Abstract
Toll-like receptor 4 (TLR4) initiates both myeloid differentiation factor 88 (MyD88)-dependent and Toll/interleukin (IL)-1R domain–containing adapter, inducing interferon (IFN)-β–dependent signaling, leading to production of proinflammatory mediators and type I interferon (IFN) to eliminate pathogens. However, uncontrolled TLR4 activation may contribute to pathogenesis of autoimmune and inflammatory diseases. TLR4 is transported from the plasma membrane to the endosome for ubiqutination and to the lysosome for degradation, and downregulation of TLR4 expression or promotion of TLR4 degradation are important ways for negative regulation of TLR4 signaling. We previously identified a lysosome-associated small guanosine triphosphatase (GTPase) Rab7b that may be involved in lysosomal trafficking and degradation of proteins. Here we demonstrate that Rab7b can negatively regulate lipopolysaccharide (LPS)-induced production of tumor necrosis factor (TNF)-α, IL-6, nitric oxide, and IFN-β, and potentiate LPS-induced activation of mitogen-activated protein kinase, nuclear factor κB, and IFN regulatory factor 3 signaling pathways in macrophages by promoting the degradation of TLR4. Rab7b is localized in LAMP-1–positive subcellular compartments and colocalized with TLR4 after LPS treatment and can decrease the protein level of TLR4. Our findings suggest that Rab7b is a negative regulator of TLR4 signaling, potentially by promoting the translocation of TLR4 into lysosomes for degradation.
Introduction
Toll-like receptors (TLRs) play a critical role in innate immunity by recognizing structurally conserved bacterial and viral components termed pathogen-associated molecular patterns.1-3 TLRs activate signaling through the Toll/IL-1R (TIR) domain found in the cytoplasmic tails of these proteins, which in turn triggers the binding of the adaptor protein myeloid differentiation factor 88 (MyD88) to the TIR domain, allowing for interaction and phosphorylation of IL-1R–associated kinases and subsequent activation of tumor necrosis factor receptor (TNF)-associated factor 6 (TRAF-6), leading to the activation of nuclear factor κB (NF-κB) and mitogen-activated protein kinase (MAPK) pathways and the induction of proinflammatory cytokines.1-3 However, recent studies have identified molecules involved in the MyD88-independent signaling for TLR3 and TLR4, including the TIR domain-containing adaptor protein (TIRAP/MAL), as a second adaptor instead of MyD88 for activation of NF-κB and MAPK,4,5 and the TIR domain-containing adapter inducing interferon (IFN)-β (TRIF) as a critical MyD88-independent adaptor used by TLR3 and TLR4 to regulate type I IFN production.6,7 The signaling receptor for lipopolysaccharide (LPS) is TLR4/MD-2, which receives LPS from CD14. LPS delivered by CD14 to TLR4/MD-2 initiates a signaling cascade through the TIR domain containing adaptors MyD88, TIRAP/MAL, TRIF, and TRAM (TRIF-related adaptor molecule), which eventually leads to activation of MAPK (including extracellular signal-regulated kinase [ERK], c-Jun N-terminal kinase [JNK], and p38), NF-κB, and IFN regulatory factor 3 (IRF-3), resulting in the production of proinflammatory mediators such as TNF-α, IL-6, nitric oxide (NO), and IFN-β.1-3
TLR activation is a double-edged sword.1-3 Members of the TLR family are involved in the pathogenesis of autoimmune, chronic inflammatory, and infectious diseases.1-3 Prolonged or excessive production of LPS-induced mediators may result in the lethal disease known as septic shock. Scientists are therefore investigating how to negatively regulate TLR4 signaling and have described more than 10 negative regulators of TLR4 pathways.3 A20 and Triad3A regulate TLR4 ubiquitination and proteolytic degradation,8-10 whereas the TIR-containing proteins ST2 and single immunoglobulin IL-1 receptor-related also act as negative regulators of signaling by sequestering MyD88 via homotypic TIR–TIR interactions.11,12 The kinase-inactive IL-1R–associated kinase M, the splice variant of MyD88 (MyD88s), and the TLR homolog RP105 can serve as competitors for the TLR4 signaling machinery.13-15 A negative regulator for Janus kinase-signal transducer and activator of transcription (JAK-STAT) signaling, suppressor of cytokine-signaling-1 (SOCS-1), has also been demonstrated as regulating TLR4 signaling by interacting with and promoting the degradation of TIRAP/MAL.16-18 Other machineries involved in negative regulation of TLR4 have also been described for a protein associated with TLR4 (PRAT4A), TNF-related apoptosis-inducing ligand receptor (TRAIL-R), and FLN29/TRAFD1.19-21 Our previous studies show that Src homology 2 domain-containing inositol-5-phosphatase 1 (SHIP-1) is a potent phosphoinositide-3 kinase (PI-3K)-independent negative regulator of TLR4 signaling,22 and protein tyrosine phosphatase 2 (SHP-2) is a negative regulator of TRIF-dependent TLR signaling by directly binding and dephosphorylating tumor necrosis factor receptor-associated factor (TRAF)-associated activator of NF-kB (TANK)-binding kinase 1 (TBK1).23 Given the complexity of the negative regulation network, identifying novel regulatory proteins may contribute to a better understanding of the control and mechanisms of TLR signaling.
Rab (Ras related in brain) proteins are small guanosine triphosphatases (GTPases) belonging to the Ras superfamily that regulate vesicular formation, movement, and fusion processes.24,25 Despite the known functions for Rab's in membrane trafficking, it is emerging that Rab's are also involved in signal transduction by regulating the membrane trafficking of several hormone, cytokine, and chemokine receptors.26 For example, Rab5 has been shown to play a role in epidermal growth factor receptor-mediated signal transduction.27,28 Recently it has been suggested that Rab7, a small GTPase that is localized to late endosomes and regulates the later stages of the endocytic pathway, is involved in trafficking and lysosomal degradation of several kinds of receptors,29-32 such as the angiotensin II type 1A receptor, epidermal growth factor receptor, and the nerve growth receptor TrkA.33-35 It has been revealed that TLR4 can be localized in subcellular compartments such as the plasma membrane, the Golgi apparatus, endosomes, and lysosomes,36-40 which may play crucial roles in the temporal and spatial regulation of TLR4 signaling. We previously have identified a small GTPase homologous to Rab7, namely Rab7b, which is localized to lysosome-associated compartments and is selectively expressed in monocytic cells.41 Our preliminary investigations demonstrated that Rab7b can regulate LPS-induced cytokine production in macrophages, suggesting that Rab7b may be involved in the control of TLR4 signaling. Therefore, herein we investigated the roles of Rab7b in LPS-initiated TLR4 signaling in macrophages. We found that Rab7b could serve as a negative regulator of TLR4 signaling in macrophages by accelerating lysosomal degradation of TLR4 and decreasing the plasma membrane TLR4 expression level, thus providing the first evidence for a role of Rab proteins in TLRs signaling.
Materials and methods
Mice and reagents
C57BL/6 mice (5-6 weeks old) were purchased from SIPPR-BK Experimental Animal (Shanghai, China). Experiments and animal care were performed in accordance with the guidelines of Second Military Medical University. LPS derived from Escherichia coli 0111:B4 was obtained from Sigma (St Louis, MO) and repurified as described previously.22,23 Antibodies (Abs) specific for total and phosphorylated forms of ERK1/2 (Thr202/Tyr204), JNK1/2 (Thr183/Tyr185), p38 (Thr180/Tyr182), interferon regulatory factor (IRF)-3 (Ser396), and IκBα (Ser32/36), and Abs against TLR4 and hemagglutinin epitope (HA) tag were obtained from Cell Signaling Technology (Beverly, MA). Abs against β-actin, transferrin receptor (TfR/CD71), early endosome antigen 1 (EEA-1), LAMP-1, trans Golgi network 38 (TGN38), cathepsin D (Cath D), Lamin B, hexa-His tag, and horseradish peroxidase-coupled secondary antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). The enzyme-linked immunoassay kits for IL-6 and TNF-α were from R&D Systems (Minneapolis, MN), and the kit for IFN-β was from Biosource (Carlsbad, CA). The pGL3.5X-B-luciferase plasmid was a kind gift from S. J. Martin (Trinity College, Dublin, Ireland),42 and the pRL-TK-Renilla-luciferase plasmid was obtained from Promega (Madison, WI). IRF3 reporter plasmids were kind gifts from Dr T. Fujita (Tokyo Metropolitan Institute of Medical Science, Japan).43 Fluorescent dyes and fluorescent Abs used in confocal microscopy were from Molecular Probes (Eugene, OR).
Construction of expression plasmids
The recombinant vector encoding mouse Rab7b (mRab7b, GenBank Accession number NM_145509) was constructed by polymerase chain reaction (PCR)-based amplification, and then subcloned into the pcDNA3.1 eukaryotic expression vector (Invitrogen, San Diego, CA). All expression vectors were constructed via PCR amplification. Vectors encoding hexa-His–tagged or green fluorescent protein (GFP)-tagged Rab7b (termed as His-Rab7b and GFP-Rab7b, respectively), as well as vectors without tags, were constructed and prepared as described previously.41 Tags were placed at the N-terminal of Rab7b cDNA to avoid interfering with Rab7b localization. Mutant forms of mRab7b, namely mRab7bT22N (mutation of Thr22 into Asn, GTP-binding deficiency) and mRab7bΔCC (truncation of the C-terminal 2 cysteines), were prepared using Mutanbest Kit according to manufacturer's instructions (TaKaRa, Shiga, Japan). All the clones were confirmed by DNA sequencing. Primer sequences used in cloning are available on request.
Cell preparation, cell culture, and transfection
C57BL/6J mice were used for the preparation of primary mouse macrophages.22,23 Mouse macrophage cell line RAW 264.7 was obtained from the American Type Culture Collection (Manassas, VA) and cultured as described previously.22,23 Transfections using Lipofactamine 2000 (Invitrogen) were performed as described.22,23 Stable cell lines overexpressing Rab7b and its mutants were selected in 600 μg/mL G418 for 3-4 weeks.
Reverse-transcription PCR and quantitative PCR
Total cellular RNA was extracted using Trizol reagent (Invitrogen). Reverse-transcription PCR (RT-PCR) was performed as described previously.22,23 Specific primers used for RT-PCR assays were 5′-CGAGGAATACCAGACCACACT-3′ (sense) and 5′-GGCTGGCCAGAACCTCAAAGG-3′ (antisense) for mRab7b, and 5′-AGTGTGACGTTGACATCCGT-3′ (sense) and 5′-GCAGCTCAGTAACAGTCCGC-3′ (antisense) for β-actin. Quantitative PCR was performed on a MJR Chromo4 Continuous Fluorescence detector (Bio-Rad, Hercules, CA) according to the manufacturer's protocol and as described.22,23
RNA interference assay
For transient transfection, 21-nucleotide sequences of mRab7b siRNA were synthesized as follows: 5′-AGGUUCCGAUGGCUGUAUCTT-3′ (sense), and 5′-GAUACAGCCAUCGGAACCUTT-3′ (antisense). The sequence 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense), and 5′-ACGUGACACGUUCGGAGAATT-3′ (antisense) were used as a scrambled RNA interference control. SiRNA duplexes were transfected into peritoneal macrophages using Genesilencer Transfection Reagent (Genlantis, San Diego, CA) according to the standard protocol. For stable knockdown of mRab7b, an expression vector (psilencer-U6 neo; Ambion, Austin, TX) with insertion of the specific siRNA duplexes of mRab7b or the scrambled siRNA duplexes were transfected into Raw 264.7 cells.
Measurement of cytokines and NO production
Peritoneal macrophages (1.5 × 105/mL) or Raw 264.7 cells (1 × 105/mL) were stimulated with 100 ng/mL LPS for indicated time periods. IL-6, TNF-α, and IFN-β concentrations in culture supernatants were measured by enzyme-linked immunoassay. NO was detected by the Griess reaction as described previously.44
Luciferase reporter assay
Flow cytometric analysis
To detect cell surface expression of TLR4, cells were incubated with phycoerythrin-labeled antibody against mouse TLR4 (eBioscience, San Diego, CA) or mouse IgG2a for 30 minutes on ice, washed, and analyzed by using a FACScalibur flow cytometer (Becton Dickinson, San Jose, CA) as described.45 For analysis of TLR4 internalization after LPS stimulation, Raw 264.7 cells were treated with or without 100 ng/mL LPS for 5 to 30 minutes, and immediately cooled on ice. The remaining TLR4 on plasma membrane was evaluated by fluorescence-activated cell sorting (FACS). To eliminate possible TLR4 loss during the assays, TLR4 levels were evaluated by FACS assay of TLR4 expression after fixation with 4% paraformaldehyde and permeabilization of the cells with 0.1% Triton X-100. Data were processed by Cell Quest Software (Becton Dickinson) as described.45
Western blotting and extraction of nuclear proteins
Total cell lysates were prepared as described previously45 and protein concentration determined by the bicinchoninic acid (BCA) protein assay (Pierce, Rockford, IL). Nuclear proteins were extracted by NE-PER Protein Extraction Reagent (Pierce) according to the manufacturer's instructions. Cell extracts were subjected to SDS-PAGE, transferred onto nitrocellulose membrane, and blotted as described previously.45
GTP-binding, GTPase activity, and GTP loading assays
Purification of recombinant GST fusion proteins were performed as described previously.41 In vitro GTP binding and GTPase assays for GST fusion proteins were performed as described previously.46 The in vivo GTP loading assay was performed as described after labeling Raw 264.7 cells with 0.5 mCi/mL [32P]-H3PO4 for 3-4 hours at 37°C.46
Immunofluorescence staining and confocal microscopy
Raw 264.7 cells transiently transfected with plasmids encoding GFP-Rab7b and/or HA-tagged TLR4 (Invitrogen) were cultured on coverslips for 48 hours. For the colocalization analysis of GFP-Rab7b or TLR4-HA with LysoTracker (an acidotropic dye), cells were stained with 100 nM LysoTracker Red (Molecular Probes) for 30 minutes, and then cells were directly examined under confocal microscopy (for GFP-Rab7b) or visualized after sequential immunostaining with anti-HA Ab and secondary Ab (for TLR4-HA) as described previously.41 For the colocalization analysis of Rab7b with organelle markers and TLR4-HA, transfected Raw 264.7 cells were sequentially immunostained first with Ab against EEA-1, LAMP-1, TGN38, or HA, and then with proper Alexa Fluo 555-conjugated or Orange Green 488-conjugated secondary Abs. The immunostaining process was performed as described.41 Briefly, cells were washed with phosphate-buffered saline, fixed with 4% paraformaldehyde in phosphate-buffered saline for 15 minutes at room temperature, permeabilized with 0.1% Triton X-100 in phosphate-buffered saline for 5 minutes, and then blocked for 1 hour in phosphate-buffered saline containing 5% bovine serum albumin and 5% fetal calf serum. Staining with primary antibodies was performed for 1 hour at room temperature in blocking buffer. After washing, samples were incubated with appropriate secondary antibodies (1:500). Slides were finally examined under a Zeiss LSM 510 confocal microscope (Carl Zeiss, Heidelberg, Germany). Images were acquired under 40×/0.75 NA oil objective and processed using Zeiss LSM Image Browser Version 3.1.0.99 (Carl Zeiss).41
Membrane preparation, subcellular fractionation, and immunoisolation of membrane organelles
For the preparation of membrane fractions, Raw 264.7 cells stably overexpressing His-Rab7b were collected by scraping into phosphate-buffered saline containing 1 mM phenyl methane sulphonyl fluoride. After washing, cells were homogenized on ice using 10 strokes of a ball-bearing homogenizer in buffer containing 10 mM Tris.HCl (pH 7.6), 0.25 M sucrose, 1 mM MgCl2, 5 mM CaCl2, and a mixture of protease inhibitors (Calbiochem, San Diego, CA). Nuclei were pelleted at 800g for 15 minutes and the postnuclear supernatant was subjected to centrifugation at 100 000g for 1 hour at 4°C to generate a cytosol fraction and a crude membrane fraction.
For subcellular fractionation, Percoll gradient fractionation was performed as described.30 Then 1-mL fractions were collected from the top of the gradient. For Western blotting assay of membrane components in each fraction, 10 mM CHAPS detergent was added to each fraction and incubated for 1 hour on ice. Subsequently, the Percoll was removed by centrifugation for 1 hour at 100 000g in a TLA 100.2 rotor (Beckman, Palo Alto, CA).
For immunoisolation of early endosomes and late endosomes/lysosomes, the corresponding fraction pools after removal of Percoll were incubated with antibodies against C-terminal EEA1 or LAMP-1 at 4°C for 4 hours. Subsequently, bovine serum albumin-blocked protein A agarose (Pierce) was added and followed by a further incubation for 3 hours. The sedimented beads were washed 4 times in a buffer containing 0.25 M sucrose, 20 mM Hepes (pH 8.0), and 1 mM EDTA, and bound organelles were solubilized with Laemmli sample buffer for the blotting.
Statistical analysis
All experiments were independently performed 3 times in triplicate. Results are given as means plus or minus the standard error (SE) or standard deviation (SD). Comparisons between 2 groups were performed using Student t test, whereas comparisons between multiple groups were done using Kruskal-Wallis tests. Statistical significance was determined as P values less than .05.
Results
Silencing of Rab7b expression promotes the production of LPS-induced proinflammatory mediators in macrophages
We cloned the mouse homolog of human Rab7b from Raw 264.7 macrophages under the GenBank number NM_145509. The murine clone shares 92% similarity with human Rab7b protein and also encodes a protein of 199-residues, designated as mRab7b.41 There was a moderate expression of mRab7b in peritoneal macrophages (Figure 1A) and Raw 264.7 macrophages (Figure 1B). After 4 to 6 hours of exposure to LPS, the accumulation of mRab7b mRNA (Figure 1A,B) and protein (Figure S1A,B, available on the Blood website; see the Supplemental Materials link at the top of the online article) in macrophages was significantly reduced, which, however, was not significantly altered by 50 ng/mL TNF-α treatments (data not shown), suggesting that Rab7b was potentially involved in regulation of TLR4 signaling.
Silencing of Rab7b expression promotes LPS-induced production of proinflammatory mediators in macrophages. Rab7b expression was regulated by LPS. (A) Peritoneal macrophages and (B) Raw 264.7 cells were treated with 100 ng/mL LPS as indicated. Expression level of Rab7b was examined by both RT-PCR (upper panels) and quantitative PCR (lower panels). For quantitative PCR, the results were presented as folds expression of Rab7b mRNA to that of β-actin. (C) Efficiency of transient Rab7b silencing. 1.5 × 105 mouse peritoneal macrophages were transfected with 0.1 nM control small RNAs (Ctrl siRNA1 and Ctrl siRNA2) or Rab7b siRNA (Rab7b siRNA1 and Rab7b siRNA2); 48 hours later, the efficiency of silencing was examined by either RT-PCR (upper panel) or quantitative PCR (lower panel). Results were presented as described for panels A and B. (D) Production of proinflammatory mediators by Rab7b-silenced peritoneal macrophages and (E) Raw 264.7 cells after LPS treatments. Cells were silenced with small RNAs as described for panel C, and the proinflammatory mediators were measured either by enzyme-linked immunoassay (for TNF-α, IL-6, and IFN-β), or Griess reaction assay (for NO). Similar results were obtained in 3 independent experiments. Data are presented as means (± SE). ▴P < .05; *P < .05; **P < .01; ***P < .001.
Silencing of Rab7b expression promotes LPS-induced production of proinflammatory mediators in macrophages. Rab7b expression was regulated by LPS. (A) Peritoneal macrophages and (B) Raw 264.7 cells were treated with 100 ng/mL LPS as indicated. Expression level of Rab7b was examined by both RT-PCR (upper panels) and quantitative PCR (lower panels). For quantitative PCR, the results were presented as folds expression of Rab7b mRNA to that of β-actin. (C) Efficiency of transient Rab7b silencing. 1.5 × 105 mouse peritoneal macrophages were transfected with 0.1 nM control small RNAs (Ctrl siRNA1 and Ctrl siRNA2) or Rab7b siRNA (Rab7b siRNA1 and Rab7b siRNA2); 48 hours later, the efficiency of silencing was examined by either RT-PCR (upper panel) or quantitative PCR (lower panel). Results were presented as described for panels A and B. (D) Production of proinflammatory mediators by Rab7b-silenced peritoneal macrophages and (E) Raw 264.7 cells after LPS treatments. Cells were silenced with small RNAs as described for panel C, and the proinflammatory mediators were measured either by enzyme-linked immunoassay (for TNF-α, IL-6, and IFN-β), or Griess reaction assay (for NO). Similar results were obtained in 3 independent experiments. Data are presented as means (± SE). ▴P < .05; *P < .05; **P < .01; ***P < .001.
To test whether Rab7b was involved in the regulation of TLR4 signaling, we silenced the expression of Rab7b in peritoneal macrophages and Raw 264.7 cells by transient transfection of silencing RNA duplexes (siRNA1 and siRNA2) and corresponding scrambled controls (Ctrl siRNA1 and Ctrl siRNA2). We found that mRab7b was significantly downregulated by siRNA1 and to a lesser extent (≈70%) by siRNA2, but not by corresponding scrambled controls (Figure 1C). Similar results were obtained in Raw 264.7 cells (data not shown). siRNA1 was then used to determine the effects of mRab7b knockdown on the production of proinflammatory mediators by peritoneal macrophages and Raw 264.7 cells treated with 100 ng/mL LPS for 4 and 8 hours. We found that the production of TNF-α, IL-6, NO, and IFN-β in peritoneal macrophages was significantly increased after LPS treatments both at the mRNA level as determined by quantitative RT-PCR (data not shown) and at the protein level (Figure 1D and data not shown) by enzyme-linked immunoassay after mRab7b silencing. Similar changes were observed in Raw 264.7 cells (Figure 1E and data not shown) except in TNF-α, which was not significantly high at 8 hours after LPS treatment. To exclude the possible off-targets effects of RNA silencing, we transfected siRNA1-silenced Raw264.7 cells with Rab7b full-length expression vector and found that Rab7b overexpression could rescue Rab7b silence-induced enhancement of pro-inflammatory cytokine production by macrophages (data not shown). The LPS-induced production of TNF-α, IL-6, and NO has been shown previously to be related to LPS-initiated MyD88-dependent NF-κB or MAPK activation, whereas the LPS-induced IFN-β production is primarily mediated by the MyD88-independent and TRIF-dependent activation of IRF-3 pathway.1-3 Therefore, these results suggest that mRab7b may be involved in the negative regulation of LPS-initiated MyD88-dependent and TRIF-dependent signaling pathways.
Silencing of Rab7b expression potentates LPS-initiated signaling pathways in macrophages
To verify whether Rab7b was a negative regulator of TLR4 signaling, we stably silenced Rab7b expression in Raw 264.7 macrophages after stable overexpression of a plasmid encoding Rab7b silencing duplexes corresponding to the sequence for transient silencing used in Figure 1C. The efficiency of stable silencing was confirmed by quantitative RT-PCR assay to be more than 90% (Figure 2A). The Rab7b-stably silenced Raw 264.7 cells also demonstrated increased production of IL-6 and IFN-β to a similar extent as that induced by transient Rab7b silencing after TLR4 ligation (data not shown). Then the Rab7b-stably silenced Raw 264.7 cells were used in the following assays of LPS-initiated TLR4 signaling.
Silencing of Rab7b expression promotes LPS-initiated signaling pathways in macrophages. (A) Stable silencing of Rab7b expression in Raw 264.7 cells. Raw 264.7 cells were transfected with plasmids encoding small RNAs (Ctrl siRNA1 and Rab7b siRNA2), and selected under 600 μg/mL neomycin. The efficiency of silencing was evaluated by quantitative PCR as described in Figure 1A-C. (B) Western blotting assay of MAPK and (C) NF-κB activation after LPS treatments. Control (Ctrl siRNA1)-stably silenced or Rab7b (Rab7b siRNA1)-stably silenced Raw 264.7 cells were treated with 100 ng/mL LPS as indicated. Then cell lysates were prepared and blotted with indicated Abs. (D) Western blotting assay of IRF-3 activation after LPS treatments. Cells in panels B and C were extracted for lysates and nuclear proteins. Then p-IRF-3 contained in lysate (lysate) and total IRF-3 contained in the nuclei (nuclear) were examined by Western blotting. In panels B-D, total ERK1/2, p38, and β-actin contained in lysates as well as Lamin B contained in nuclear fractions were probed as quantitative controls. (E) NF-κB and (F) IFN-β luciferase reporter assay. RAW264.7 macrophages in 24-well plates were transiently cotransfected with 10 ng of pTK–Renilla-luciferase, and indicated amounts of GL3.5XκB-luciferase or IFN-β luciferase reporter plasmids and silencing vectors (pCtrl siRNA1 or pRab7b siRNA1). Total amounts of plasmid DNA were equalized using corresponding control vector (mock). After 24 hours of culture, the cells were stimulated with 100 ng/mL LPS for 8 hours. NF-κB luciferase activity (E) and IFN-β luciferase activity (F) were measured using the Dual-Luciferase Reporter Assay System, normalized by Renilla luciferase activity, expressed as fold induction relative to the activity in unstimulated cells transfected with control vector. In panels E and F, data are shown as mean (± SE) of triplicate samples. Similar results were obtained in 3 independent experiments. ▴P < .05; *P < .05; **P < .01; ***P < .001.
Silencing of Rab7b expression promotes LPS-initiated signaling pathways in macrophages. (A) Stable silencing of Rab7b expression in Raw 264.7 cells. Raw 264.7 cells were transfected with plasmids encoding small RNAs (Ctrl siRNA1 and Rab7b siRNA2), and selected under 600 μg/mL neomycin. The efficiency of silencing was evaluated by quantitative PCR as described in Figure 1A-C. (B) Western blotting assay of MAPK and (C) NF-κB activation after LPS treatments. Control (Ctrl siRNA1)-stably silenced or Rab7b (Rab7b siRNA1)-stably silenced Raw 264.7 cells were treated with 100 ng/mL LPS as indicated. Then cell lysates were prepared and blotted with indicated Abs. (D) Western blotting assay of IRF-3 activation after LPS treatments. Cells in panels B and C were extracted for lysates and nuclear proteins. Then p-IRF-3 contained in lysate (lysate) and total IRF-3 contained in the nuclei (nuclear) were examined by Western blotting. In panels B-D, total ERK1/2, p38, and β-actin contained in lysates as well as Lamin B contained in nuclear fractions were probed as quantitative controls. (E) NF-κB and (F) IFN-β luciferase reporter assay. RAW264.7 macrophages in 24-well plates were transiently cotransfected with 10 ng of pTK–Renilla-luciferase, and indicated amounts of GL3.5XκB-luciferase or IFN-β luciferase reporter plasmids and silencing vectors (pCtrl siRNA1 or pRab7b siRNA1). Total amounts of plasmid DNA were equalized using corresponding control vector (mock). After 24 hours of culture, the cells were stimulated with 100 ng/mL LPS for 8 hours. NF-κB luciferase activity (E) and IFN-β luciferase activity (F) were measured using the Dual-Luciferase Reporter Assay System, normalized by Renilla luciferase activity, expressed as fold induction relative to the activity in unstimulated cells transfected with control vector. In panels E and F, data are shown as mean (± SE) of triplicate samples. Similar results were obtained in 3 independent experiments. ▴P < .05; *P < .05; **P < .01; ***P < .001.
First, we examined the activation status of MAPK (ERK1/2, JNK1/2, and p38) in Rab7b-silenced Raw 264.7 cells after LPS stimulation. We found that the activation of both ERK1/2 and JNK1/2 was remarkably enhanced in Rab7b-silenced cells (Rab7b siRNA1) after LPS treatments, compared with that of the control siRNA-transfected Raw 264.7 cells (Ctrl siRNA1), whereas activation of p38 MAPK by LPS was not significantly altered by Rab7b silencing (Figure 2B). Second, we examined the effects of Rab7b silencing on the phosphorylation and degradation of IκBα. We found that mRab7b knockdown enhanced LPS-induced phosphorylation of I-κBα and the degradation of total I-κBα (Figure 2C). Third, we examined the effects of Rab7b silence on the activation of IRF-3 in macrophages by LPS. We found that mRab7b silence increased the phosphorylation of IRF-3 and the nuclear presence of IRF-3 (Figure 2D). As further evidence for Rab7b-mediated negative regulation of LPS-induced NF-κB and IRF-3 activation in macrophages, wild-type Raw 264.7 cells were transiently transfected with a NF-κB–luciferase construct and an IFN-β–luciferase construct, along with either a construct encoding interfering RNA of mRab7b (pRab7b siRNA1) or corresponding scrambled control vector (pCtrl siRNA1). The cells were left untreated or treated with LPS, and the activity of luciferase reporter gene was then detected. Rab7b knockdown increased the LPS-induced transcriptional activity of NF-κB dose-dependently (Figure 2E) and potentate the transactivation of IFN-β promoter (Figure 2F) in Raw 264.7 macrophages. Taken together, these data (Figures 1,2) convincingly demonstrated that Rab7b could negatively regulate LPS-initiated activation of MAPK, NF-κB, and IRF-3 pathways and LPS-induced production of proinflammatory mediators in macrophages, suggesting that Rab7b is a negative regulator in LPS-initiated Myd88-dependent and TRIF-dependent signaling pathways.
Overexpression of Rab7b inhibits LPS-induced cytokine production and LPS-initiated signaling pathways in macrophages
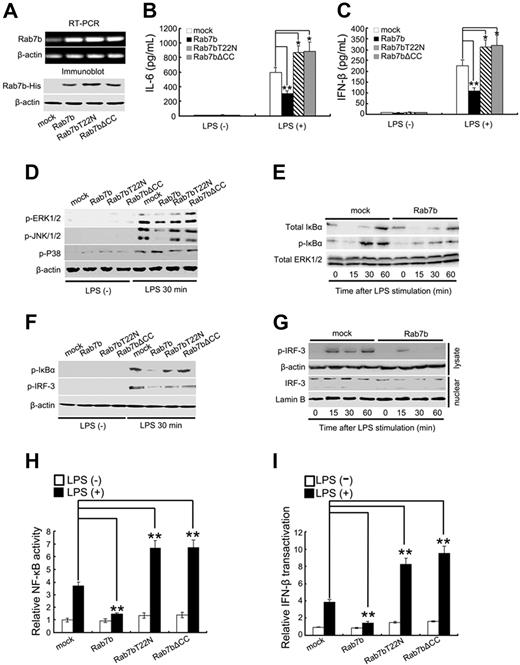
As further evidence for the roles of Rab7b in negative regulation of TLR4 signaling, we stably overexpressed His-tagged Rab7b in Raw 264.7 cells. The expression of Rab7b was confirmed by both RT-PCR and Western blot (Figure 3A). After overexpression of His-tagged full-length Rab7b in Raw264.7 cells, we found that both the LPS-induced MyD88-dependent production of IL-6 (Figure 3B) and the LPS-induced TRIF-dependent production of IFN-β (Figure 3C) were significantly inhibited, indicating that Rab7b overexpression inhibited the LPS-initiated Myd88-dependent and TRIF-dependent TLR4 signaling in macrophages.
Overexpression of Rab7b inhibits LPS-induced production of proinflammatory mediators and LPS-initiated signaling pathways in macrophages. (A) Stable overexpression of His-tagged Rab7b and Rab7b mutants in Raw 264.7 cells. Raw 264.7 cells were transfected with plasmids encoding His-tagged Rab7b, Rab7bT22N, and Rab7b▵CC, and selected under 600 μg/mL neomycin. The stable transfectants were confirmed by RT-PCR (top 2 panels) and Western blotting (bottom 2 panels) using anti-His Ab. (B) Enzyme-linked immunoassay assay of IL-6 and (C) IFN-β production by the indicated Rab7b transfectants after LPS treatments. Cells (2 × 105/well) were treated with 100 ng/mL LPS for 8 hours. The supernatants were then collected and the concentration of (B) IL-6 and (C) IFN-β evaluated by enzyme-linked immunoassay. (D) Western blotting assay of MAPK (E,F), NF-κB, and (F,G) IRF-3 activation after LPS treatments. Raw 264.7 stable transfectants were treated with 100 ng/mL LPS as described for Figure 1B-D. Cell lysates and nuclear proteins were then prepared and blotted with indicated Abs. Total ERK1/2 and β-actin contained in lysates as well as Lamin B contained in nuclear fractions were probed as quantitative controls. (H) NF-κB and (I) IFN-β luciferase reporter assay. RAW264.7 macrophages in 24-well plates were transiently cotransfected with 10 ng of pTK–Renilla-luciferase, and indicated amounts of GL3.5XκB-luciferase or IFN-β luciferase reporter plasmids and Rab7b (mutants)-overexpressing vectors. Luciferase activity was then determined and presented as that described for Figure 2E,F. In panels B, C, H, and I, data are shown as mean (± SE) of triplicate samples. Similar results were obtained in 3 independent experiments. *P < .05; **P < .01.
Overexpression of Rab7b inhibits LPS-induced production of proinflammatory mediators and LPS-initiated signaling pathways in macrophages. (A) Stable overexpression of His-tagged Rab7b and Rab7b mutants in Raw 264.7 cells. Raw 264.7 cells were transfected with plasmids encoding His-tagged Rab7b, Rab7bT22N, and Rab7b▵CC, and selected under 600 μg/mL neomycin. The stable transfectants were confirmed by RT-PCR (top 2 panels) and Western blotting (bottom 2 panels) using anti-His Ab. (B) Enzyme-linked immunoassay assay of IL-6 and (C) IFN-β production by the indicated Rab7b transfectants after LPS treatments. Cells (2 × 105/well) were treated with 100 ng/mL LPS for 8 hours. The supernatants were then collected and the concentration of (B) IL-6 and (C) IFN-β evaluated by enzyme-linked immunoassay. (D) Western blotting assay of MAPK (E,F), NF-κB, and (F,G) IRF-3 activation after LPS treatments. Raw 264.7 stable transfectants were treated with 100 ng/mL LPS as described for Figure 1B-D. Cell lysates and nuclear proteins were then prepared and blotted with indicated Abs. Total ERK1/2 and β-actin contained in lysates as well as Lamin B contained in nuclear fractions were probed as quantitative controls. (H) NF-κB and (I) IFN-β luciferase reporter assay. RAW264.7 macrophages in 24-well plates were transiently cotransfected with 10 ng of pTK–Renilla-luciferase, and indicated amounts of GL3.5XκB-luciferase or IFN-β luciferase reporter plasmids and Rab7b (mutants)-overexpressing vectors. Luciferase activity was then determined and presented as that described for Figure 2E,F. In panels B, C, H, and I, data are shown as mean (± SE) of triplicate samples. Similar results were obtained in 3 independent experiments. *P < .05; **P < .01.
To verify the effects of Rab7b overexpression on LPS-initiated MAPK, NF-κB, and IRF-3 activation, we examined the activation status of ERK1/2, JNK1/2, p38, IκBα, and IRF-3 by Western blotting. We found that Rab7b overexpression could inhibit LPS-induced activation of ERK1/2 and JNK1/2 (Figure 3D), phosphorylation of IκB-α (Figure 3E,F), and activation of IRF-3 (Figure 3G). Furthermore, both the NF-κB activity (Figure 3H) and the transactivation of IFN-β promoter (Figure 3I) induced by LPS were significantly inhibited by Rab7b overexpression, consistent with the Western blotting results (Figure 3D-G). Therefore, we could convincingly conclude that Rab7b is a negative regulator in TLR4-initiated signaling pathways.
It was interesting to find that the mutants of Rab7b, including Rab7bT22N and mRab7bΔCC, could serve as dominant-negative forms of Rab7b in LPS-initiated MAPK, NF-κB, and IRF-3 activation (Figure 3D-I) and in LPS-induced IL-6 and IFN-β production in macrophages (Figure 3B,C). It has been shown that mutation of the corresponding Thr22 into Asn in Rab7 can result in GTP-binding deficiency29-31 and that the deletion of C-terminal cysteines in Rab proteins can lead to misdistribution of Rab proteins.24-26 Therefore, we examined the GTP-binding activity and the GTP-bound status of Rab7b in macrophages. We found that the recombinant GST fusion proteins of Rab7b and Rab7bΔCC, but not Rab7bT22N, could specifically bind radioactive GTP (Figure S2A) and hydrolyze the bound GTP into guanosine diphosphate (GDP) (Figure S2B) in vitro. Furthermore, we found that Rab7bT22N demonstrated decreased level of GTP-bound form in vivo in Raw 264.7 cells (Figure S2C), and LPS treatments could increase the levels of Rab7b in GTP-bound form (Figure S2D). Therefore, the roles of Rab7b in negative regulation of TLR4 signaling were related to the native GTPase activity of Rab7b.
Rab7b negatively regulates the expression of TLR4
We went further to investigate the mechanisms by which Rab7b negatively regulates TLR4 signaling in macrophages. It has been recently shown that TLR4 can be transported from cell membrane to endosome for ubiqutination and to lysosome for degradation.40 As a small GTPase, Rab7b is predicted to regulate the membrane transport of proteins.24-26,41 Previous studies have revealed that negative regulation of TLR4 signaling is mostly dependent on the regulation of TLR4 expression levels or TLR4 degradation.8-23,40 We therefore wondered whether Rab7b negatively regulates TLR4 signaling, possibly through its effects on the membrane transport of TLR4 signaling components. We first examined TLR4 expression in macrophages after Rab7b overexpression or silencing. We found that Rab7b overexpression did not affect the mRNA level of TLR4 expression (data not shown) but downregulated the overall protein level of TLR4 in peritoneal macrophages (Figure 4A) and Raw 264.7 macrophages (Figure 4B). Accordingly, we found that the expression of TLR4 in peritoneal macrophages (Figure 4C) and Raw 264.7 macrophages (Figure 4D) was increased after Rab7b silencing. Furthermore, the TLR4 expression level on the plasma membrane of Rab7b-silenced peritoneal macrophages (Figure 4E) and Raw 264.7 cells (Figure 4F) was also increased. To further examine the effects of Rab7b on TLR4 expression, we observed the dynamic changes of TLR4 after LPS treatments of Raw 264.7 cells. We found that the overall TLR4 expression was significantly decreased 15 minutes and 30 minutes after LPS treatments in control vector-transfected Raw 264.7 cells, whereas that in Rab7b-silenced Raw 264.7 macrophages remain at a high level after LPS treatments (Figure 4G), indicating that Rab7b was possibly involved in the degradation of TLR4. Because the cells used here were stably transfected with Rab7b plasmid, our data suggest that TLR4 may be recycled automatically under normal conditions.
Rab7b negatively regulates the TLR4 expression. (A,B) Western blotting assay of TLR4 expression in Rab7b-overexpressed or (C,D) Rab7b-silenced peritoneal macrophages (A,C) and Raw 264.7 cells (B,D). Stable Raw 264.7 transfectants (B,D) and murine peritoneal macrophages (A,C) transiently transfected with indicated vectors were prepared for cell lysates and TLR4 expression examined by Western blotting using anti-TLR4 Ab. β-actin was used as quantitative control. The expression level of Rab7b by these transfectants was determined by quantitative PCR, and the mean folds of Rab7b mRNA level to that of β-actin were shown at the top of each panel. (E,F) FACS assay of TLR4 expression on plasma membrane of Rab7b-silenced (lower panels) or control vector-silenced (upper panels) murine peritoneal macrophages and Raw 264.7 cells. TLR4 expression on plasma membrane was evaluated by labeling cells with phycoerythrin-conjugated TLR4 Ab. The mean fluorescence intensity (MFI) was also shown at the upper right corner of each panel. (G) Dynamic changes of TLR4 expression in Rab7b-silenced peritoneal macrophages and Raw 264.7 cells after LPS treatments. Cells were transiently transfected with control silencing small RNAs (Ctrl siRNA1) or Rab7b silencing small RNAs (Rab7b siRNA1); 48 hours later, cells were treated with 100 ng/mL LPS. TLR4 contained in cell lysates were examined by Western blotting using TLR4 Ab. β-actin was used as quantitative control. Similar results were obtained in 3 independent experiments.
Rab7b negatively regulates the TLR4 expression. (A,B) Western blotting assay of TLR4 expression in Rab7b-overexpressed or (C,D) Rab7b-silenced peritoneal macrophages (A,C) and Raw 264.7 cells (B,D). Stable Raw 264.7 transfectants (B,D) and murine peritoneal macrophages (A,C) transiently transfected with indicated vectors were prepared for cell lysates and TLR4 expression examined by Western blotting using anti-TLR4 Ab. β-actin was used as quantitative control. The expression level of Rab7b by these transfectants was determined by quantitative PCR, and the mean folds of Rab7b mRNA level to that of β-actin were shown at the top of each panel. (E,F) FACS assay of TLR4 expression on plasma membrane of Rab7b-silenced (lower panels) or control vector-silenced (upper panels) murine peritoneal macrophages and Raw 264.7 cells. TLR4 expression on plasma membrane was evaluated by labeling cells with phycoerythrin-conjugated TLR4 Ab. The mean fluorescence intensity (MFI) was also shown at the upper right corner of each panel. (G) Dynamic changes of TLR4 expression in Rab7b-silenced peritoneal macrophages and Raw 264.7 cells after LPS treatments. Cells were transiently transfected with control silencing small RNAs (Ctrl siRNA1) or Rab7b silencing small RNAs (Rab7b siRNA1); 48 hours later, cells were treated with 100 ng/mL LPS. TLR4 contained in cell lysates were examined by Western blotting using TLR4 Ab. β-actin was used as quantitative control. Similar results were obtained in 3 independent experiments.
Rab7b is a small GTPase localized to lysosome-associated subcellular compartments
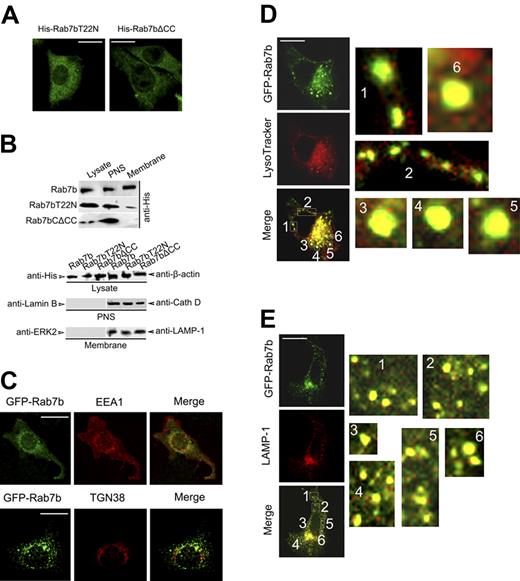
Most of the Rab proteins function via regulating protein transport between different subcellular compartments. And the results in Figures 3D-I indicated that mutations that were expected to affect the subcellular localization of Rab7b could influence the activities of Rab7b. Therefore, we examined the subcellular localization of Rab7b and its mutants in Raw 264.7 cells. We transiently or stably overexpressed these constructs in Raw 264.7 cells, and examined their distribution by confocal microscopy or by Western blotting after subcellular fractionation. As expected, we found that Rab7b was localized into vesicular and perinuclear membrane organelles while Rab7bT22N and Rab7bΔCC showed dispersed pattern in cytoplasm (Figure 5A-E). Although Rab7b was detected in the membrane fractions in Raw 264.7 cells (Figure 5B), the specific membrane compartments had not been identified. Therefore, we investigated the subcellular colocalization of Rab7b with different compartments markers in Raw 264.7 cells by confocal microscopy. We found that Rab7b was mainly localized into LysoTracker (acidic compartment)–positive (Figure 5D) and LAMP-1 (late endosomes/lysosomes)–positive (Figure 5E) compartments, but was not detected in EEA1 (early endosomes)–positive and TGN38 (Golgi apparatus)–positive (Figure 5C) compartments in Raw 264.7 macrophages, suggesting that Rab7b was a late endosome/lysosome-associated small GTPase.
Rab7b is a small GTPase localized to late endosomes and lysosomes. (A) Distribution of Rab7b mutants in Raw 264.7 cells. Raw 264.7 cells were transiently transfected with His-tagged Rab7bT22N or Rab7b▵CC; 48 hours later, cells were immunostained with anti-His Ab and Orange Green 488-conjugated secondary Ab. (B) Examination of the subcellular localization of full-length Rab7b and Rab7b mutants in stable Raw 264.7 transfectants by Western blotting of the lysate, postnuclear supernatant, and membrane factions using anti-His Ab (upper panel). The lysate, postnuclear supernatant, and membrane fractions were also probed with indicated Abs (lower panel) to show the loading quantity and the fractionation specificity. (C-E) Confocal analysis of Rab7b subcellular localization. Raw 264.7 cells were transiently transfected with GFP-Rab7b plasmid; 48 hours later, cells were labeled with LysoTracker Red (D), or immunostained with first Ab against EEA-1 (C), TGN38 (C), or LAMP-1 (E) and Alexa Fluo 555-conjugated secondary Ab. The slides were finally examined under confocal microscopy, and representative images were presented. In panels D and E, the numerated regions were magnified and shown correspondingly at the right. (Bar = 100 μm).
Rab7b is a small GTPase localized to late endosomes and lysosomes. (A) Distribution of Rab7b mutants in Raw 264.7 cells. Raw 264.7 cells were transiently transfected with His-tagged Rab7bT22N or Rab7b▵CC; 48 hours later, cells were immunostained with anti-His Ab and Orange Green 488-conjugated secondary Ab. (B) Examination of the subcellular localization of full-length Rab7b and Rab7b mutants in stable Raw 264.7 transfectants by Western blotting of the lysate, postnuclear supernatant, and membrane factions using anti-His Ab (upper panel). The lysate, postnuclear supernatant, and membrane fractions were also probed with indicated Abs (lower panel) to show the loading quantity and the fractionation specificity. (C-E) Confocal analysis of Rab7b subcellular localization. Raw 264.7 cells were transiently transfected with GFP-Rab7b plasmid; 48 hours later, cells were labeled with LysoTracker Red (D), or immunostained with first Ab against EEA-1 (C), TGN38 (C), or LAMP-1 (E) and Alexa Fluo 555-conjugated secondary Ab. The slides were finally examined under confocal microscopy, and representative images were presented. In panels D and E, the numerated regions were magnified and shown correspondingly at the right. (Bar = 100 μm).
TLR4 is internalized and transported to Rab7b-positive compartments after LPS ligation
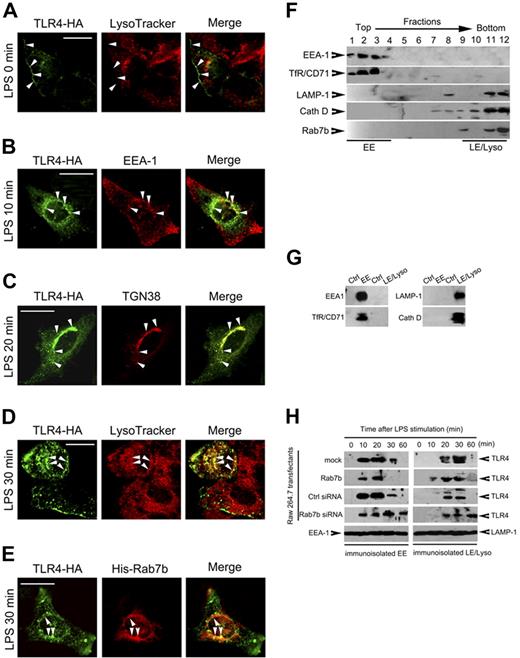
To elucidate the mechanisms of Rab7b in regulating TLR4 signaling, we then investigated the roles of Rab7b in TLR4 internalization and subcellular transport by FACS and confocal microscopy. Raw 264.7 cells were transiently transfected with TLR4-HA (Figure 6A-D) or TLR4-HA plus GFP-Rab7b (Figure 6E) plasmid(s) and were then treated with LPS as indicated. We found that Rab7b silencing or overexpression did not affect the internalization of TLR4 (data not shown). TLR4 was mainly localized on plasma membrane before LPS treatment (Figure 6A). After LPS treatments, TLR4 was transported into EEA1-positive compartments (Figure 6B) and TGN38-positive compartments (Figure 6C) approximately 10-20 minutes after LPS treatment, and was colocalized with LysoTracker (Figure 6D) and Rab7b (Figure 6E) approximately 30 minutes after LPS treatment. These data (Figure 6A-E) suggested a direct linkage between Rab7b localization and TLR4 transport.
TLR4 is internalized and transported to Rab7b-positive compartments after LPS treatment. (A-D) TLR4 translocation after LPS treatments. Raw 264.7 cells were transiently transfected with TLR4-HA vector; 48 hours later, the cells were left untreated or treated with 100 ng/mL LPS for indicated period. The subcellular localization of TLR4 was examined after immunostaining for TLR4 and indicated compartments markers under confocal microscopy. (E) Colocalization of TLR4 with Rab7b after LPS treatment. Raw 264.7 cells were transiently cotransfected with TLR4-HA with His-Rab7b plasmids, and treated with 100 ng/mL LPS for 30 minutes to 1 hour. Then the cells were immunostained with anti-HA Ab and anti-His Ab, manifested with Orange Green 488-conjugated or Alexa Fluo 555-conjugated secondary Ab. Representative images are shown in (A-E). (Bar = 100 μm) (F) Subcellular fractionation of Rab7b-overexpressed Raw 264.7 cells. Raw 264.7 cells stably overexpressing Rab7b were fractionated for early endosomes (EE) and late endosomes/lysosomes (LE/Lyso). Distribution of specific markers as well as Rab7b in each fraction was examined by Western blotting. (G) Immunoisolation of EE and LE/Lyso. Fractions obtained in (F) were pooled for immunoisolation of EE (fractions 1-4) and LE/Lyso (fractions 8-12). The EE was enriched by using anti-TfR/CD71 Ab while LE/Lyso was enriched by using anti-LAMP-1 Ab. Rabbit IgG was used as negative control. The immunoisolated organelles were examined for indicated markers, showing the specificity of immuno-isolated EE and LE/Lyso. (H) Dynamic translocation of TLR4 from EE to LE/Lyso after LPS treatments. The indicated transfectants were treated with 100 ng/mL LPS for indicated time periods, and the EE and LE/Lyso organelles were immunoisolated and subjected to Western blot assay of TLR4 expression. EEA-1 and LAMP-1 were examined to determine the loading quantity of the immunoisolated organelles. Similar results were obtained in 3 independent experiments.
TLR4 is internalized and transported to Rab7b-positive compartments after LPS treatment. (A-D) TLR4 translocation after LPS treatments. Raw 264.7 cells were transiently transfected with TLR4-HA vector; 48 hours later, the cells were left untreated or treated with 100 ng/mL LPS for indicated period. The subcellular localization of TLR4 was examined after immunostaining for TLR4 and indicated compartments markers under confocal microscopy. (E) Colocalization of TLR4 with Rab7b after LPS treatment. Raw 264.7 cells were transiently cotransfected with TLR4-HA with His-Rab7b plasmids, and treated with 100 ng/mL LPS for 30 minutes to 1 hour. Then the cells were immunostained with anti-HA Ab and anti-His Ab, manifested with Orange Green 488-conjugated or Alexa Fluo 555-conjugated secondary Ab. Representative images are shown in (A-E). (Bar = 100 μm) (F) Subcellular fractionation of Rab7b-overexpressed Raw 264.7 cells. Raw 264.7 cells stably overexpressing Rab7b were fractionated for early endosomes (EE) and late endosomes/lysosomes (LE/Lyso). Distribution of specific markers as well as Rab7b in each fraction was examined by Western blotting. (G) Immunoisolation of EE and LE/Lyso. Fractions obtained in (F) were pooled for immunoisolation of EE (fractions 1-4) and LE/Lyso (fractions 8-12). The EE was enriched by using anti-TfR/CD71 Ab while LE/Lyso was enriched by using anti-LAMP-1 Ab. Rabbit IgG was used as negative control. The immunoisolated organelles were examined for indicated markers, showing the specificity of immuno-isolated EE and LE/Lyso. (H) Dynamic translocation of TLR4 from EE to LE/Lyso after LPS treatments. The indicated transfectants were treated with 100 ng/mL LPS for indicated time periods, and the EE and LE/Lyso organelles were immunoisolated and subjected to Western blot assay of TLR4 expression. EEA-1 and LAMP-1 were examined to determine the loading quantity of the immunoisolated organelles. Similar results were obtained in 3 independent experiments.
To further determine the roles of Rab7b in regulating TLR4 transport and lysosome degradation, we immunoisolated the early endosomes and late endosomes/lysosomes fractionations after subcellular fractionation. We found that His-tagged Rab7b was mainly localized in fractions positive for late endosome/lysosome markers LAMP-1 and cathepsin D (fractions 9-12), but not in fractions positive for early endosome markers EEA1 and TfR (fractions 1-4; Figure 6F), which were consistent with the confocal microscopy analysis of GFP-Rab7b and suggested that the subcellular fraction method used here was effective and specific. Then we analyzed the TLR4 translocation in Raw 274.7 cells stably transfected with His-tagged Rab7b or Rab7b silencing plasmids after LPS treatments by immunoisolation of early endosomes and late endosomes/lysosomes (Figure 6G). We found that 10-20 minutes after LPS treatments, TLR4 in these cells could be detected in early endosomes (Figure 6H); 30 minutes after LPS treatments, TLR4 in control cells and in Rab7b-overexpressing cells began to accumulate in late endosome/lysosome compartments. However, stable overexpression of Rab7b-silencing plasmid could impair the location of TLR4 in late endosome/lysosome, as evidenced by the persistence of TLR4 in EEA-1–positive compartments and the prolonged presence of TLR4 in LAMP-1–positive compartments (Figure 6H), suggesting that Rab7b may be involved in a process of translocation of TLR4 from early endosomes to late endosomes/lysosomes.
Discussion
LPS-initiated MyD88-dependent signaling pathways can activate MAPK and NF-κB via sequential activation of MyD88/IL-1R–associated kinase/TRAF6, resulting in production of proinflammatory cytokines (IL-1, IL-6, TNF-α, etc), whereas LPS can also initiate MyD88-independent and TRIF-dependent activation of IRFs to regulate the IFN-inducible genes (CXCL10, IFN-α/β etc).1-3 Excessive or uncontrolled TLR activation may contribute to the pathogenesis of autoimmune, chronic inflammatory, and infectious diseases. One of the most severe diseases is sepsis caused by TLR4 ligation of LPS. So, TLR4 signaling must be regulated strictly so that immune system can be activated to eliminate pathogens but not cause damages of the host. Many negative regulators of TLR4 have been identified during the recent years.8-23 In this study, we demonstrate that Rab7b may serve as a potent negative regulator of Myd88-dependent and TRIF-dependent TLR4 signaling pathways by showing that Rab7b silencing promotes the activation of MAPK, NF-κB, and IRF-3, and increases the production of TNF-α, IL-6, NO, and IFN-β in macrophages after LPS treatments. Rab7b-medaited inhibition of TLR4 signaling may serve as a feedback mechanism in inflammation after infection, and Rab7b may be a novel target in the control of LPS/TLR4-mediated inflammatory and autoimmune diseases. Furthermore, our demonstration that the small GTPase Rab7b can negatively regulate the TLR4 expression level, possibly by promoting its degradation after LPS treatments in macrophages, has provided a first line of evidence for the involvement of Rab small GTPase in the regulation of TLRs signaling.
TLRs are typical type I membrane receptors.1-3 It has been shown that TLR1, TLR2, TLR4, TLR5, and TLR6 are cell surface-bound, whereas TLR3, TLR7, and TLR9 are expressed intracellularly in acidic compartment.1-3,47 There is an extensive body of data implying that TLR signaling impacts on many aspects of pathogen uptake and elimination.1-3,47 However, little is known about the subcellular transport of TLRs together with their associated ligands in TLR signaling. LPS initiates TLR4 signaling on cell surface after LPS has engaged CD14, and LPS/TLR4/MD-2 are rapidly endocytosed and can be present in endosomes, Golgi apparatus, and lysosomes,36-40 suggesting a role of TLR4 trafficking in TLR4 signaling. Recently, it has been revealed that TLR4 can be transported to the endosome for ubiquitination and to the lysosome for degradation.40 Consistent with these reports,36-40 we find that TLR4 is mainly localized on the plasma membrane before LPS treatment, and is then internalized and present in EEA-1–positive early endosomes, TGN-38–positive Golgi apparatus, and LAMP-1–positive late endosomes/lysosomes, outlining a dynamic trafficking for TLR4. Cells take up various kinds of ligands, such as nutrients, hormones, and growth factors, and deliver them to their correct destinations via the endocytic pathway.48 After internalization, endocytosed solute and receptor-bound proteins are delivered to a common early endosomal compartment, where they are sorted and targeted to other intracellular destinations, including a putative recycling compartment, a trans-Golgi network pathway, and diversion to a lysosomal degradative/storage compartment.48 Rab7b is localized into Lysotrcker Red-positive and LAMP-1–positive late endosomes/lysosomes, and is colocalized with TLR4 at a later stage after LPS treatments. We also demonstrate that Rab7b can downregulate the TLR4 expression, which, in turn, leads to hyposensitivity of macrophages to LPS signaling. Taken together, our study and the previous studies36-40 infer that TLR4 is internalized and transported to endosomes, after which TLR4 is transported to the Golgi network either for recycling or for degradation in lysosomes. Although we have not provided direct evidence for the role of Rab7b in lysosomal degradation of TLR4, we show that silencing of Rab7b can upregulate TLR4 expression similarly to the effects of the lysosome inhibitor chloroquine and the proteasome inhibitor lactacystin (data not shown), comparable with the previous report.40 Moreover, we find that Rab7b silencing can prolong the presence of TLR4 in early endosomes and delay the presence of TLR4 in late endosomes and lysosomes, suggesting that Rab7b may negatively regulate TLR4 signaling via promoting the targeting of TLR4 to lysosomes for degradation. TLR4 signaling involves many key components other than TLR4, such as MyD88, TRIF, TBK1, and TAK1. A potential explanation for negative regulation of TLR4 signaling by Rab7b may possibly involve degradation of TLR4 signaling components other than TLR4. However, little is known about the degradation of these components. Our examination of these components by Western blotting suggested that Rab7b could not affect the protein levels of MyD88, TBK1, and TAK1 (data not shown), suggesting that TLR4 was at least the major target involved in Rab7b-mediated negative regulation of TLR4 signaling in macrophages.
Ras-related Rab GTPases, in their GTP-binging status, regulate intracellular trafficking during endocytosis and secretion by specific localization to the cytoplasmic surface of specific intracellular compartments, where they regulate the individual steps of vesicular transport by controlling vesicle docking and fusion.24-26 The Rab proteins known to be associated with late endosomes are Rab7 and Rab9. Rab9 has been shown to be involved in trafficking between late endosomes and the trans-Golgi network,49 whereas Rab7 regulates the early to late endosome transport.31-35 Small GTP-binding proteins have been supposed to localize on lysosomal membranes, but the precise Rab family members have not been identified. As a homolog of Rab7, Rab7b is a typical small GTPase, as evidenced by the findings that Rab7b can specifically bind GTP both in vitro and in vivo. We suggest here that Rab7b may be a lysosome-associated small GTPase potentially involved in the regulation of protein transport by facilitating a docking and fusion process from early endosomes to late endosomes/lysosomes. Like Rab7, Rab7b is detected in late endosomes; unlike Rab9, Rab7b is not localized in the trans-Golgi network and early endosomes. Therefore, Rab7b is structurally and functionally homologous to Rab7 but not Rab9. Our data, that Rab7b is localized into LAMP-1–positive compartments containing lysosomal component cathepsin D and the finding that Rab7b silencing leads to impaired translocation of TLR4 from early endosomes to late endosomes/lysosomes together, suggest that Rab7b may be functionally related or complementary to Rab7, which mediates early endosome/late endosome docking and fusion processes.24-26,41 Because Rab's-regulated membrane trafficking is compartment-specific but not protein-specific, it can be predicted that Rab7b may also be involved in the regulation of other TLRs or membrane receptors signaling that involves endosomal–lysosomal transport. However, the detailed membrane trafficking process mediated by Rab7b may need further specific experiments in the future.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Wei Chen, Lihuang Zhang, Hongmei Xu, and Qiuyan Liu for their helpful discussion. The authors thank Mei Jin, Yan Li, and Xiaoting Zuo for their excellent technical assistance.
This work was supported by grants from the National Natural Science Foundation of China (30572102, 30490240, 30121002), National High Biotechnology Development Program of China (2006AA02A305), 973 National Key Basic Research Program of China, and Shanghai Committee of Science and Technology (06DJ14011).
Authorship
Contribution: Y. W. and D.H. cloned the full-length mouse Rab7b cDNA. Y.W., N.L., and D.H. constructed all the vectors. Y.W., T.C., C.H., and H.L. analyzed the effects of Rab7b on LPS-induced cytokine production and signaling pathways. T.C., C.H., and H.L. performed the immunofluorescent assays. Y.W., T.C., and Z.C. collected all the data. Y.W. and T.C. prepared the manuscript. X.C. supervised the overall project and edited the manuscript. All authors checked the final version of the manuscript.
Y. W. and T. C. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Xuetao Cao, Institute of Immunology, Zhejiang University, 388 Yuhangtang Road, Hangzhou 310058, PR China; or Institute of Immunology and National Key Laboratory of Medical Immunology, Second Military Medical University, 800 Xiangyin Road, Shanghai 200433, PR China; e-mail: caoxt@public3.sta.net.cn.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal