Abstract
The oncogene c-maf is frequently overexpressed in multiple myeloma cell lines and patient samples and contributes to increased cellular proliferation in part by inducing cyclin D2 expression. To identify regulators of c-maf, we developed a chemical screen in NIH3T3 cells stably overexpressing c-maf and the cyclin D2 promoter driving luciferase. From a screen of 2400 off-patent drugs and chemicals, we identified glucocorticoids as c-maf–dependent inhibitors of cyclin D2 transactivation. In multiple myeloma cell lines, glucocorticoids reduced levels of c-maf protein without influencing corresponding mRNA levels. Subsequent studies demonstrated that glucocorticoids increased ubiquitination-dependent degradation of c-maf and up-regulated ubiquitin C mRNA. Moreover, ectopic expression of ubiquitin C recapitulated the effects of glucocorticoids, demonstrating regulation of c-maf protein through the abundance of the ubiquitin substrate. Thus, using a chemical biology approach, we identified a novel mechanism of action of glucocorticoids and a novel mechanism by which levels of c-maf protein are regulated by the abundance of the ubiquitin substrate.
Introduction
Multiple myeloma appears to arise from initiating chromosomal translocations and duplications in postgerminal center plasma cells with subsequent secondary mutations contributing to disease progression.1,2 Six unique myeloma variants can be identified by gene expression profiling with 3 of the 6 groups anchored by an initiating chromosome translocation. It is noteworthy that a unifying event of cyclin D dysregulation is identified in all subgroups.1,2 One of the initiating chromosomal abnormalities in myeloma involves members of the Maf family of oncogenes.1 Maf proteins are basic leucine zipper transcription factors in the AP1 family and regulate gene transcription by binding DNA sequences known as MAF responsive elements (MAREs).3,4 Of the Maf family, c-maf was the first endogenous member identified and is probably the best characterized with respect to function.5 Genes up-regulated by c-maf include cyclin D2, β-integrin 7, ARK 5 and CCR1, all of which are important in the pathogenesis of multiple myeloma.6-8
c-maf is dysregulated in multiple myeloma. For example, approximately 25% of myeloma cell lines have a t(14;16) translocation.6 In patients with myeloma, approximately 6% have a t(14;16) or t(14;20), translocations that juxtapose IgH with c-maf and its homolog mafB, respectively.9,10 The frequency of c-maf overexpression in patients who lack the t(14;16) has varied from study to study, depending on the method to assess its overexpression and ranges from 5% to 50%.6,11,12 In malignant cell lines, including multiple myeloma, overexpression of c-maf augments cell proliferation and increases tumor formation in xenograft models.6,7 Conversely, inhibition of c-maf with dominant-negative constructs decreases cell proliferation, impairs adhesion to marrow stroma, and delays tumor growth.6 Overexpression of c-maf is also clinically relevant in that patients with myeloma and the t(14;16) c-maf translocation have a shorter overall survival.13 Although the functional importance of c-maf has been described, the mechanisms that govern its regulation have not been fully elucidated. Therefore, molecules that decrease c-maf and subsequently its downstream targets, particularly cyclin D2, could further our understanding of the regulation of c-maf. Improved understanding of c-maf will help develop therapies that target this protein.
To this end, we developed a high-throughput chemical genomics screen to identify compounds that inhibit c-maf–dependent transactivation of the cyclin D2 promoter. With this assay, we screened libraries of off-patent drugs and chemicals and were surprised to identify glucocorticoids as inhibitors of c-maf–dependent cyclin D2 transactivation. Subsequent studies demonstrated that glucocorticoids decrease c-maf protein by promoting its ubiquitination through the up-regulation of ubiquitin C mRNA. Thus, this chemical biology approach has provided insights into a novel mechanism of c-maf regulation.
Materials and methods
Cell culture, constructs, and transduction
Mouse fibroblast NIH3T3 cells were maintained in Dulbecco modified Eagle medium plus 10% calf serum (Hyclone, Logan, UT). Human myeloma cell lines were grown in Iscove modified essential medium plus 10% fetal bovine serum (Hyclone). All media were supplemented with 1 mmol/L glutamate and antibiotics. Cells were cultured at 37°C with 5% CO2 in a humid incubator.
Full-length c-maf cDNA was subcloned into an internal ribosome entry site (IRES)-green fluorescent protein (GFP)-MIEV retroviral vector. NIH3T3 cells were infected with this construct, and stable cells expressing GFP and c-maf were selected by flow cytometry and immunoblotting, respectively. The full-length c-maf was also subcloned into a pcDNA3.1 vector under the control of a cytomegalovirus (CMV) promoter.
The promoter of cyclin D2 (−894 to −4), containing c-maf responsive element sequence (MARE), was cloned from HeLa cell genomic DNA and subcloned into the pGL2 luciferase reporter vector (Promega, Madison, WI). This construct was cotransfected with pcDNA3.1 containing a neomycin resistance gene into NIH3T3 wild-type cells and NIH3T3 cells stably overexpressing c-maf-IRES-GFP. Cells stably expressing c-maf, GFP, and luciferase were selected for further application.
Myeloma cells were transfected with cDNA corresponding to cyclin D2, ubiquitin or GFP using Amaxa Nucleofection System (Amaxa, Gaithersburg, MD) per the manufacturer's instructions.
High-throughput screen for inhibitors of cyclin D2 transactivation
NIH3T3 cells stably expressing c-maf and the cyclin D2 promoter driving luciferase (13 000 cells per well) were plated in 96-well plates by the Biomek FX liquid handler (Beckman Coulter, Fullerton, CA). The same workstation was used for plate formatting and reagent distribution. After the cells had adhered (6 hours after plating), they were treated with aliquots of molecules from the LOPAC (Sigma, St Louis, MO) and Prestwick (Prestwick Chemical, Illkirch, France) libraries. The final concentration of LOPAC compounds was 5 μmol/L (0.05% dimethyl sulfoxide [DMSO]), whereas for the Prestwick library, 10 ng of each sample was added, resulting in an average final concentration of approximately 5 μmol/L in 0.1% DMSO. Control wells, treated with vehicle alone, containing consistent levels of DMSO, were distributed in the first and last columns of the plate to monitor signal variability. Cells were incubated with the molecules at 37°C for 20 hours. After incubation, cyclin D2 transactivation was assessed by the luciferase assay and viability was assessed by the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay.
Luciferase assay
Luciferase activity was measured according to the manufacturer's instructions (Promega, Madison, WI). In brief, the cell culture medium was removed using an Embla plate washer (Molecular Devices, Sunnyvale, CA) and 1× Glo Lysis buffer (Promega, Madison, WI) was added by the robotic liquid handler. After a 10-minute incubation, an equal volume of Bright-Glo Luciferase substrate (Promega) was added, and the luminescence signal was detected with a 96-well Luminoskan luminescence plate reader (Thermo Fisher Scientific, Waltham, MA) with a 5-second integration.
MTS assay
Cell viability was assessed with the CellTiter96 aqueous nonradioactive assay kit according to the manufacturer's instructions (Promega) and as described previously.14
Immunoblotting
Cytosolic extracts were prepared from NIH3T3 cells and myeloma cells as described previously.14 Cells were washed with phosphate-buffered saline, pH 7.4, and suspended in lysis buffer (10 mmol/L Tris, pH 7.4, 150 mmol/L NaCl, 0.1% Triton X-100, 0.5% sodium deoxycholate, and 5 mmol/L EDTA) containing protease inhibitors (Complete tablets; Roche, Indianapolis, IN). Protein concentrations were determined by the Bradford assay.15 Immunoblot assays were performed as described previously.16 In brief, equal amounts of protein were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gels followed by transfer to polyvinylidene difluoride membranes. Membranes were probed with polyclonal rabbit anti–human c-maf (0.5 μg/mL) or rabbit anti–human cyclin D2 (0.5 μg/mL), both from Santa Cruz Biotechnology (Santa Cruz, CA), or with monoclonal mouse anti–human X-linked inhibitor of apoptosis protein (XIAP, 0.25 μg/mL; BD Transduction Laboratories, Lexington, KY), monoclonal mouse anti–Bcl-2 (a gift from JC Reed, Burnham Institute, CA), and monoclonal mouse anti-β–actin (1:10 000 [v/v]) (Sigma). Secondary antibodies (GE Healthcare, Chalfont St Giles, United Kingdom) were horseradish peroxidase-conjugated goat anti mouse IgG (1:10 000 [v/v]) and antirabbit (1:5000 [v/v]). Detection was performed by the enhanced chemical luminescence method (Pierce, Rockford, IL).
Immunoprecipitation
NIH3T3 cells were transfected with c-maf along with ubiquitin C or GFP cDNA. Twenty-four hours after transfection, cells were lysed in radioimmunoprecipitation assay buffer in the presence of protease inhibitors. After clarification by centrifugation, cell lysates were incubated with anti- hemagglutinin (HA) beads (Roche) overnight at 4°C. The precipitated proteins were analyzed by SDS-polyacrylamide gels, and c-maf was detected as above.
Assessment of β-integrin expression
Myeloma cell lines were treated with dexamethasone or buffer control. After incubation, cells were harvested, and β-integrin 7 surface expression was measured by staining cells with anti-β–integrin 7-fluorescein isothiocyanate (FITC) and flow cytometric analysis.
Quantitative real-time polymerase chain reaction
The cDNAs encoding the c-maf, ubiquitin C, proteasome subunit C3, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were amplified using the following primer pairs: c-maf, forward, 5′-AAAAAGGAACCGGTGGAGAC-3′; reverse, 5′-GGTAGCCGGTCATCCAGTAG-3′; ubiquitin C, forward, 5′-CTTTCCAGAGAGCGGAACAG-3′; reverse, 5′-ATCACAGCGATCCACAAACA-3′, proteasome subunit C3, forward, 5′-TGGAATCTGCAATGAAGCTG-3′; reverse, 5′-TGCAAAAAGTCTGCAAAACAA-3′-3′; and GAPDH, forward, 5′-GAAGGTGAAGGTCGGAGTC-3′; reverse, 5′-GAAGATGGTGATGGGATTTC-3′. Equal amounts of cDNA for each sample were added to a prepared master mix (SYBR Green PCR Master mix; Applied Biosystems, Foster City, CA). Quantitative real-time polymerase chain reaction (RTPCR) reactions were performed on an ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, CA) as described previously.14 The relative abundance of a transcript was represented by the threshold cycle of amplification (CT), which is inversely correlated to the amount of target RNA/first-strand cDNA being amplified. To normalize for equal amounts of the latter, we assayed the transcript levels of the putative housekeeping gene GAPDH. The comparative CT method was calculated per the manufacturer's instructions. The expression level of c-maf relative to the baseline level was calculated as 2−ΔΔCT(c-maf), where ΔCT is (average c-maf CT − average GAPDH CT) and is ΔΔCT (average ΔCT-untreated sample − average ΔCT-treated sample.
Results
High-throughput assay for inhibitors of c-maf–dependent transactivation of cyclin D2
c-maf and its target gene cyclin D2 are frequently overexpressed in multiple myeloma and contribute to the pathogenesis and chemoresistance of this disease. To better understand the regulation of c-maf and cyclin D2, we sought to identify c-maf–dependent inhibitors of transactivation of the cyclin D2 promoter. To identify such inhibitors, we developed a high-throughput chemical genomics screen using NIH3T3 cells stably overexpressing a c-maf-IRES-GFP cassette in an MIEV retroviral vector and the cyclin D2 promoter driving firefly luciferase. In the optimized automated assay, NIH3T3 cells overexpressing c-maf and the cyclin D2 promoter driving luciferase were plated in 96-well plates by a robotic liquid handler. Six hours after plating, when the cells had adhered to the plates, they were treated with aliquots of molecules from the compound libraries at a final concentration of approximately 5 μmol/L and less than 0.1% DMSO. As a control, cells were treated with buffer alone. Cells were incubated with the molecules at 37°C for 20 hours. After incubation, cyclin D2 transactivation was assessed by luciferase assay. In parallel, we also assessed cell viability with an MTS assay. Thus, each compound from the library was tested in a luciferase assay for cyclin D2 activity and an MTS assay for viability (Figure 1).
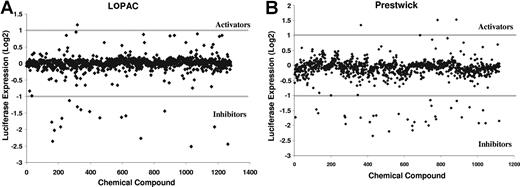
Identification of small-molecule inhibitors of the cyclin D2 promoter. NIH3T3 cells overexpressing c-maf and the cyclin D2 promoter driving luciferase were plated in 96-well plates (13 000 cells per well) by a robotic liquid handler. After the cells had adhered to the plates, they were treated with aliquots of molecules from the LOPAC and Prestwick libraries of off-patent drugs and chemicals at a final concentration of approximately 5 μmol/L and approximately 0.1% DMSO. As a control, cells were treated with DMSO alone. Cells were incubated with the molecules at 37°C in a humid atmosphere for 20 hours. After incubation, cyclin D2 transactivation was assessed by the luciferase assay. In parallel, we also assessed viability with an MTS assay. The results of the screens are shown. The activity of the compound is expressed as log2((sample luciferase RFU/control luciferase RFU)/(sample MTS OD/control MTS OD)). Compounds with an activity less than −1 were considered inhibitors.
Identification of small-molecule inhibitors of the cyclin D2 promoter. NIH3T3 cells overexpressing c-maf and the cyclin D2 promoter driving luciferase were plated in 96-well plates (13 000 cells per well) by a robotic liquid handler. After the cells had adhered to the plates, they were treated with aliquots of molecules from the LOPAC and Prestwick libraries of off-patent drugs and chemicals at a final concentration of approximately 5 μmol/L and approximately 0.1% DMSO. As a control, cells were treated with DMSO alone. Cells were incubated with the molecules at 37°C in a humid atmosphere for 20 hours. After incubation, cyclin D2 transactivation was assessed by the luciferase assay. In parallel, we also assessed viability with an MTS assay. The results of the screens are shown. The activity of the compound is expressed as log2((sample luciferase RFU/control luciferase RFU)/(sample MTS OD/control MTS OD)). Compounds with an activity less than −1 were considered inhibitors.
With this approach, we screened the LOPAC (n = 1280) and Prestwick (n = 1120) libraries of pharmacologically active chemicals and off-patent drugs, respectively, to identify inhibitors of the cyclin D2 promoter. To assess the robustness of the screen, we calculated the Z score of the assay, which is defined as 1 − ((3SD sample + 3SD control)/(mean sample − negative mean control)).17 The Z factor of the combined luciferase and MTS assays was 0.58, where a Z factor of 1 is ideal and a Z factor above 0.5 denotes a very good screening quality.17
Hits from the screening assay were defined as compounds that preferentially reduced luciferase activity over reductions in viability. Hits were empirically defined mathematically as log 2((sample luciferase/control luciferase)/(sample MTS OD/control MTS OD)) < −1, which corresponds to a 50% reduction in relative luciferase expression over viability.
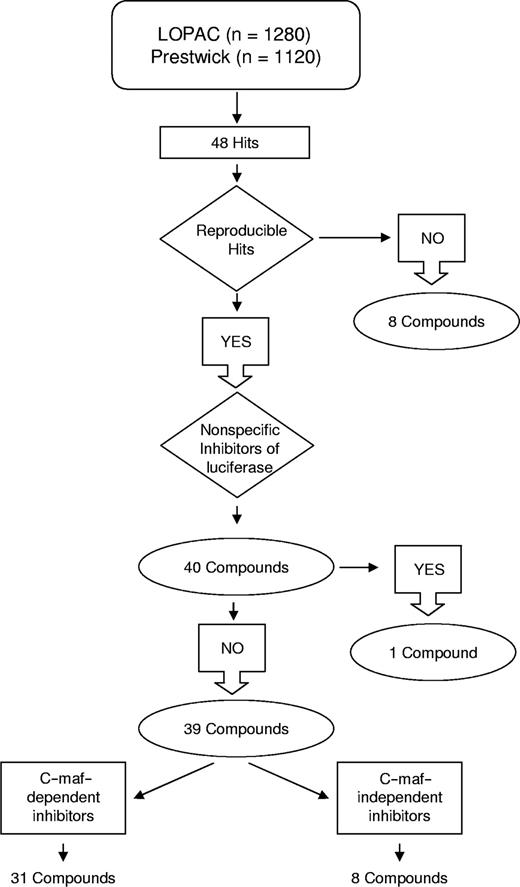
This screen identified 48 hits, of which 40 were reproducible on repeat testing (hit rate = 1.67%; Figures 2 and 3). The work flow of the screen is shown in Figure 2. Thirty-two of the reproducible hits were unique to either library, whereas 8 hits were present in both libraries. Using Cluster and Treeview gene expression algorithms,18 we determined that 24 of the 32 unique hits belonged to the corticosteroid family of drugs (Figure 3A). Indeed, the combined libraries contained 26 corticosteroids, of which 24 were identified as hits; the remaining 2 corticosteroids in the libraries were weakly positive.
Workflow of the screen for inhibitors of the cyclin D2 promoter. Hits from the high-throughput screen were assessed for reproducibility and selectivity for c-maf. Of the 31 c-maf–dependent inhibitors identified, 24 were unique to one of the libraries tested, and 7 were detected in the screens of both libraries. Of the 8 c-maf–independent inhibitors, 7 were unique to one of the libraries, and 1 was identified in both screens.
Workflow of the screen for inhibitors of the cyclin D2 promoter. Hits from the high-throughput screen were assessed for reproducibility and selectivity for c-maf. Of the 31 c-maf–dependent inhibitors identified, 24 were unique to one of the libraries tested, and 7 were detected in the screens of both libraries. Of the 8 c-maf–independent inhibitors, 7 were unique to one of the libraries, and 1 was identified in both screens.
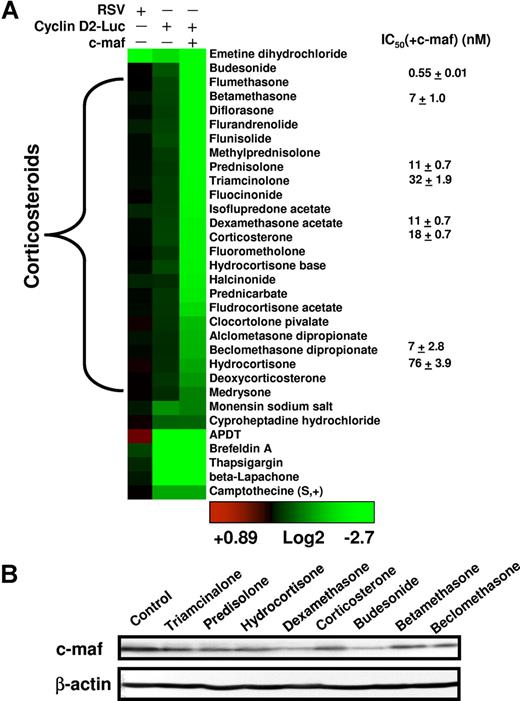
Clustering and characterization of small-molecule inhibitors of the cyclin D2 promoter. (A) The numeric value of the inhibition of cyclin D2 transactivation was represented colorimetrically. Drugs were assigned into families based on the annotation from the LOPAC and Prestwick libraries and clustered using the Cluster and Treeview algorithms.18 The results of the representative reproducible hits are shown. To determine whether molecules identified in the screen were nonspecific inhibitors of luciferase, preferential inhibitors of c-maf–dependent cyclin D2 transactivation, or c-maf–independent inhibitors of cyclin D2 transactivation, hits were tested in NIH3T3 cells overexpressing the RSV promoter driving luciferase, NIH3T3 cells overexpressing c-maf and the cyclin D2 promoter driving luciferase, and NIH3T3 cells without c-maf but overexpressing the cyclin D2 promoter driving luciferase. Molecules were tested at a final concentration of 5 μmol/L. Selected glucocorticoids were tested in dose-response studies in NIH3T3 cells overexpressing c-maf and the cyclin D2 promoter driving luciferase. The mean IC50(± SD) is shown, where the IC50 represents the concentration of the compound required to reduce luciferase activity by 50% from untreated control cells. (B) NIH3T3 cells overexpressing c-maf were treated with selected glucocorticoids (final concentration 5 μmol/L). Twenty-four hours after treatment, cell lysates were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotted using antibodies specific for c-maf and β-actin.
Clustering and characterization of small-molecule inhibitors of the cyclin D2 promoter. (A) The numeric value of the inhibition of cyclin D2 transactivation was represented colorimetrically. Drugs were assigned into families based on the annotation from the LOPAC and Prestwick libraries and clustered using the Cluster and Treeview algorithms.18 The results of the representative reproducible hits are shown. To determine whether molecules identified in the screen were nonspecific inhibitors of luciferase, preferential inhibitors of c-maf–dependent cyclin D2 transactivation, or c-maf–independent inhibitors of cyclin D2 transactivation, hits were tested in NIH3T3 cells overexpressing the RSV promoter driving luciferase, NIH3T3 cells overexpressing c-maf and the cyclin D2 promoter driving luciferase, and NIH3T3 cells without c-maf but overexpressing the cyclin D2 promoter driving luciferase. Molecules were tested at a final concentration of 5 μmol/L. Selected glucocorticoids were tested in dose-response studies in NIH3T3 cells overexpressing c-maf and the cyclin D2 promoter driving luciferase. The mean IC50(± SD) is shown, where the IC50 represents the concentration of the compound required to reduce luciferase activity by 50% from untreated control cells. (B) NIH3T3 cells overexpressing c-maf were treated with selected glucocorticoids (final concentration 5 μmol/L). Twenty-four hours after treatment, cell lysates were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotted using antibodies specific for c-maf and β-actin.
Exclusion of nonspecific inhibitors
In our assay, drugs that decreased the luciferase signal without affecting viability may have reduced transactivation of cyclin D2 or may have acted nonspecifically on the activity of the luciferase enzyme. To distinguish between these 2 possibilities, hits were retested in dose-response studies in NIH3T3 cells overexpressing luciferase driven by a Rous sarcoma virus (RSV) promoter. One compound decreased luciferase nonspecifically, whereas the other 39 hits (31 unique compounds), including all corticosteroids, preferentially inhibited the cyclin D2 promoter (Figure 3A).
c-maf–dependent versus independent inhibitors of cyclin D2
Hits that decreased cyclin D2 transactivation may have acted through c-maf–dependent or independent mechanisms. To distinguish between these possibilities, hits were tested in NIH3T3 cells that did and did not have detectable levels of c-maf by immunoblotting but expressed the cyclin D2 promoter-Luciferase construct. Seven compounds reduced cyclin D2 transactivation equally in cells with or without c-maf expression. We concluded that these compounds inhibited cyclin D2 transactivation independently of c-maf. Conversely, 31 compounds (24 unique) preferentially inhibited cyclin D2 transactivation in the presence of c-maf (Figures 2 and 3A), suggesting that these may represent c-maf–dependent inhibitors of cyclin D2.
Glucocorticoids inhibited c-maf–dependent cyclin D2 transactivation
All 24 compounds identified as c-maf–dependent cyclin D2 inhibitors were corticosteroid derivatives. The most potent inhibitors were glucocorticoids such as dexamethasone. Mineralocorticoids such as fludrocortisone were weak hits, which probably reflects their weak glucocorticoid activity at higher concentrations. Glucocorticoids inhibited cyclin D2 transactivation in NIH3T3 cells in a c-maf–dependent manner (Figure 3A). For example, the IC50 of dexamethasone was 11 (± 0.7) nM in NIH 3T3 cells overexpressing c-maf, whereas in NIH3T3 cells without c-maf expression, the IC50 was more than 10 μmol/L (Figure 3A). Furthermore, when we assessed c-maf protein levels, we observed that glucocorticoids decreased levels of c-maf protein in c-maf overexpressing NIH3T3 cells (Figure 3B). Glucocorticoids did not reduce the viability of c-maf expressing of c-maf–deficient NIH3T3 cells at concentrations up to 50 μmol/L (data not shown). These data suggest that glucocorticoids may inhibit c-maf–dependent transactivation of cyclin D2 by regulating c-maf levels.
Glucocorticoid-mediated decreases in c-maf may be due to inhibition of the retroviral promoter, driving expression of c-maf expression from the c-maf-IRES-GFP cassette. To exclude this possibility, GFP expression was measured in the NIH3T3 cells by flow cytometry after treatment with glucocorticoids. No changes in GFP expression were detected up to 72 hours after treatment (data not shown). In addition, when c-maf was introduced into NIH3T3 cells under the control of a CMV promoter in a pcDNA3.1 vector, glucocorticoids continued to inhibit c-maf–dependent cyclin D2 transactivation (data not shown). Thus, these results together with the studies outlined below in myeloma cells (Figure 4) indicate that the effects of glucocorticoids on c-maf and the cyclin D2 promoter are not due to artifactual inhibition of the viral LTR promoter driving the c-maf-IRES-GFP cassette.
Glucocorticoids reduced c-maf protein and its targets in multiple myeloma cells
Given the effects of glucocorticoids on c-maf in NIH3T3 cells, we extended our investigation to multiple myeloma cell lines and tested the effects of glucocorticoids on c-maf and its targets in human myeloma cell lines. For these experiments, we focused on dexamethasone, given that it is used therapeutically for the treatment of multiple myeloma. In myeloma cell lines (such as OCI-MY5, KMS11, and RPMI 8226 cells, which harbor the c-maf t(14;16) translocation, LP-1 cells that overexpresses c-maf but lack the t(14;16) translocation, and OPM-2 cells that have the t(14;20) mafB translocation19 ) dexamethasone reduced levels of c-maf and its target cyclin D2 but did not alter levels of unrelated proteins such as XIAP and Bcl-2 (Figure 4A-C and data not shown). The effects of dexamethasone were also evaluated in an expanded panel of myeloma cells. Dexamethasone decreased levels of cyclin D2 in cells that expressed c-maf, but no change was observed in cells such as U266, KHM11, and KMS12 that lacked detectable levels of c-maf by immunoblotting (Figure 4C). In U266 and KMS12 cells, a second band that migrated higher than cyclin D2 was detected and may be cyclin D1, given that the antibodies cross-react.
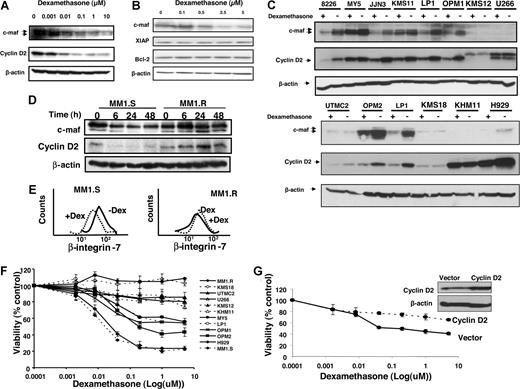
Dexamethasone decreases expression of c-maf and its target genes in multiple myeloma cell lines. LP-1 myeloma cells that overexpress c-maf but lack the t(14;16) translocation (A) and RPMI 8226 myeloma cells that have the t(14;16) translocation (B) were treated with increasing concentrations of dexamethasone. After treatment, cell lysates were immunoblotted using antibodies specific for c-maf, cyclin D2, XIAP, Bcl-2, and β-actin. (C) Myeloma cell lines were treated with dexamethasone (5 μmol/L) or buffer control for 24 hours. Total cell lysates were immunoblotted using antibodies specific for c-maf, cyclin D2, and β-actin. (D) MM1.S and the paired cell line MM1.R that lacks the glucocorticoid receptor were treated with dexamethasone (5 μmol/L). At increasing times after treatment, cell lysates were immunoblotted using antibodies specific for c-maf, cyclin D2, and β-actin. (E) MM1.S (glucocorticoid sensitive) and the paired cell line glucocorticoid-resistant MM1.R were treated with dexamethasone (DEX; 5 μmol/L) for 48 hours. After incubation, β-integrin-7 surface expression was measured by staining cells with anti-β–integrin-7-FITC, and flow cytometric analysis was performed. (F) Myeloma cell lines were treated with increasing concentrations of dexamethasone. Cell viability was measured 48 hours later by MTS assay. Cell viability is expressed as a mean percentage (± SD; n = 3) relative to untreated cells. (G) MM1.S cells were transfected with cDNA corresponding to cyclin D2 or empty vector. Twenty-four hours after transfection, cells were treated with increasing concentrations of dexamethasone for 24 hours. Cell viability was measured by MTS assay and expressed as a mean percentage (± SD; n = 3) relative to control cells. Inset: expression of cyclin D2 and β-actin by immunoblotting cell lysates from MM1.S cells transfected with cyclin D2 or empty vector.
Dexamethasone decreases expression of c-maf and its target genes in multiple myeloma cell lines. LP-1 myeloma cells that overexpress c-maf but lack the t(14;16) translocation (A) and RPMI 8226 myeloma cells that have the t(14;16) translocation (B) were treated with increasing concentrations of dexamethasone. After treatment, cell lysates were immunoblotted using antibodies specific for c-maf, cyclin D2, XIAP, Bcl-2, and β-actin. (C) Myeloma cell lines were treated with dexamethasone (5 μmol/L) or buffer control for 24 hours. Total cell lysates were immunoblotted using antibodies specific for c-maf, cyclin D2, and β-actin. (D) MM1.S and the paired cell line MM1.R that lacks the glucocorticoid receptor were treated with dexamethasone (5 μmol/L). At increasing times after treatment, cell lysates were immunoblotted using antibodies specific for c-maf, cyclin D2, and β-actin. (E) MM1.S (glucocorticoid sensitive) and the paired cell line glucocorticoid-resistant MM1.R were treated with dexamethasone (DEX; 5 μmol/L) for 48 hours. After incubation, β-integrin-7 surface expression was measured by staining cells with anti-β–integrin-7-FITC, and flow cytometric analysis was performed. (F) Myeloma cell lines were treated with increasing concentrations of dexamethasone. Cell viability was measured 48 hours later by MTS assay. Cell viability is expressed as a mean percentage (± SD; n = 3) relative to untreated cells. (G) MM1.S cells were transfected with cDNA corresponding to cyclin D2 or empty vector. Twenty-four hours after transfection, cells were treated with increasing concentrations of dexamethasone for 24 hours. Cell viability was measured by MTS assay and expressed as a mean percentage (± SD; n = 3) relative to control cells. Inset: expression of cyclin D2 and β-actin by immunoblotting cell lysates from MM1.S cells transfected with cyclin D2 or empty vector.
To determine whether glucocorticoid-mediated reductions in c-maf were dependent on the glucocorticoid receptor, we tested the effects of dexamethasone in the myeloma cell lines MM1.S and its glucocorticoid-resistant counterpart MM1.R, which lacks a functional glucocorticoid receptor. Dexamethasone decreased c-maf and its downstream targets cyclin D2 in MM1.S myeloma cells within 6 hours of treatment, but did not reduce these proteins in the MM1.R that lacks the glucocorticoid receptor up to 48 hours after treatment (Figure 4D). We also examined another known target of c-maf, β-integrin, and observed that dexamethasone decreased surface β-integrin expression in MM1.S but not in MM1.R cells (Figure 4E).
To investigate the relationship between reductions in c-maf and loss of viability by glucocorticoids, myeloma cell lines were treated with increasing concentrations of dexamethasone and cell viability was measured 48 hours later by the MTS assay. Dexamethasone reduced viability of cells such as LP-1, MM1.S, and MY-5, cells at concentrations associated with reductions in c-maf. In contrast, U266 and KHM11 cells with undetectable levels of c-maf and KMS18 cells whose levels of c-maf did not decrease after dexamethasone treatment were resistant to dexamethasone-induced death at 48 hours (Figure 4F). In addition, in dexamethasone-sensitive MM1.S myeloma cells, transfection and overexpression of cyclin D2 cDNA partially inhibited dexamethasone-mediated cell death (Figure 4G). Taken together, decreases in c-maf and cyclin D2 by dexamethasone may be one of several mechanisms by which dexamethasone exerts its antimyeloma effect. However, reductions in c-maf may merely reflect the biologic activity of glucocorticoids and may not necessarily relate to their mechanism of toxicity.
Glucocorticoids promoted the ubiquitination of c-maf
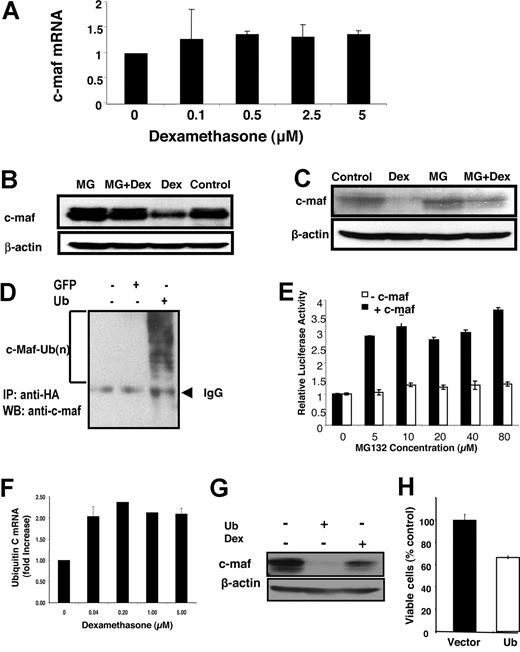
To determine the mechanism by which glucocorticoids reduce c-maf, we tested the effects of glucocorticoids on c-maf protein and mRNA levels. In myeloma cell lines such as OCI-MY5, LP-1, and RPMI 8226, dexamethasone reduced c-maf protein within 8 hours of treatment but did not alter expression of c-maf mRNA up to 48 hours after treatment (Figure 5A and data not shown). Therefore, we concluded that glucocorticoids reduce c-maf through post-transcriptional regulation.
Glucocorticoids induce c-maf degradation through the ubiquitination-proteasome pathway. (A) RPMI 8226 myeloma cells were treated with increasing concentrations of dexamethasone for 48 hours. After treatment, mRNA was extracted. Levels of c-maf and GAPDH were detected by quantitative real-time PCR. Levels of c-maf were normalized for GAPDH expression and expressed as a mean fold change (± SD) over buffer-treated cells. (B) NIH3T3 cells overexpressing c-maf were treated with dexamethasone (DEX; 2 μmol/L) in the absence or presence of the proteasome inhibitor MG132 (MG; 10 μmol/L). After treatment, cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for c-maf and β-actin. (C) LP-1 myeloma cells overexpressing c-maf were treated with MG132 (10 μmol/L) and/or dexamethasone (2 μmol/L). After treatment, cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for c-maf and b-actin. (D) NIH3T3 cells were cotransfected with c-maf along with HA-ubiquitin (Ub) or GFP cDNA. Twenty-four hours after transfection, cells were lysed. Cell lysates were incubated with anti-HA beads overnight. The resultant precipitates were analyzed by SDS-PAGE/immunoblotting using antibodies specific for c-maf. IP indicates immunoprecipitation; and WB, Western blotting. (E) NIH3T3 cells overexpressing c-maf and the cyclin D2 promoter driving luciferase, and NIH3T3 cells without c-maf but overexpressing the cyclin D2 promoter driving luciferase were treated with dexamethasone (2 μmol/L) and increasing concentrations of MG132. After incubation, cyclin D2 transactivation was measured by luciferase assay and viability measured by MTS assay. The relative luciferase expression represents the mean of 3 independent experiments. (F) LP-1 myeloma cells were treated with increasing concentrations of dexamethasone for 24 hours. After treatment, total mRNA was extracted. Levels of ubiquitin C and GAPDH were measured by quantitative RT-PCR. Ubiquitin C expression was normalized for GAPDH and expressed as a mean fold change (± SD) over buffer-treated cells. (G) NIH3T3 cells overexpressing c-maf were transfected with ubiquitin cDNA or vector control, or treated with dexamethasone (5 μmol/L). Forty-eight hours after transfection, whole-cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for c-maf and β-actin. (H) LP-1 cells were transfected with cDNA corresponding to ubiquitin or empty vector. Twenty-four hours after transfection, cell viability was measured by MTS assay and expressed as a mean percentage (± SD; n = 3) relative to control cells.
Glucocorticoids induce c-maf degradation through the ubiquitination-proteasome pathway. (A) RPMI 8226 myeloma cells were treated with increasing concentrations of dexamethasone for 48 hours. After treatment, mRNA was extracted. Levels of c-maf and GAPDH were detected by quantitative real-time PCR. Levels of c-maf were normalized for GAPDH expression and expressed as a mean fold change (± SD) over buffer-treated cells. (B) NIH3T3 cells overexpressing c-maf were treated with dexamethasone (DEX; 2 μmol/L) in the absence or presence of the proteasome inhibitor MG132 (MG; 10 μmol/L). After treatment, cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for c-maf and β-actin. (C) LP-1 myeloma cells overexpressing c-maf were treated with MG132 (10 μmol/L) and/or dexamethasone (2 μmol/L). After treatment, cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for c-maf and b-actin. (D) NIH3T3 cells were cotransfected with c-maf along with HA-ubiquitin (Ub) or GFP cDNA. Twenty-four hours after transfection, cells were lysed. Cell lysates were incubated with anti-HA beads overnight. The resultant precipitates were analyzed by SDS-PAGE/immunoblotting using antibodies specific for c-maf. IP indicates immunoprecipitation; and WB, Western blotting. (E) NIH3T3 cells overexpressing c-maf and the cyclin D2 promoter driving luciferase, and NIH3T3 cells without c-maf but overexpressing the cyclin D2 promoter driving luciferase were treated with dexamethasone (2 μmol/L) and increasing concentrations of MG132. After incubation, cyclin D2 transactivation was measured by luciferase assay and viability measured by MTS assay. The relative luciferase expression represents the mean of 3 independent experiments. (F) LP-1 myeloma cells were treated with increasing concentrations of dexamethasone for 24 hours. After treatment, total mRNA was extracted. Levels of ubiquitin C and GAPDH were measured by quantitative RT-PCR. Ubiquitin C expression was normalized for GAPDH and expressed as a mean fold change (± SD) over buffer-treated cells. (G) NIH3T3 cells overexpressing c-maf were transfected with ubiquitin cDNA or vector control, or treated with dexamethasone (5 μmol/L). Forty-eight hours after transfection, whole-cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for c-maf and β-actin. (H) LP-1 cells were transfected with cDNA corresponding to ubiquitin or empty vector. Twenty-four hours after transfection, cell viability was measured by MTS assay and expressed as a mean percentage (± SD; n = 3) relative to control cells.
Rapid decreases in protein levels can be related to increased destruction through the ubiquitination-proteasome pathway. Therefore, we tested whether glucocorticoids promote the ubiquitination of c-maf. NIH3T3 overexpressing c-maf and the endogenously c-maf expressing multiple myeloma cell line LP-1 were treated with dexamethasone with or without the proteasome inhibitor MG132. After treatment, levels of c-maf protein were measured by immunoblotting. MG132 abrogated dexamethasone-mediated reductions in c-maf (Figure 5B,C). Conversely, the MEK inhibitor U0126 had no effect on dexamethasone-mediated reductions in c-maf protein accumulation (data not shown). In keeping with these findings, we confirmed that c-maf is ubiquitinated in LP-1 cells (Figure 5D) and that pretreatment with MG132 also blocked the inhibition of cyclin D2 transactivation by dexamethasone (Figure 5E).
Glucocorticoids can up-regulate genes in the ubiquitin pathway of proteasomal degradation. Therefore, we measured changes in ubiquitin-related genes in LP-1 myeloma cells after treatment with increasing concentrations of dexamethasone. As measured by quantitative real-time PCR, dexamethasone increased expression of ubiquitin C mRNA at concentrations of the compound that were associated with decreases in c-maf protein (Figure 5F). Conversely, no change in expression of the proteasome subunit C3 mRNA was detected after dexamethasone treatment (data not shown). To determine whether increased levels of ubiquitin C are functionally important in regulating c-maf levels, we transfected the ubiquitin C cDNA into c-maf overexpressing NIH3T3 cells. Overexpression of ubiquitin but not vector control decreased expression of c-maf protein (Figure 5G). It is noteworthy that transfection of LP-1 cells with ubiquitin cDNA increased the abundance of ubiquitinated proteins by immunoblotting and decreased cell viability compared with cells transfected with vector control (Figure 5H and data not shown). Thus, taken together, these results suggest a novel finding that levels of c-maf protein can be regulated by ubiquitination and the abundance of the ubiquitin substrate.
Discussion
Dysregulation of c-maf and its target genes such as cyclin D2 occur frequently in multiple myeloma and contribute to the pathogenesis of this disease.6,11,13,20 Molecules that inhibit c-maf and its targets could be useful chemical probes to better understand the regulation of these proteins. To identify such molecules, we developed a cell-based assay to identify c-maf regulators based on the ability of c-maf to transactivate the cyclin D2 promoter. We conducted our screen in NIH3T3 cells because their viability is not decreased after inhibiting c-maf or cyclin D2 (data not shown). With this assay, we screened 2400 off-patent drugs and chemicals representing a relatively large variety of bioactive and pharmacologically active pharmacophores and identified glucocorticoids as c-maf–dependent inhibitors of cyclin D2 transactivation. Demonstrating the robustness of the screen, the assay identified 26 of the 28 corticosteroids in the libraries as hits in the screen, and the remaining 2 corticosteroids were weak hits in the assay.
The results from the NIH3T3 cells were then extended to myeloma cell lines. In myeloma cell lines, glucocorticoids such as dexamethasone also decreased c-maf and 2 of its principal target genes, including cyclin D2 and β-integrin. Reductions in c-maf target genes occurred at concentrations and times of incubation associated with reductions in c-maf. Why some cell lines are more sensitive to dexamethasone-mediated reductions in c-maf is unknown; this finding will be investigated in future studies. It is possible that the difference in response may relate to the differences in the sensitivity to up-regulate ubiquitin or the amount of ubiquitin needed to increase the proteasomal degradation of c-maf.
Few prior studies have investigated the mechanisms governing the regulation of c-maf and its related family members. These studies have demonstrated that expression of maf-family members can be regulated at the transcriptional level.21,22 In addition, phosphorylation of the c-maf homologues MafA and L-maf is required for their function.23,24 Prior studies have also demonstrated that phosphorylation of L-maf increases its stability and decreases its proteasomal degradation.24 Our study has identified a novel mechanism for the regulation of c-maf protein levels. We have demonstrated that c-maf proteasomal degradation can be enhanced by increasing levels of the ubiquitin substrate. Typically, the rate limiting step in the ubiquitination pathway is the activity of the E3 ligase. To our knowledge this report is the first to describe a specific protein whose levels can be regulated by the abundance of the ubiquitin substrate. Further studies will also be required to determine the extent to which other proteins are also regulated by levels of the ubiquitin substrate.
The mechanism of action of glucocorticoids is complex and not fully understood (reviewed in Frankfurt and Rosen25 and Greenstein et al26 ). Glucocorticoids passively enter cells where they bind the soluble glucocorticoid receptor. After ligand binding, the glucocorticoid receptor and its ligand translocate to the nucleus.27 In the nucleus, the glucocorticoid receptor binds DNA and regulates transcription of genes including activator protein-1 (AP-1), nuclear factor of activated T cells, and nuclear factor-κB.28-30 In this study, we demonstrated that glucocorticoids reduced c-maf protein but not c-maf mRNA. We showed subsequently that glucocorticoids increase the ubiquitination of c-maf, potentially by increasing the expression of ubiquitin C mRNA.
Our work is consistent with a previous study in which up-regulation of ubiquitin mRNA was detected in a microarray analysis of genes altered by dexamethasone treatment.31 However, the effect of increased ubiquitin C in myeloma cells was not reported. Future studies will explore the mechanism by which dexamethasone increases ubiquitin expression.
Some of our results suggest that the antimyeloma effects of glucocorticoids could be related to the reductions in c-maf and cyclin D2. It is noteworthy that, in a recent randomized study, 40% of patients with newly diagnosed myeloma achieved at least a partial response after treatment with pulsed dexamethasone alone.32 In comparison, 6%–50% of myeloma patients overexpress c-maf through translocation-dependent and -independent mechanisms depending on the method used to assess its overexpression.6,9-12 The randomized study did not report levels of c-maf, so one cannot relate responses to dexamethasone to c-maf expression. However, glucocorticoids have multiple mechanisms of actions, and it is probably premature to conclude that reductions in c-maf are functionally important for its ability to induce cell death. Rather, reductions in c-maf and its targets may be markers of the biologic activity of glucocorticoids and not functionally important for its toxicity in myeloma cells. It is possible that resistance to glucocorticoids may have reflected increased cyclin D1 expression, because glucocorticoid-resistant cell lines such as KMS12 carry a t(11;14) translocation that activates cyclin D1.19 Even if reductions in c-maf do not contribute to the mechanism of glucocorticoid-induced death, these studies are important to understand the regulation of c-maf expression, an important biologic target in myeloma.
Our screen also identified c-maf–independent inhibitors of cyclin D2 transactivation. Overexpression of cyclin D2 independent of c-maf is another initiating event in the pathogenesis of multiple myeloma.1 Furthermore, increased levels of cyclin D2 contribute to the proliferation and chemoresistance of malignant cells.33-37 Therefore, these c-maf–independent inhibitors of cyclin D2 could be useful tools to better understand the regulation of cyclin D2. In addition, some of these molecules could be novel therapeutic agents for the treatment of multiple myeloma.
In summary, a high-throughput screen seeking inhibitors of cyclin D2 promoter identified glucocorticoids as c-maf–dependent inhibitors of cyclin D2 transactivation. Additional studies demonstrated a novel mechanism for the regulation of c-maf by glucocorticoids. We demonstrated that the ubiquitination and degradation of c-maf can be increased by the up-regulation of ubiquitin C mRNA. Thus, this work used a chemical biology approach to provide insights into a novel mechanism of regulation of c-maf in myeloma cells.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Multiple Myeloma Research Foundation, the Canadian Institutes of Health Research (CIHR), the Leukemia and Lymphoma Society of Canada, and the McLaughlin Centre for Molecular Medicine. A.D.S. is a CIHR Clinician Scientist.
Authorship
Contribution: X.M. designed the research, analyzed data, performed research, and wrote the manuscript. A.K.S., S.T., and S.E designed the research and analyzed data. R.H., A.D., X.Z., Y.Z., C.S., K.L., R.T., Y.E., S.L., and L.C.G. performed research and analyzed data. S.J.C. and J.L.W. contributed vital new reagents. D.L.B. designed the research. A.D.S. supervised research, designed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Aaron D. Schimmer, Princess Margaret Hospital, Rm 9–516, 610 University Ave, Toronto, ON, M5G 2M9 Canada; e-mail: aaron.schimmer@utoronto.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal