Abstract
Signal transducer and activator of transcription 3 (Stat3) is implicated in the pathogenesis of many malignancies and essential for IL-6–dependent survival and growth of multiple myeloma cells. Here, we demonstrate that the gene encoding oncogenic microRNA-21 (miR-21) is controlled by an upstream enhancer containing 2 Stat3 binding sites strictly conserved since the first observed evolutionary appearance of miR-21 and Stat3. MiR-21 induction by IL-6 was strictly Stat3 dependent. Ectopically raising miR-21 expression in myeloma cells in the absence of IL-6 significantly reduced their apoptosis levels. These data provide strong evidence that miR-21 induction contributes to the oncogenic potential of Stat3.
Introduction
Signal transducer and activator of transcription 3 (Stat3), the major mediator of IL-6 signaling,1,2 participates in cellular transformation and oncogenesis.3,4 Its activation is essential for the transforming potential of many oncogenes. Previously, we could show that Stat3 activation is required for IL-6–dependent growth and survival of the human myeloma cell lines INA-6 and XG-1.5 Although microarray analyses revealed that in these cells the vast majority of IL-6 target genes is controlled through Stat3, differential expression of known apoptosis regulators (eg, members of the Bcl-2 family) could not sufficiently account for the observed Stat3-mediated survival effect.5 We therefore searched for additional mechanisms through which Stat3 may suppress apoptosis in cancer cells.
MicroRNAs (miRNAs) are an abundant class of short non-protein-coding RNAs (ncRNAs) mediating posttranscriptional down-regulation of target genes. They have been shown to play a fundamental role in diverse physiologic and pathologic processes, including proliferation, apoptosis, differentiation, and cell fate determination.6,7 Recent evidence indicates that miRNAs can function as tumor suppressors and oncogenes.8 Hence, we reasoned that miRNAs might be implicated in the Stat3-mediated survival of myeloma cells.
Materials and methods
A detailed description of the bioinformatic procedures and the vectors and primer sequences used is given in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
INA-6 and XG-1 myeloma and HepG2 hepatocellular carcinoma cells were cultivated, and reporter gene assays were performed as recently described.5,9
Chromatin immunoprecipitation was performed using a kit (EZ ChIP) from Upstate (Lake Placid, NY) according to the manufacturer's instructions. Coimmunoprecipitated DNA was amplified using primers specific for the miR-21 upstream region and analyzed on an agarose gel. Anti-Stat3 serum (PA-ST3) was from R&D Systems (Wiesbaden, Germany).
RNA interference knock-down of Stat3 and overexpression of miR-21 in myeloma cells was performed by electroporation with pSUPER vectors as described.9 In short, 1 μg green fluorescent protein expression vector pEGFP-N1 (Clontech, Mountain View, CA) and 19 μg expression vectors encoding either a small hairpin RNA targeting Stat3 mRNA (pSUPER-siStat3)9 or precursor miR-21 (pSUPER-miR-21), or a pSUPER control vector were electroporated into 5 × 106 cells. For each experiment, 3 × 107 cells were combined and successfully transfected, and green fluorescent cells were sorted 48 hours after transfection using a FACS-Vantage Sorter (BD Biosciences, Heidelberg, Germany).
For real-time polymerase chain reaction (PCR) of Stat3 mRNA and primary miR-21 transcripts (pri-miR-21), total RNA was prepared using the mirVana miRNA Isolation Kit (Ambion, Austin, TX) and quantified by the RNA 6000 LabChip Kit (2100 Bioanalyzer; Agilent Technologies, Waldbronn, Germany). Reverse transcription was done with the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, Karlsruhe, Germany). Real-time PCR was performed using a LightCycler (Roche, Mannheim, Germany). An optimal PCR reaction was established with the FastStart DNA Master SYBR Green I Kit (Roche), and it was taken care that only 1 PCR product was generated. Specificity of amplification was validated by confirming the identity of PCR products by sequencing. Mature miR-21 was detected by stem-loop reverse transcription followed by real-time PCR.10 Signals were normalized to values obtained for glyceraldehyde-3-phosphate dehydrogenase.
Apoptosis was detected by flow cytometry using annexin-V staining as previously described.5
Results and discussion
To investigate whether Stat3 is involved in the regulation of miRNA genes, we analyzed the genomic distribution of Stat3 binding sites and their correlation with known miRNAs by phylogenetic footprinting (for details, see Document S1). Among the miRNA genes associated with evolutionarily conserved putative Stat3 binding sites, miR-21 has been described as an oncogenic miRNA exhibiting antiapoptotic activity in various carcinomas.11–13 The putative regulatory region of the miR-21 gene is located within an intron of the overlapping transmembrane protein 49 (TMEM49) gene, and contains 2 consensus Stat3 binding sites approximately 800 bp upstream of the transcription start site. It is evident from Figure 1A that the Stat3 sites and the surrounding region, spanning 300 bp, are highly conserved in vertebrates at least since the common ancestor of tetrapods and actinopterygian fish (ie, since the first evolutionary appearance of miR-21 and Stat3 in species for which genome-wide data are available).
A highly conserved enhancer is responsible for the Stat3-mediated responsiveness of the miR-21 promoter to IL-6. (A) 130-bp regions containing 2 predicted Stat3 binding sites upstream of the miR-21 genes of various vertebrate species are aligned. T, G, C, and A nucleotides are colored green, orange, blue, and red, respectively. Stat3 sites are highlighted by yellow boxes. *Sequences from organisms where no genomic sequence was available were obtained from the Ensembl trace repository (Table S2). (B) XG-1 cells were deprived of IL-6 for 72 hours or restimulated for 30 minutes and subjected to a chromatin immunoprecipitation (ChIP) assay using anti-Stat3 or IgG isotype control. Coimmunoprecipitated DNA was amplified by PCR with primers specific for the miR-21 upstream enhancer. (C) Reporter gene assays were performed in HepG2 cells transfected either with a luciferase vector driven by the miR-21 promoter/enhancer alone (−) or in the presence of a vector encoding a small hairpin RNA silencing Stat3 expression (siStat3), or with a miR-21 promoter/enhancer construct containing point mutations in both Stat3 sites (ΔStat3). Values represent the mean luciferase activities ± SD of 3 independent experiments relative to samples from unmanipulated IL-6–treated cells.
A highly conserved enhancer is responsible for the Stat3-mediated responsiveness of the miR-21 promoter to IL-6. (A) 130-bp regions containing 2 predicted Stat3 binding sites upstream of the miR-21 genes of various vertebrate species are aligned. T, G, C, and A nucleotides are colored green, orange, blue, and red, respectively. Stat3 sites are highlighted by yellow boxes. *Sequences from organisms where no genomic sequence was available were obtained from the Ensembl trace repository (Table S2). (B) XG-1 cells were deprived of IL-6 for 72 hours or restimulated for 30 minutes and subjected to a chromatin immunoprecipitation (ChIP) assay using anti-Stat3 or IgG isotype control. Coimmunoprecipitated DNA was amplified by PCR with primers specific for the miR-21 upstream enhancer. (C) Reporter gene assays were performed in HepG2 cells transfected either with a luciferase vector driven by the miR-21 promoter/enhancer alone (−) or in the presence of a vector encoding a small hairpin RNA silencing Stat3 expression (siStat3), or with a miR-21 promoter/enhancer construct containing point mutations in both Stat3 sites (ΔStat3). Values represent the mean luciferase activities ± SD of 3 independent experiments relative to samples from unmanipulated IL-6–treated cells.
Genomic data suggest that miR-21 and its associated Stat3 binding sites are functionally independent of TMEM49 (Table S1). A stable distance between miR-21 and the Stat3 sites throughout all vertebrates strongly suggests their functional correlation (Figure S1).
Chromatin immunoprecipitation revealed that Stat3 is indeed recruited to the miR-21 upstream region in XG-1 human myeloma cells in response to IL-6 (Figure 1B). Furthermore, a genomic DNA fragment extending from −1120 to +25 bp relative to the miR-21 transcription start site14 and containing the putative Stat3 enhancer region proved to be IL-6–responsive in a reporter assay (Figure 1C). IL-6–induced miR-21 promoter activity was abolished when the Stat3 motifs were eliminated by site-directed mutagenesis. Likewise, knocking down Stat3 by RNA interference inhibited miR-21 promoter activity. The effects of Stat3 knockdown could be rescued by expression of an siRNA-resistant variant of Stat3 (Figure S2). These data demonstrate the existence of a functional Stat3-regulated enhancer upstream of the miR-21 gene, as predicted by the bioinformatic analysis.
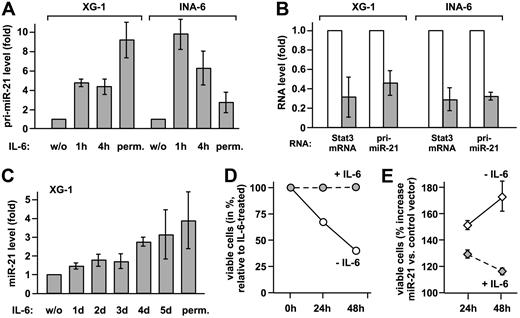
To verify that IL-6 induces the transcription of the miR-21 gene, pri-miR-21 and mature miR-21 were quantified by real-time PCR in XG-1 and INA-6 myeloma cells. IL-6 increased pri-miR-21 expression substantially in either cell line (Figure 2A). Upon Stat3 knockdown, comparable reductions of Stat3 mRNA and pri-miR-21 levels were observed (Figure 2B), proving that miR-21 induction by IL-6 is mediated by Stat3. Induction of miR-21 expression by IL-6 is not restricted to multiple myeloma, as it is observed in other tumor cell lines as well (Figure S3). Taken together, our data demonstrate that miR-21 gene transcription is controlled by IL-6 and requires Stat3. In contrast to the rapid induction of pri-miR-21, mature miR-21 levels raised slowly in XG-1 cells, eventually reaching a 4-fold increase (Figure 2C). The same was observed in prostate carcinoma cells (Figure S3), indicating a rather slow processing rate of pri-miR-21.
miR-21 is induced by IL-6 via Stat3 activation and promotes survival of myeloma cells. (A) XG-1 and INA-6 cells were either restimulated with IL-6 for the times indicated after cytokine withdrawal for 72 and 12 hours, respectively, or continuously cultured with IL-6 (perm.). Levels of pri-miR-21 were determined by real-time PCR. Values obtained for cells deprived of IL-6 were set to 1. (B) XG-1 and INA-6 cells cultured in the presence of IL-6 were transiently transfected with an expression plasmid for a small hairpin RNA-silencing Stat3 (▩) or a scrambled sequence RNA (□) together with a vector-encoding enhanced GFP. Successful down-regulation of Stat3 protein levels under these conditions has been demonstrated by us previously.9 After 48 hours, green fluorescent cells were sorted, and Stat3 and pri-miR-21 transcript levels were determined by real-time PCR. Values for the control samples were taken as reference. (C) In XG-1 cells treated as were INA-6 cells, mature miR-21 was quantified by stem-loop reverse transcription followed by real-time PCR.10 (D,E) INA-6 cells were transiently transfected by electroporation with a control (D) or a miR-21 expression vector (E). An expression plasmid encoding enhanced GFP was cotransfected. At 1 day after transfection, cell culture was continued either in the presence or absence of IL-6. After an additional 24 or 48 hours, apoptosis was studied by flow cytometric annexin-V assay, with the transfected cells gated on the basis of green fluorescence. In panel D, the reduced levels of viable (annexin-V−) cells after cytokine withdrawal are represented relative to those observed in IL-6–treated cells. Panel E shows the relative percentage of viable cells with miR-21 expression vector relative to control vector transfection. Data represent mean values plus or minus SD from 3 independent experiments.
miR-21 is induced by IL-6 via Stat3 activation and promotes survival of myeloma cells. (A) XG-1 and INA-6 cells were either restimulated with IL-6 for the times indicated after cytokine withdrawal for 72 and 12 hours, respectively, or continuously cultured with IL-6 (perm.). Levels of pri-miR-21 were determined by real-time PCR. Values obtained for cells deprived of IL-6 were set to 1. (B) XG-1 and INA-6 cells cultured in the presence of IL-6 were transiently transfected with an expression plasmid for a small hairpin RNA-silencing Stat3 (▩) or a scrambled sequence RNA (□) together with a vector-encoding enhanced GFP. Successful down-regulation of Stat3 protein levels under these conditions has been demonstrated by us previously.9 After 48 hours, green fluorescent cells were sorted, and Stat3 and pri-miR-21 transcript levels were determined by real-time PCR. Values for the control samples were taken as reference. (C) In XG-1 cells treated as were INA-6 cells, mature miR-21 was quantified by stem-loop reverse transcription followed by real-time PCR.10 (D,E) INA-6 cells were transiently transfected by electroporation with a control (D) or a miR-21 expression vector (E). An expression plasmid encoding enhanced GFP was cotransfected. At 1 day after transfection, cell culture was continued either in the presence or absence of IL-6. After an additional 24 or 48 hours, apoptosis was studied by flow cytometric annexin-V assay, with the transfected cells gated on the basis of green fluorescence. In panel D, the reduced levels of viable (annexin-V−) cells after cytokine withdrawal are represented relative to those observed in IL-6–treated cells. Panel E shows the relative percentage of viable cells with miR-21 expression vector relative to control vector transfection. Data represent mean values plus or minus SD from 3 independent experiments.
Since miR-21 is involved in the control of tumor cell apoptosis,11–13 we reasoned that it might contribute to the Stat3-dependent survival of myeloma cells. Due to relatively high basal miR-21 expression levels, a miR-21 knockdown might cause effects that go beyond Stat3 regulation. Therefore, we chose to ectopically elevate miR-21 expression in the absence of IL-6 to levels comparable with those observed in response to Stat3 activation. Upon electroporation of INA-6 cells with a miR-21 expression vector and sorting of the transfected cells, we determined a 2.4-fold increase in mature miR-21 expression over endogenous levels by real-time PCR analysis (mean of 3 independent experiments). As reported earlier,5 IL-6 withdrawal rapidly induced apoptosis in INA-6 cells (Figure 2D). Raising miR-21 expression, however, yielded significantly higher levels of nonapoptotic cells (Figure 2E). This effect was most dramatic in the absence of IL-6, reducing the cytokine dependency of the INA-6 cells.
Equivalent observations were made in XG-1 cells (data not shown). In summary, these data provide strong evidence that induction of miR-21 contributes to the antiapoptotic function of Stat3.
Stat3 controls the expression of a number of antiapoptotic proteins on the transcriptional level, including survivin, Bcl-2, and Mcl-1,15–18 thereby providing an important fast-track survival signal for myeloma and presumably other tumor cells. However, recent data suggest that this regulatory route does not sufficiently explain the full antiapoptotic potential of Stat3.5,19 As demonstrated here, miR-21 induction by Stat3 represents a rather slow-acting yet long-lasting survival stimulus, which appears as an ideal complementation of the immediate induction of antiapoptotic proteins. Overexpression of miR-21 has been reported for numerous cancers,11,20,21 all of which contain constitutively activated Stat3 or even rely on Stat3 with respect to cell survival or growth.11,12,15,16,22 Taken together, our observations strongly suggest a pivotal relevance of miR-21 for the oncogenic potential of Stat3 and thereby for its involvement in the pathogenesis of multiple myeloma and other malignancies.
Since miRNAs function as negative regulators of the stability and/or translation of specific target mRNAs, an important step to unravel the molecular mechanism of miR-21 action will be to identify its mRNA targets. Recently, miR-21 was reported to repress the expression of the tumor suppressor PTEN.12 Yet, we did not detect up-regulation of PTEN expression or AKT phosphorylation in INA-6 and XG-1 cells in response to IL-6 (data not shown).
Stat3 is regarded a promising target for tumor therapy.23–25 However, its involvement in the control of numerous cellular processes may cause adverse side effects of such treatments. The identification of miR-21 as a Stat3 target gene may provide an even more selective and promising target for future therapeutic strategies.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Stefan Rose-John (University of Kiel) for recombinant IL-6, Reuven Agami (The Netherlands Cancer Institute, Amsterdam) for the pSuper vector, and Viola Döbel (Fluorescence Technologies Core Unit of the IZKF Leipzig) for cell sorting.
This work was supported in part by the Deutsche Forschungsgemeinschaft, grant nos. SFB610, project C2 (to D.L., C.B., H.C., and F.H.) and BIZ 6-1/3 (to C.F and P.F.S).
Authorship
Contribution: F.H., K.B.-H., J.H., A.K.K., M.G., and P.F.S designed research; D.L., K.B.-H., G.P., C.F., C.B., K.B., and H.C. performed research; R.B. and M.G. contributed vital new reagents or analytical tools; A.K.U. analyzed data; and F.H., K.B.-H., and P.F.S wrote the paper. D.L. and K.B.-H. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Friedemann Horn, Institute of Clinical Immunology and Transfusion Medicine, University of Leipzig, Johannisallee 30, 04103 Leipzig, Germany; e-mail: friedemann.horn@medizin.uni-leipzig.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal