+ cells were sorted from MigR1 or MigR1-Lef1–infected cultures and treated with DMSO or GSI for 18 hours prior to addition of BrdU and cell-cycle analysis. The percentage of cells in G1 (light gray), S (white), G2/M (dark gray) and sub-G1 (black) phases of the cell cycle are represented. One of 2 experiments is shown.
Lef1 delays apoptosis after inhibition of Notch1 signaling. E2A−/− lymphoma lines 0531 (A) and 1.F9 (B) were infected with MigR1-Lef1 for 24 hours (□), divided in 2, and treated with DMSO (▩) or GSI (■). The percentage of cells expressing GFP was measured at the indicated time by flow cytometry. One of 2 experiments for 0531 and 1 of 3 experiments for 1.F9 are shown. P < .03, paired Student t test, for the percentage of GFP in DMSO versus GSI-treated Lef1-expressing cultures at 72 (0531) or 96 (1.F9) hours. (C) MigR1 and MigR1-Lef1–infected cultures were divided in 2 and treated with DMSO (light gray histogram) or GSI (black histogram); 48 hours later, expression of GFP (top row) or forward light scatter on GFP+ cells (bottom row) was measured by flow cytometry. (D) GFP+ cells were sorted from MigR1 or MigR1-Lef1–infected cultures and treated with DMSO or GSI for 18 hours prior to addition of BrdU and cell-cycle analysis. The percentage of cells in G1 (light gray), S (white), G2/M (dark gray) and sub-G1 (black) phases of the cell cycle are represented. One of 2 experiments is shown.
Discussion
We have identified the transcription factor Lef1 as a direct target of Notch1 that promotes survival of murine T-cell lymphomas. We show that Lef1 expression in T-cell lymphomas is dependent on Notch1 signaling both in vitro and in vivo, and that ICN1 is associated with sequences in the Lef1 gene. Moreover, expression of Lef1 is important for survival and expansion of T-cell lymphoma lines, and ectopic expression of Lef1 in these lines delays death after inhibition of Notch signaling. Taken together with our observation that Lef1 expression is not Notch dependent in primary T-cell progenitors, our data indicate that heightened Notch1 signaling during the process of transformation co-opts Lef1 to deregulate survival in T-lineage cells.
Our observation that Lef1 is required for survival of T-cell lymphomas is surprising because Lef1 is not essential for normal T-cell development.33 Indeed, Lef1 and Tcf1 perform redundant functions in T-lymphocyte progenitors, with Tcf1 being more essential than Lef1.43 However, we observed that Lef1 but not Tcf1 mRNA and protein are elevated in Notch1-dependent T-cell lymphomas when compared with normal thymocytes. Interestingly, Lef1 mRNA has previously been reported to be elevated in numerous patients with acute lymphoblastic leukemia, indicating that deregulated Lef1 expression may play a role in transformation in this disease.44 A sustained increase in Lef1 expression may lead to “addiction” of lymphomas to this transcription factor, as has been observed for known oncogenes.45 In this case, Lef1 may regulate essential survival genes that are normally regulated by both Lef1 and Tcf1. However, it is also feasible that heightened expression of Lef1 leads to activation of low-affinity targets that are not essential in primary thymocytes but are required for survival of transformed cells. While there are not many known targets of Lef1, it was reported previously that Lef1 can promote Notch1 signaling by regulating expression of DL1 in somitic mesoderm.46 However, DL1 is not expressed in E2A−/− T-cell lymphomas, indicating that regulation of DL1 by Lef1 may be cell-type specific and does not explain the requirement for Lef1 in Notch1-dependent lymphomas (unpublished observation, B.K., June 2006). Moreover, we did not find evidence for regulation of any Notch1 ligands by Lef1 in E2A−/− lymphomas. Therefore, the targets of Lef1 that promote survival of Notch1-dependent lymphomas remain to be identified.
Lef1 can regulate gene expression through multiple different mechanisms. Like other members of the high mobility group family of transcription factors, Lef1 is able to bend DNA, allowing distant regulatory proteins on a gene to come into proximity. Such an architectural role for Lef1 has been characterized on the TCRα promoter, although direct binding to coactivator proteins has also been implicated in Lef1-mediated regulation of TCRα.47,48 Lef1 can also act as a transcriptional activator in response to Wnt signaling. Wnt signaling leads to stabilization of β-catenin, which can physically interact with the amino-terminal domain of Lef1 and function as a coactivator to allow Lef1-dependent gene transcription.38 In the absence of Wnt signaling, Lef1 can function as a transcriptional repressor through recruitment of Groucho-related corepressor proteins.49,50 It is not presently known whether the transcriptional activating, repressing, or architectural functions of Lef1 are required downstream of Notch1 for survival of T-cell lymphomas. Interestingly, however, a recent study demonstrated that constitutive activation of β-catenin in T-lymphocyte progenitors resulted in T-cell lymphoma, and the resulting lymphomas expressed high levels of c-myc but not Notch1 or Notch1 target genes.51 Therefore, prolonged activation of Lef1/Tcf1 target genes may predispose thymocytes to malignant transformation. Taken together, these observations raise the possibility that Notch1 could influence Wnt-dependent gene expression in T-cell lymphomas through regulation of Lef1; however, the role of Wnt signaling in these cells remains to be determined.
We conclude that regulation of Lef1 by Notch1 is a consequence of the increased expression of ICN1 that occurs after acquisition of mutations in the Notch1 PEST domain. This conclusion is based on our observation that Lef1 expression in T-lymphocyte progenitors cultured in vitro on OP9-DL1 stromal cells is not dependent on Notch signaling. Ectopic expression of ICN1 results in increased expression of the Notch target gene Deltex1 as well as Lef1 in primary T-lymphocyte progenitors, indicating that high levels of ICN1 can deregulate Lef1. While ICN1 could be found associated with the Lef1 gene, our EMSA experiments indicate that CSL has a low affinity for both the Lef1p +11-bp and Lef1p −2-kb CSL-binding sites. Therefore, it is plausible that high concentrations of ICN1 are necessary for stabilization of the ICN1/CSL/MAML complex on the Lef1 promoter. A recent study demonstrated that the CSL/ICN1/MAML complex functions optimally on closely spaced inverted CSL sites that allow for formation of higher-order complexes.52 In the case of the Lef1 promoter, the low-affinity CSL-binding sites may allow for optimal activation only when sufficient ICN1 is present to allow interaction with neighboring proteins or distant CSL-binding sites that stabilize the promoter-bound activating complexes.52 Alternatively, CSL may have to compete with Ikaros for binding to these sites, and increasing concentrations of ICN1 may increase the likelihood of transcriptional activation after infrequent binding by CSL.11 Future studies will address the mechanism by which heightened ICN1 co-opts Lef1 transcription.
Our findings reveal an important role for Lef1 downstream of Notch1 in murine T-cell lymphomas. While the precise mechanism of Lef1 function remains to be determined, the possibility of an essential role of Lef1 in T-cell lymphomas but not in normal thymopoiesis may make Lef1 an attractive target for therapeutic intervention in Notch1-dependent T-cell malignancies.
An Inside Blood analysis of this article appears at the front of this article.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Iannis Aifantis for discussion and comments on the manuscript, Ryan Duggan and David LeClerc for cell sorting, and Ximin Li for screening of gene arrays.
This work was supported in part by an award to the University of Chicago's Division of Biological Sciences under the Research Resources Program for Medical Schools of the Howard Hughes Medical Institute, a New Investigator Award from the Leukemia Research Foundation, the Concern Foundation and the American Cancer Society (B.L.K), a National Institutes of Health (NIH)/National Cancer Institute (NCI) training grant (E.J.R.), and the NIH (W.S.P.).
National Institutes of Health
Authorship
Contribution: C.S., E.J.R., Y.Y.-O., and L.J.B. designed and performed experiments; D.E.Z. provided technical assistance; A.C. and W.S.P. provided essential reagents and contributed to the interpretation of data; and B.L.K., designed and performed experiments, coordinated and supported the work, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Barbara L. Kee, Department of Pathology, University of Chicago, 5841 S Maryland Ave, MC 1089, Chicago, IL 60637; e-mail:bkee@bsd.uchicago.edu.



![Figure 5. Regulation of Lef1 by ICN1 in primary T-cell progenitors and T-cell lymphoma. (A) QPCR analysis of Lef1 and Deltex1 expression in primary T-cell progenitors cultured in vitro on OP9-DL1 in the presence of DMSO or GSI for 12 hours or after infection with MigR1 or MigR1-ICN1 retrovirus. mRNA expression was standardized to HPRT. Comparison of the mouse and human Lef1 gene sequence surrounding the mouse Lef1p + 11-bp CSL site (B) and the Lef1p −2179-bp CSL site (C). The CSL-binding sites are shaded. (D) EMSA using the Lef1p +11-bp and Lef1p −2179-bp CSL sites. Complexes were competed with self-competitor, a CSL consensus sequence (Jκ3), or nonspecific competitor (Oct). The position of the free oligonucleotides and shifted CSL complex are shown. Vertical lines have been inserted to indicate where a gel was cut. All lanes are from the same gel. (E) ChIP analysis. DNA precipitated with IgG or anti-Notch1 from 0531 cells treated with GSI (squlf]) or DMSO (▩) for 15 hours was analyzed by QPCR using primers specific for sequences near the Lef1p −2-kb and Lef1p +11-bp CSL-binding sites, in Lef1 intron 11, and the Hes1 promoter. Results are representative of 4 precipitations. Hes1 promoter P < .001; Lef1 intron P < .25; and Lef1p −2 kb P < .05 using a paired Student t test. The Lef1p +11-bp site was precipitated specifically with anti-Notch1 in 3 of 4 experiments, resulting in P < .25. (F) QPCR analysis of DNA precipitated with IgG or antiacetylated histone H4 (AcH4) from the same samples as in panel E. Histograms represent the means (± SE) of triplicate QPCR amplifications.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/7/10.1182_blood-2007-04-084202/2/m_zh80190707940005.jpeg?Expires=1769848035&Signature=h21-0fKNkyjp3qxspQVtsvbPuVK7RFxJr7EKSRvcgxeeInOKk0MNrWhoFpNeY1NqLw54UUdlwKc-kXJLGyhqnCVOPLdpTiVP0ooUBiJN1ndNNOQ94BfkYeUVEfaPAIO6-EyPgv3RV-Y-i4d2nk80xU670exUTz6D0d0WoUj61AdR5NWS7WipYWwq-tuqa7Ckc5eyGfbBSxnNY0LsSa0NaRMDTskQ2gvG~O4MZhnRWkBN59zCcWUYhZQ1VZXaLdU6NuV1cg8ZuOsM7YT28nFCd-ND8ZBMSYXpjilBtkAJRV8b-opQz4y~KkNvy0t~01XLduZ6ELtWy~1TKmL9y-nKSg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal