Plasma cells producing high levels of paraprotein are dependent on the unfolded protein response (UPR) and chaperone proteins to ensure correct protein folding and cell survival. We hypothesized that disrupting client–chaperone interactions using heat shock protein 90 (Hsp90) inhibitors would result in an inability to handle immunoglobulin production with the induction of the UPR and myeloma cell death. To study this, myeloma cells were treated with Hsp90 inhibitors as well as known endoplasmic reticulum stress inducers and proteasome inhibitors. Treatment with thapsigargin and tunicamycin led to the activation of all 3 branches of the UPR, with early splicing of XBP1 indicative of IRE1 activation, upregulation of CHOP consistent with ER resident kinase (PERK) activation, and activating transcription factor 6 (ATF6) splicing. 17-AAG and radicicol also induced splicing of XBP1, with the induction of CHOP and activation of ATF6, whereas bortezomib resulted in the induction of CHOP and activation of ATF6 with minimal effects on XBP1. After treatment with all drugs, expression levels of the molecular chaperones BiP and GRP94 were increased. All drugs inhibited proliferation and induced cell death with activation of JNK and caspase cleavage. In conclusion, Hsp90 inhibitors induce myeloma cell death at least in part via endoplasmic reticulum stress and the UPR death pathway.
Introduction
Despite the advent of several new therapies for myeloma, none is curative, and there remains an urgent need to develop new treatments. Heat shock protein 90 (Hsp90) inhibition is one such novel targeted approach that is being explored as the disruption of client proteins from their molecular chaperone proteins affects many pathways important for tumor cell survival. However, to develop the treatment clinically, a more detailed understanding of the effects of Hsp90 inhibition in myeloma is required
One of the key characteristics that distinguish myeloma plasma cells as a therapeutic target is the large quantity of monoclonal paraprotein they synthesize and secrete. These immunoglobulins are folded into their tertiary structures within the endoplasmic reticulum (ER), where the unfolded protein response (UPR) maintains the equilibrium between the rate of protein production and the capacity for nascent protein folding. Activation of the UPR results in a bias of translation toward the synthesis of chaperone proteins involved in protein folding, an increase in disposal of misfolded proteins via the ubiquitin proteasome pathway, and the delivery of a survival signal. If the UPR is unable to correct the balance, then an ER stress signal is generated and apoptosis ensues. The potential ability of Hsp90 inhibitors to disrupt normal protein folding, in this case the folding of nascent immunoglobulin in the ER, may contribute a novel therapeutic strategy especially relevant for the killing of plasma cells.
The key players in the UPR are well characterized. BiP is associated with the ER luminal domains of the kinases PERK (ER resident kinase) and IRE1, and also activating transcription factor 6 (ATF6).1,–3 This interaction is destabilized in the presence of misfolded proteins, resulting in the release of PERK and IRE1 and their autocatalytic activation.4,5 The endoribonuclease activity of IRE1 regulates the activation of XBP1 by the cleavage of XBP1 mRNA to form XBP1s, which acts as a positive feedback signal to the ER, allowing it to handle more unfolded protein.6 XBP1 is of particular relevance to myeloma, where it has been shown to mediate a checkpoint in plasma-cell development6,7 and has been shown to be dysregulated.8 PERK phosphorylation inhibits the general translation initiation factor eIF2a, resulting in a general shutdown of nonchaperone protein synthesis.9,10 In the presence of unfolded proteins the precursor form of ATF6 is also released from BiP and translocates to the nucleus, where it mediates the expression of key genes involved in the UPR response, including BiP and XBP1.11,–13
In addition to BiP, other molecular chaperones are also essential for the correct folding of nascent immunoglobulin, including members of the Hsp family, some of which are located within the ER. One such protein, grp94, is an Hsp90 isoform that stabilizes the interactions of the cytoplasmic domains of IRE1, ATF6, and PERK with BiP.14,15 In addition, Hsps also have a broader antiapoptotic role within the cell, mediating the response of both the intrinsic and extrinsic apoptotic pathways.16 Thus, while inhibiting Hsp90 kills cells via the modulation of apoptosis, it may also induce cell death via the generation of an ER stress signal and a death signal mediated by the UPR.14
ER stress can be triggered by a number of stimuli, including calcium depletion from the ER lumen, inhibition of N-linked glycosylation of proteins, reduction of disulphide bonds, and glucose deprivation.17,18 Initially, the cell will resist death while attempts at correct protein folding are performed. To do this, the UPR and cell survival signals are triggered in parallel, resulting in the activation of AKT, ERK, and IAPs, which antagonize apoptosis and allow time for the UPR to function.19 However, if this mechanism fails, an apoptotic signal is delivered via a number of mechanisms. First, it is delivered via the induction of CHOP,20 which is activated by PERK and ATF6, with consequent suppression of bcl-2 transcription together with the induction of other proapoptotic genes and cell-cycle arrest.21 Second, it is delivered via the activation of IRE1, resulting in ASK1 and JNK activation.22 Finally, it is delivered via activation of the intrinsic apoptotic pathway,23 resulting in the direct activation of caspase 4 as well as release of cytochrome c from the mitochondria and its subsequent binding to Apaf1, with activation of caspase 9 and subsequent activation of caspase 3.24
In this study we have investigated the mechanism of action of Hsp90 inhibition on the UPR in myeloma cells. Our results demonstrate that a proportion of the ability of Hsp90 inhibitors to kill myeloma plasma cells is mediated via the induction of ER stress with the downstream initiation of all 3 branches of the UPR.
Materials and methods
Cell lines
Multiple myeloma cell lines (U266, H929, MM1s) were grown in RPMI1640 supplemented with 10% heat-inactivated fetal calf serum and GlutaMAX-I (Invitrogen, Paisley, United Kingdom), 60 μg/mL penicillin, and 100 μg/mL streptomycin. The colon cancer cell line HCT116 was grown in Dulbecco modified Eagle medium (DMEM) with 10% fetal calf serum, 2 mM glutamine, and nonessential amino acids. Cultures were maintained in exponential growth at 37°C in a humidified atmosphere of 95% air/5% carbon dioxide. To induce ER stress, myeloma cells were treated with a number of compounds, including tunicamycin (100 μM), thapsigargin (10 μM), 17-(allylamino)-17-demethoxygeldanamycin (17-AAG, 5 μM), radiciciol (10 μM), dexamethasone (10 μM), and melphalan (30 μM) (all from Sigma-Aldrich Company, St. Louis, MO), and bortezomib (8 nM; Millenium Pharmaceuticals, Cambridge, MA). All experiments were performed in triplicate.
Cell growth and survival assays
The in vitro toxicology assay (methyl-thiazol-tetrazolium, MTT based) was performed according to manufacturers' instructions (Sigma-Aldrich) in 96-well microculture plate format. Cells were seeded at 106 cells/mL and plates were incubated in a humidified incubator in 5% CO2 for 24 hours at 37°C. Plates were read after 24 hours on a Dynatech Laboratories (Billingshurst, United Kingdom) MRX plate reader. Cell death was determined using 2 methods: trypan blue exclusion and flow cytometric analysis of Annexin V and propidium iodide using the Becton Dickinson (Oxford, United Kingdom) FACSCalibar.
Detection of cellular inclusions
The formation of cellular inclusions was detected by staining with May-Grunwald-Giemsa stain (MGG). Immunoglobulin light chain was detected using polyclonal κ and λ antibodies (Dako, Ely, United Kingdom) with a labeled streptavidin biotin secondary antibody (Ventana Medical Systems, Illkirch, France). Samples were processed using the Ventana benchmark XT immunostaining machine and the iView DAB detection kit.
RNA extraction, quantification, and amplification
RNA was extracted using commercially available kits (Abgene, Epsom, United Kingdom) according to the manufacturers' instructions and quantitated using a Nanodrop spectrophotometer (Labtech, East Sussex, United Kingdom). cDNA was synthesized from equal quantities of RNA using the Superscript III First-strand synthesis kit (Invitrogen).
Real-time polymerase chain reaction
SYBR Green real-time polymerase chain reaction (Applied Biosystems) was performed on cDNA extracted from cells after inhibitor treatment. Primer sequences were designed for intron/exon boundaries: BiP, forward 5′-CAATCAAGGTCTATGAAGGTGAAAGA-3′ and reverse 5′-CACATCTATCTCAAAGGTGACTTCAATC-3′; CHOP, forward 5′-TGGAAATGAAGAGGAAGAATCAAAA-3′ and reverse 5′-CAGCCAAGCCAGAGAAGCA-3′; GRP94, forward 5′-TCGCCTCAGTTTGAACATTGAC-3′ and reverse 5′-CTTCTGCTGTCTCTTCAGGTTCTTC-3′; and Edem1, forward 5′-ACTCCAGCTCCAACTGCAATC-3′ and reverse 5′-GGTCAATCTGTCGCATGTAGATG-3′. Thermal cycling conditions were 10 minutes at 95°C, 40 cycles at 95°C for 15 seconds, followed by 1 minute at 60°C. Data analysis was completed using the 7500 Sequence Detection software (Applied Biosystems).
Polymerase chain reaction
To determine relative expression levels of XBP1/XBP1s within a sample, polymerase chain reaction was performed in a 50-μL platinum Taq reaction containing 4 pmol of primers (forward 5′-CCTTGTAGTTGAGAACCAGG-3′ and reverse 5′-GGGGCTTGGTATATATGTGG-3′; Invitrogen), 1 unit of platinum Taq DNA polymerase, and 200 μM dNTP. The temperature profile was at 94°C for 2 minutes, followed by 30 cycles of 94°C for 15 seconds, 60°C for 1 minute, and 72°C for 30 seconds. Products were run on 2% agarose gels containing ethidium bromide.
Immunoblotting and peptide-blocking studies
After inhibitor treatment, cells were centrifuged at 1200 rpm for 5 minutes, washed with ice-cold phosphate-buffered saline, pelleted, and resuspended in 1 mL of ice-cold phosphate-buffered saline. The suspension was spun at 1200g (3500 rpm) for 15 minutes at 4°C and the pellet resuspended in lysis buffer (1% NP-40, 20 mM Tris, pH 8, 100 mM NaCl, 1 mM EDTA[ethylenediaminetetraacetic acid]) supplemented with 1 × protease inhibitor cocktail 50 mM NaF, 200 mM phenylmethylsulfonyl fluoride (PMSF), and 1 mM Na3VO4 (Sigma-Aldrich). Nuclear and cytoplasmic fractions of protein lysates harvested for the detection of ATF6 were prepared using the NE-PER Nuclear and Cytoplasmic Extraction Reagent (Pierce Biotechnology, Rockford, IL). Proteins were resolved by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred to polyvinylidene difluoride (PVDF) membranes, blocked with either 5% milk or 5% bovine serum albumin, and incubated with primary antibody. Primary antibodies to Hsp90, Hsp70, Grp94 (Stressgen, Bioreagents, Victoria, BC), Grp78 (Santa Cruz Biotechnology, Heidelberg, Germany), phosphor-SAPK/JNK, caspases 3, 8, and 9 antibodies (Cell Signaling Technology, Danvers, MA); anti-ATF6 (Stratagene, La Jolla, CA), anti-β actin antibody (Sigma-Aldrich), and CBP(A-22) (Santa Cruz Biotechnology), and secondary antibodies, antimouse, antirabbit, or antirat conjugated to horseradish peroxidase (Amersham Biosciences, Amersham, United Kingdom) were used. ECL-Plus (Amersham Biosciences) was used for detection.
Results
Activation of the UPR induces myeloma cell death
Thapsigargin (TG) acts to deliver a stress response by inhibiting Ca2+ uptake by the ER, whereas tunicamycin (TM) induces stress by inhibiting N-linked glycosylation resulting in the accumulation of misfolded proteins. Exposure of myeloma cells to TM and TG results in apoptosis as demonstrated by trypan blue staining, flow cytometric detection of annexin V/PI staining, and a decrease in cell proliferation by MTT assays (data not shown). Importantly, exposure of myeloma cells to 2 other drug types, proteasome inhibitors (bortezomib) and Hsp90 inhibitors (17-AAG, and radicicol), which, we hypothesize, would induce the UPR, also results in myeloma cell apoptosis (data not shown).
TG and TM induce the UPR in myeloma cells
To determine the nature of the intracellular signal response and mode of action of known UPR stressors in myeloma cells against which we could compare the response generated by Hsp90 inhibitors, we analyzed the key UPR response genes by real-time polymerase chain reaction, and by Western blotting after exposure to TG and TM. Under normal conditions BiP, the most abundant ER chaperone, associates with IRE1, PERK, and ATF6. Accumulation of misfolded proteins results in the dissociation of this complex and activation of its individual components. A central player in this pathway is IRE1, which mediates a proportion of its activity via XBP1. Activation of IRE1 results in splicing of XBP1 to its active form XBP1s and activation of the UPR, resulting in a pro-survival signal allowing stress to be resolved. Consistent with these observations and activation of this branch of the pathway, induction of ER stress in myeloma cells by both TG and TM rapidly induced splicing of XBP1 to XBP1s after a 2-hour incubation period (Figure 1A).
The ER stress response is initiated in myeloma cells by tunicamycin (TM) and thapsigargin (TG). (A) Treatment of myeloma cells over a 24-hour period with TM- or TG-induced splicing of XBP1, as determined by reverse-transcription polymerase chain reaction (PCR). (B) Levels of the proapoptotic factor CHOP were determined by real-time Taqman PCR. (C) Immunoblotting of nuclear fractions of cell lysates demonstrated an increase in cleaved ATF6α. Representative cell line data on U266 are shown. All experiments were repeated in triplicate.
The ER stress response is initiated in myeloma cells by tunicamycin (TM) and thapsigargin (TG). (A) Treatment of myeloma cells over a 24-hour period with TM- or TG-induced splicing of XBP1, as determined by reverse-transcription polymerase chain reaction (PCR). (B) Levels of the proapoptotic factor CHOP were determined by real-time Taqman PCR. (C) Immunoblotting of nuclear fractions of cell lysates demonstrated an increase in cleaved ATF6α. Representative cell line data on U266 are shown. All experiments were repeated in triplicate.
One component of the proapoptotic signal generated by ER stress is delivered via PERK; however, directly measuring activation of PERK is difficult and consequently CHOP, a proapoptotic transcription factor downstream of PERK, is used as a surrogate marker. Exposure of myeloma cells to ER stress mediated by either TG or TM results in an increase in the level of CHOP transcripts (7-22-fold) within 2 hours (Figure 1B).
We also investigated the activation of the third branch of the UPR pathway, which is initiated after cleavage of ATF6 by S1 and S2 proteases. The resulting fragment then translocates to the nucleus, where it induces genes with an ER stress response element (ERSE) within their promoter, allowing the cell to better cope with stress. Within 4 hours of treatment with either TM or TG, the cleaved fragment could be detected in both the cytoplasmic and nuclear fraction, consistent with activation of this pathway (Figure 1C).
Therefore, after ER stress in myeloma cells there is an early activation of the IRE1 branch associated with the induction of splicing of XBP1 to XBP1s, with cleavage of ATF6 and the induction of its target genes, and activation of PERK with the delivery of a proapoptotic signal via CHOP. This pattern is induced rapidly within 2 hours of exposure and occurs simultaneously in all 3 pathways. These results provide a baseline against which to compare the antimyeloma agents 17-AAG, radicicol, and bortezomib, which may also induce the UPR as a component of their mode of action. Importantly, the steroid dexamethasone and the DNA-damaging agent melphalan failed to activate these pathways (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article), consistent with the mechanism of action of these drugs not being dependent on the UPR.
Hsp90 inhibition also induces an ER stress pattern in myeloma cells
As a number of key mediators of the UPR are Hsp client proteins, we have investigated the ability of 2 known Hsp90 inhibitors to induce ER stress. We demonstrate that 17-AAG and radicicol are capable of inducing splicing of XBP1 to XBP1s, indicating that Hsp90 inhibition initiates the UPR and generates an ER stress response (Figure 2A). Although the early effects (within 2 hours) on XBP1 splicing after exposure to both Hsp90 inhibitors were similar to that seen with TM or TG, prolonged incubation (up to 24 hours) failed to induce further splicing. Hsp90 inhibition induced CHOP expression, indicative of PERK activation. This response also occurred rapidly after 2 hours with 9- to 30-fold changes in its expression level being detected in response to the 2 reagents, respectively (Figure 2B). The effect of ER stress induction on ATF6 was also investigated, and the cleaved p50 fragment could be detected within the cytoplasmic and nuclear fractions after 4 hours (Figure 2C).
The Hsp90 inhibitor 17-AAG and radicicol induce ER stress in myeloma cells. (A) Treatment of myeloma cells over a 24-hour period with 17-AAG or radicicol-induced splicing of XBP1, as demonstrated by reverse-transcription PCR. (B) Levels of the proapoptotic factor CHOP were determined by real-time Taqman PCR. (C) Immunoblotting of nuclear fractions of cell lysates demonstrated an increase in cleaved ATF6α. Representative cell line data on U266 are shown. All experiments were repeated in triplicate.
The Hsp90 inhibitor 17-AAG and radicicol induce ER stress in myeloma cells. (A) Treatment of myeloma cells over a 24-hour period with 17-AAG or radicicol-induced splicing of XBP1, as demonstrated by reverse-transcription PCR. (B) Levels of the proapoptotic factor CHOP were determined by real-time Taqman PCR. (C) Immunoblotting of nuclear fractions of cell lysates demonstrated an increase in cleaved ATF6α. Representative cell line data on U266 are shown. All experiments were repeated in triplicate.
Taken together, these results suggest that Hsp90 inhibitors are able to induce ER stress and activate all 3 branches of the UPR. The activation of the UPR occurs rapidly, within 2 hours, and occurs simultaneously for all of the 3 branches of the UPR. However, not all pathways are activated equally with distinct differences being seen in comparison to TM and TG. After treatment with Hsp90 inhibitors, a strong activation of PERK and ATF6 is seen with only a transient effect on IRE1 compared with that seen with TM and TG, with XBP1 splicing being seen early but not being subsequently maintained. This may explain why 17-AGG is more potent in killing myeloma cells than TM or TG as the proapoptotic effects of PERK and ATF6 are maintained after drug treatment, whereas the pro-survival response stimulated by the IRE1 pathway is only transient.
Proteasome inhibition in myeloma cells induces ER stress but not via activation of the IRE1 pathway
Previous reports have suggested that inhibition of the ubiquitin–proteasome pathway results in the accumulation of defective proteins within the ER, inducing ER stress, activating the UPR, and consequently inducing apoptosis. We tested this hypothesis by treating myeloma cells with bortezomib, a selective and potent inhibitor of the 26S proteasome, and determined its effect on a number of key molecules involved in the UPR. In contrast to the effects induced by TM and TG, treatment with bortezomib did not lead to early XBP1 splicing and only minimal splicing of XBP1 to XBP1s was seen at the 24-hour time point. This pattern of splicing was also associated with a time-dependent degradation of unspliced XBP1 (Figure 3A). In contrast to these effects on XBP1, we clearly demonstrate large increases in the levels of the proapoptotic transcription factor CHOP, with a 25-fold increase being noted after a 12-hour incubation period with bortezomib (Figure 3B), together with cleavage of ATF6 within 4 hours (Figure 3C). These results suggest that while TM, TG, and Hsp90 inhibition generate ER stress and activate all 3 branches of the UPR equally, treatment with bortezomib seems to induce an unbalanced response, with the predominant effect of activating the proapoptotic PERK and ATF6 branches and with little effect on the induction of the pro-survival IRE1/XBP1 pathway.
Bortezomib, in contrast to Hsp90 inhibitors or TM and TG, exhibits a delayed ability to splice XBP1 and induces a progressive transcriptional upregulation of CHOP. (A) Treatment of myeloma cells over a 24-hour period with bortezomib induced late splicing of XBP1, as demonstrated by reverse-transcription PCR. (B) Levels of the proapoptotic factor CHOP were determined by real-time Taqman PCR. (C) Immunoblotting of nuclear fractions of cell lysates demonstrated the increase in cleaved ATF6α. Representative cell line data on U266 are shown. All experiments were repeated in triplicate.
Bortezomib, in contrast to Hsp90 inhibitors or TM and TG, exhibits a delayed ability to splice XBP1 and induces a progressive transcriptional upregulation of CHOP. (A) Treatment of myeloma cells over a 24-hour period with bortezomib induced late splicing of XBP1, as demonstrated by reverse-transcription PCR. (B) Levels of the proapoptotic factor CHOP were determined by real-time Taqman PCR. (C) Immunoblotting of nuclear fractions of cell lysates demonstrated the increase in cleaved ATF6α. Representative cell line data on U266 are shown. All experiments were repeated in triplicate.
Effect of TM, TG, Hsp90, and proteasome inhibition on the ER-associated degradation pathway
After the induction of ER stress and activation of the UPR, the endoplasmic reticulum associated degradation pathway is activated stimulating the clearance of the unfolded proteins from the ER lumen and their subsequent degradation.25,–27 The transcript levels of EDEM1, an ER stress–induced membrane protein that accelerates the degradation of misfolded proteins in the ER,28,29 were determined as a measure of activity of this pathway. ER stress induction after treatment with TM/TG gives rise to a similar pattern of ER-associated degradation pathway response to that induced by Hsp90 and proteasome inhibition with a 5-fold increase in the transcript levels of EDEM1 after 24 hours (Figure 4A). These results suggest that after treatment with ER stressors, Hsp90 inhibitors, and proteasome inhibitors, a feedback loop is present resulting in activation of the ER-associated degradation pathway, an increase in the expression of EDEM1, and the strengthening of the ER-associated degradation pathway machinery.
Cellular effects of ER stress induced by TM and TG, Hsp90, and proteasome inhibitors. (A) Transcriptional upregulation of EDEM-1, known to accelerate the degradation of misfolded proteins, demonstrated by real-time Taqman PCR. (B) The extent of cytoplasmic inclusions exhibited by U266 in response to treatments (24 hours, IC50 doses: TM, 100 μM; TG, 10 μM; 17-AGG, 5 μM; Bortezomib, 8 nM) was determined using May-Grunwald-Giemsa (MGG) staining and light microscopy (Olympus BH-2 microscope [Olympus, London, United Kingdom] 100×/1.40 oil; images acquired using JVC KY-F1030 digital camera and JVC Scan Rate Converter [Victor, Tokyo, Japan]). (C) Immunoglobulin light chain staining demonstrates a build-up of λ light chain within the inclusions. Representative cell line data on U266 are shown. All experiments were repeated in triplicate.
Cellular effects of ER stress induced by TM and TG, Hsp90, and proteasome inhibitors. (A) Transcriptional upregulation of EDEM-1, known to accelerate the degradation of misfolded proteins, demonstrated by real-time Taqman PCR. (B) The extent of cytoplasmic inclusions exhibited by U266 in response to treatments (24 hours, IC50 doses: TM, 100 μM; TG, 10 μM; 17-AGG, 5 μM; Bortezomib, 8 nM) was determined using May-Grunwald-Giemsa (MGG) staining and light microscopy (Olympus BH-2 microscope [Olympus, London, United Kingdom] 100×/1.40 oil; images acquired using JVC KY-F1030 digital camera and JVC Scan Rate Converter [Victor, Tokyo, Japan]). (C) Immunoglobulin light chain staining demonstrates a build-up of λ light chain within the inclusions. Representative cell line data on U266 are shown. All experiments were repeated in triplicate.
Exposure of myeloma cells to Hsp90 inhibitors results in the formation of intracellular inclusion bodies
Twenty-four hours after the exposure of myeloma cells to 17-AAG or bortezomib, the presence of intracellular inclusions was noted. These inclusions were not present after treatment with dexamethasone, an agent known to induce apoptosis via the caspase pathway, or after exposure to the DNA damaging agent melphalan. A smaller number of similar inclusions were seen after exposure to the classic ER stressor TG, but none was seen after treatment with TM (Figure 4B). Morphologically these inclusions are small, round, and located throughout the cytoplasm. They appear after 4 hours of treatment, and after 6 hours of treatment there were approximately 5 to 10 inclusions per cell and the appearances remained stable until 48 hours. At 48 hours the inclusions increased slightly in size; however, they remained as discrete inclusions and did not coalesce. At this time point the chromatin within the nucleus was pyknotic-compatible with early apoptosis. Because treatment with 17-AAG potentially impairs the correct folding of nascent immunoglobulin within the cell, we hypothesized that these inclusions would contain immunoglobulin light chain. After treatment with 17-AAG for 24 hours, U266 cells, which are known to secrete λ light chain, showed strong λ staining, whereas staining with the alternate κ light chain antibody was negative (Figure 4C), confirming the hypothesis that they contain immunoglobulin.
TM and TG exert different effects on molecular chaperones in comparison to Hsp90 and proteasome inhibition
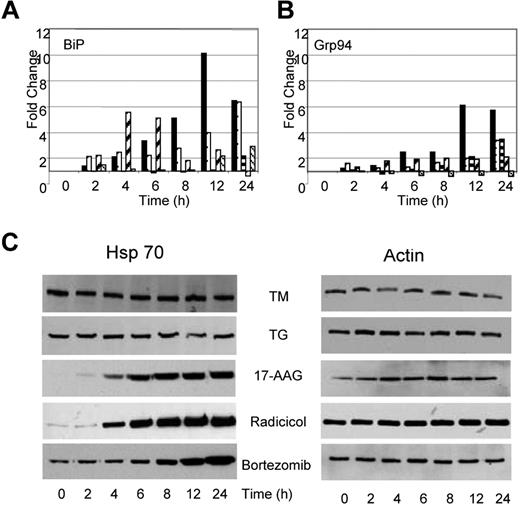
Under conditions of cellular stress, adaptive responses aimed at refolding the misfolded proteins and resolving the cellular stress take place. Consequently, it should be anticipated that stress will result in an increase in proteins central to protein folding and the UPR, particularly of the molecular chaperones. After treatment with TM or TG, we demonstrate that the transcript levels of ER resident chaperones change. BiP mRNA levels increased by 6- to 10-fold after TG and TM exposure (Figure 5A); however, protein levels remained constant throughout this time period (data not shown). Similarly, GRP94, the ER resident isoform of Hsp90, showed a 3- to 6-fold increase in expression over the 24-hour period after treatment with TG and TM (Figure 5B), with protein levels over the same time period remaining stable (data not shown). The total cellular Hsp90 (data not shown) and Hsp70 protein levels remained stable over the 24-hour period (Figure 5C).
Drugs capable of inducing ER stress have contrasting effects on Hsp70 expression. (A) Alterations in expression levels of BiP and Grp94 transcripts occur in response to all drugs (■, TM; □, TG;  , 17-AAG;
, 17-AAG;  , Radicicol;
, Radicicol;  , Bortezomib). (B) Upregulation of Hsp70 occurs in response to Hsp90 and proteasome inhibitors but not in response to inhibitors of protein glycosylation (TM) or the SERCA pump (TG). U266 cells treated, as indicated, with the Ic50 doses of drugs were lysed and analyzed by immunoblotting.
, Bortezomib). (B) Upregulation of Hsp70 occurs in response to Hsp90 and proteasome inhibitors but not in response to inhibitors of protein glycosylation (TM) or the SERCA pump (TG). U266 cells treated, as indicated, with the Ic50 doses of drugs were lysed and analyzed by immunoblotting.
Drugs capable of inducing ER stress have contrasting effects on Hsp70 expression. (A) Alterations in expression levels of BiP and Grp94 transcripts occur in response to all drugs (■, TM; □, TG;  , 17-AAG;
, 17-AAG;  , Radicicol;
, Radicicol;  , Bortezomib). (B) Upregulation of Hsp70 occurs in response to Hsp90 and proteasome inhibitors but not in response to inhibitors of protein glycosylation (TM) or the SERCA pump (TG). U266 cells treated, as indicated, with the Ic50 doses of drugs were lysed and analyzed by immunoblotting.
, Bortezomib). (B) Upregulation of Hsp70 occurs in response to Hsp90 and proteasome inhibitors but not in response to inhibitors of protein glycosylation (TM) or the SERCA pump (TG). U266 cells treated, as indicated, with the Ic50 doses of drugs were lysed and analyzed by immunoblotting.
Changes in the transcriptional activation of chaperone molecules were also noted after the exposure of myeloma cells to Hsp90 inhibitors, with the pattern being similar to that seen after TM and TG exposure, consistent with the generation of ER stress. Radicicol upregulated transcription of BiP from 2- to 5-fold between the 2- to 6-hour time point, whereas 17-AAG required a 24-hour incubation period to achieve a 2-fold change in BiP transcription levels (Figure 5A). No change was detected in the protein level of BiP after treatment with either drug (data not shown). Minimal increases in transcription of GRP94 were seen in response to both agents, with only 2- to 3-fold changes being detected after a 24-hour incubation period (Figure 5B). No changes in protein levels were seen over the same time period (data not shown). In addition, no changes were detected in Hsp90 protein levels after treatment with either drug. In contrast, to the effect of TG and TM, which demonstrated no effects on Hsp70 levels, increases in Hsp70 protein levels were noted after a 6- to 8-hour treatment with Hsp90 inhibitors. Hsp70 is a known mediator of cell survival,30 and protein levels were induced in a time-dependent manner in response to both agents (Figure 5C).
Bortezomib, a proteasome inhibitor, induces a similar pattern of chaperone expression to that seen with Hsp90 inhibition, with a 3-fold increase in the transcript level of BiP with stable protein levels. The transcript and protein levels of other key regulators including GRP94 and Hsp90 remained stable (Figure 5A,B). Western blot analysis after treatment with bortezomib demonstrates a clear upregulation of Hsp70 that is in contrast to the results seen after treatment with TM and TG but is similar to results seen after treatment with Hsp90 inhibitors (Figure 5C).
Activation of apoptotic pathways after the induction of the UPR in myeloma cells
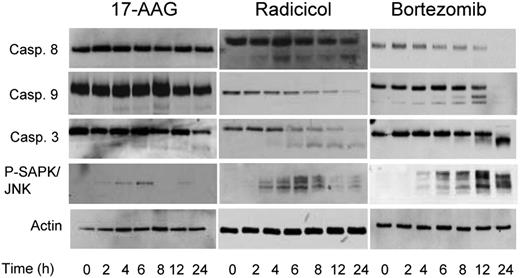
As chaperone proteins exhibit a number of important effects on cellular apoptotic pathways, treatment with Hsp90 inhibitors may modulate these responses. Treatment with TM and TG for up to 24 hours, drugs known to cause ER stress and a strong UPR response, failed to result in cleavage of caspases 8, 9, or 3 within this time frame. Hsp90 inhibition with radicicol, however, resulted in cleavage of caspases 8 and 9, whereas treatment with another Hsp90 inhibitor 17-AAG resulted in the cleavage of caspases 9 and 3 only (Figure 6). Bortezomib, in contrast, initiates the cleavage of both caspases 8 and 9, with the downstream cleavage of caspase 3 (Figure 6). Phosphorylation and activation of the stress-activated protein kinase JNK was seen within 2 to 4 hours of treatment with all the drugs tested, in keeping with the early activation of IRE1 and PERK resulting in the delivery of an apoptotic signal mediated by these pathways (Figure 6). These results suggest that differential apoptotic responses are seen with the differing Hsp90 inhibitors. In conclusion, while both Hsp90 and proteasome inhibitors caused ER stress and delivered an apoptotic signal mediated by CHOP induction and activation of the JNK pathway, they can deliver a death signal mediated via a caspase-dependent pathway, and the nature of this can differ.
Inhibitors of Hsp90 and the proteasome result in different caspase cleavage patterns. Cells were treated with the IC50 doses of each inhibitor (17-AAG, 5 μM; radicicol, 10 μM, and bortezomib, 8 nM) for a 24-hour treatment period, lysed and samples analyzed by immunoblotting. Representative cell line data on U266 are shown. All experiments were repeated in triplicate.
Inhibitors of Hsp90 and the proteasome result in different caspase cleavage patterns. Cells were treated with the IC50 doses of each inhibitor (17-AAG, 5 μM; radicicol, 10 μM, and bortezomib, 8 nM) for a 24-hour treatment period, lysed and samples analyzed by immunoblotting. Representative cell line data on U266 are shown. All experiments were repeated in triplicate.
Discussion
The main focus of this study was to investigate the impact of Hsp90 inhibition on the UPR. Our results provide evidence that Hsp90 inhibitors kill myeloma cells at least in part via the induction of ER stress. After treatment with Hsp90 inhibitors, the resultant ER stress leads to an increase in the proapoptotic molecule CHOP, with downstream activation of the JNK and intrinsic caspase pathways leading to apoptosis. In addition to such proapoptotic effects, inhibiting the action of Hsp90 results in the accumulation of misfolded proteins within cytosolic inclusions together with stimulation of the UPR, as demonstrated by the splicing of XBP1 to XBP1s and activation of ATF6. These intracellular effects are consistent with the role of molecular chaperones in promoting protein folding, which is essential for maintaining the correct structure of nascent immunoglobulins within the ER. Importantly, these effects of generating ER stress and activation of the UPR in myeloma cells are separate and additional to the previously reported mechanisms of action of Hsp90 inhibition.31,–33
The mode of action of 17-AAG has been investigated previously in myeloma, and it has been demonstrated that after drug treatment Hsp90–client protein interactions are inhibited resulting in the suppression of multiple myeloma–specific growth and survival signaling.34 Examples of this mechanism include the inhibition of the IGFR-1 and IL-6R pathways, which both induce myeloma cell death and overcome the protective effects of the bone marrow microenvironment.34 In addition to these effects, inhibition of Hsp90 can modulate the apoptotic response with many members of the intrinsic and extrinsic pathways being dependent on the Hsps for their stability and function.16
To further investigate the mechanisms by which ER stress is generated after Hsp90 inhibition, we have compared the pattern of UPR and ER stress response seen with 17-AAG and radicicol to other known ER stress inducers. TG and TM are compounds that inhibit the calcium ATPase pump and the glycosylation status of nascent proteins, respectively, resulting in a build-up of misfolded proteins within the ER, generating ER stress. We demonstrate that the effects of TG and TM seen in myeloma are similar to those seen in other cell types with activation of the 3 branches of the UPR demonstrated by splicing of XBP1 to XBP1s, the induction of CHOP, and the splicing of ATF6. Importantly, TG and TM are able to induce JNK phosphorylation quickly, resulting in myeloma cell death, whereas activation of the caspase system is much slower, occurring after the 24-hour time point.
Bortezomib, a proteasome inhibitor, had previously been thought to exert its major effect via inhibition of NF-κB–mediated pathways,35,–37 although more recent reports have shown that it can also inhibit the Ca2+ pump within the ER38 and consequently exert an effect by mediating the unfolded protein response.39 Initial reports looking at the effect of proteasome inhibition on XBP1 were generated using the myeloma cell line J558 that has a high basal level of XBP1s expression.40 In these experiments at the mRNA level, no splicing of XBP1 was demonstrated after drug exposure; however, at the protein level there was accumulation of XBP1 protein with a corresponding decrease in XBP1s. The authors suggested that XBP1 is ubiquitinated and targeted for proteasome degradation, and that inhibition of the proteasome leads to the accumulation of XBP1, which acts as a dominant-negative inhibitor of XBP1s.40 They suggested that proteasome inhibition may suppress the IRE1-mediated XBP1 mRNA splicing by blocking the phosphorylation and therefore activation of IRE1. The consequence of all of these events is that proteasome inhibition results in a heightened ER stress response, blocking one of the arms of the UPR focused on adaptation and mediating an anti-apoptotic response. More recent work performed by Obeng et al39 confirms that proteasome inhibitors induce the UPR in myeloma cells, with evidence of eIF-2a phosphorylation, activation of ATF4, and induction of CHOP. Our findings on the effect of proteasome inhibition on the UPR confirm and extend this work. We demonstrate that after treatment with bortezomib, protein accumulates because of the inhibition of the cytosolic 26S proteasome. This results in the upregulation of EDEM1, a member of the ER-associated degradation pathway, increasing the retrograde translocation of proteins from the ER into the cytoplasm, resulting in the formation of cytosolic inclusions containing immunoglobulin. In addition, the proapoptotic components of the UPR and the ER stress pathway are induced with an increase in CHOP, phosphorylation of JNK, and cleavage of caspases 8 and 9. However, survival and adaptive pathways mediated by the splicing of XBP1 to XBP1s are not activated after proteasome inhibition, with the net effect of enhanced cell death.
In contrast to the effects seen after proteasome inhibition, treatment of myeloma cells with Hsp90 inhibitors results in a more classical pattern of activation of the UPR similar to that seen with the typical ER stress inducers TG and TM. Importantly, all 3 branches of the UPR are induced, with clear evidence of IRE1 activation with associated splicing of XBP1 to XBP1s. This upregulation of XBP1s results in the induction of XBP1 target genes directing cellular efforts to the transcription and translation of molecular chaperones able to promote protein folding and turning off nonessential protein translation, with the net result being allowing the cell to resist stress. However, the effect of Hsp90 inhibition on splicing is transient, differing significantly from that seen with TM/TG. In addition, there is a time-dependent downregulation of XBP1 that may enhance the pro-apoptotic effects of these agents. Hsp90 inhibition also induces a number of apoptotic pathways, including CHOP induction, activation of JNK, and caspases 8 and/or 9 cleavage, all contributing to cell death.
Interestingly, after treatment with Hsp90 inhibitors, cytosolic inclusions containing immunoglobulin are produced. These were also seen after treatment with bortezomib and to a lesser extent after exposure to the classical ER stress–inducer thapsigargin, suggesting that an ER stress signal may contribute to their generation. A previous study of breast cancer cells has demonstrated the presence of vacuoles within the cytoplasm after treatment with a combination of both Hsp90 inhibitors and proteasome inhibitors. In that work, the presence of vacuoles was thought to represent a cellular defense mechanism alleviating stress caused by the build-up of misfolded proteins within the ER by removing misfolded proteins within the cell to an alternate location where they are less damaging.41 The inclusions we describe in myeloma cells are different to those seen in breast cancer cells because they occur after exposure to single-agent Hsp90 or proteasome inhibitors rather than to combinations. They differ morphologically, being smaller, discrete, and evenly distributed throughout the cytoplasm. The time course of their appearance is different and they do not increase in size or coalesce over time. The presence of another kind of intracellular inclusion, an aggresome, has been described after treatment with proteasome inhibitors in pancreatic cell lines42 and after treatment with HDAC6 and proteasome inhibitors in myeloma cell lines.43 However, the characteristics of aggresome formation are different from those described here, because classically aggresomes form at the cell periphery and move toward the center of the cell and coalesce. We demonstrate that the inclusions described after Hsp90 inhibition contain immunoglobulin light chain. The spherical shape we describe is characteristic of the build-up of secretory light chain and consistent with the well-described plasma cell morphologic feature of Russell bodies.44,45
BiP and grp94 are 2 Hsp homologs resident within the ER, and the effect of Hsp90 inhibitors on these molecules has not been fully considered previously. BiP is a Hsp70 homolog that ensures the correct folding of IRE1, PERK, and ATF6, and holds them in their inactive state. In addition, BiP binds to the immunoglobulin heavy chains that have not yet associated with immunoglobulin light chains and assists in immunoglobulin assembly.46 BiP and grp94, an ER resident Hsp90 homologue, are also important for stabilizing protein folding intermediates, ensuring immunoglobulin light chain folding and targeting unassembled subunits for degradation.47,–49 Our results demonstrate that Hsp90 inhibition results in an induction of an unfolded protein response with increases in BiP and grp94 mRNA levels, similar to that seen with TG and TM consistent with Hsp90 inhibition exerting effects on the unfolded immunoglobulins in the ER. Particularly the activation of IRE1 and ATF6 results in an increase in transcription of XBP1 target genes, ensuring an increase in the molecular chaperone proteins required for correct protein folding.
A well-described association with Hsp90 inhibition is the induction of Hsp70.50 The mechanism for this is partially understood because treatment with 17-AAG disrupts the association between Hsp90 and heat shock factor-1, resulting in an increase in the transcriptional activity of heat shock factor-1, mediated by its binding to the heat shock elements in the promoter of the Hsp70 gene, leading to the induction of Hsp70.51,–53 Other known stressors such as heat damage or reactive oxygen species have also been shown to induce Hsp70 levels via heat shock factor-1.54 Myeloma treatment with Hsp90 inhibitors and proteasome inhibitors initiates this induction of Hsp70, whereas TM and TG, known ER stressors, do not alter the levels of Hsp70 after treatment. These data suggest that treatment with Hsp90 inhibitors results in the induction of the heat shock response as well as the UPR, whereas treatment with TM and TG results only in the activation of the UPR. This induction of Hsp70 may act to prevent cell death induced by Hsp90 inhibition and as such may suggest the use of combinations of Hsp90 inhibitors with Hsp70 inhibitors to increase the therapeutic potential.
Designing strategies to manipulate the UPR therapeutically will most likely require a combination of drugs. Data from this present study can inform decisions about which additional compounds can be added to Hsp90 inhibitors to maximize their apoptotic effects. Ideally, the pathways inhibited by the individual drugs should be complementary to ensure synergistic/additive effects. The AKT pathway is one of the principal survival pathways activated after induction of the UPR, and its pro-survival effects have been proposed as a mediator of resistance to Hsp90 inhibitors. This suggests that a combination of inhibitors of AKT and Hsp90 may have additive effects in myeloma cells. In other cell systems, the combination of AKT inhibitors and Hsp90 inhibitors has been shown to be synergistic, supporting this hypothesis.55 We also demonstrate that both proteasome and Hsp90 inhibition results in the potent induction of Hsp70. This is in keeping with other studies demonstrating an upregulation of Hsp27, Hsp70, and Hsp72 after treatment with these drugs.56,57 These proteins have strong anti-apoptotic potential and potentially counteract the pro-apoptotic effects of the drugs, leading to the development of resistance.57 Therefore, the addition of compounds that are able to inhibit the induction of heat shock proteins may be beneficial. Hsp90 inhibition represents a potentially important strategy for the treatment of myeloma, taking advantage of the plasma cells' capacity for immunoglobulin production, to target therapy. Our data support the rapid introduction of these inhibitors into the clinical setting for the treatment of myeloma, as well as for other tumor types characterized by excess protein production, where they may have additional unique modes of action (Figure S2).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Alex Javed for his technical expertise with the immunoglobulin light chain staining. This work was funded by the Department of Health and Kay Kendall Leukaemia Fund.
Authorship
E.L.D. performed research, analyzed data, and wrote the paper. H.M., S.Y.S., and A.D. performed research and analyzed data. P.W., G.J.M., and F.E.D. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Faith Davies, Brookes Lawley Building, Institute of Cancer Research, 15 Cotswold Road, Sutton, Surrey, SM2 5NG, UK; email:faith.davies@icr.ac.uk.




![Figure 4. Cellular effects of ER stress induced by TM and TG, Hsp90, and proteasome inhibitors. (A) Transcriptional upregulation of EDEM-1, known to accelerate the degradation of misfolded proteins, demonstrated by real-time Taqman PCR. (B) The extent of cytoplasmic inclusions exhibited by U266 in response to treatments (24 hours, IC50 doses: TM, 100 μM; TG, 10 μM; 17-AGG, 5 μM; Bortezomib, 8 nM) was determined using May-Grunwald-Giemsa (MGG) staining and light microscopy (Olympus BH-2 microscope [Olympus, London, United Kingdom] 100×/1.40 oil; images acquired using JVC KY-F1030 digital camera and JVC Scan Rate Converter [Victor, Tokyo, Japan]). (C) Immunoglobulin light chain staining demonstrates a build-up of λ light chain within the inclusions. Representative cell line data on U266 are shown. All experiments were repeated in triplicate.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/7/10.1182_blood-2006-11-053728/2/m_zh80180706510004.jpeg?Expires=1769173152&Signature=dlBlyErcL8rAPE8QIF4TE6oBOJ~-5Y5Y68YNLaQ0lR5sh5cUd2uzQ99Wyc2gum0s7rxqie05OWU3~lnocaMrvXWzCDrpz91thhOSKMC6LJmWaVOcu2dKwECSvXeiulH~OQvQF4Bgjhk7-dmddC4f0rBZ4nNE6rgVaQHejY3Wd8fY2dvmvuW1C67gafsESR0hXhUMr0jaj24gbl1np-Ia1k91PhRzhW6ltlLBhv1CKSXeq2aTWaOpOaG3vfSz1IEmZjKZeOEWduf~q7jesQyq3HBBc7MB6gDCcgRHfjINiHxLMXW8YMMw6i~6ccBYGJ39q2kcBXJcsMZodYglcOcQ9w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal