Abstract
2-Cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO) induces differentiation and apoptosis of tumor cells in vitro and in vivo. Here we assessed the effects of CDDO on CCAAT enhancer–binding protein alpha (CEBPA), a transcription factor critical for granulocytic differentiation. In HL60 acute myeloid leukemia (AML) cells, CDDO (0.01 to 2 μM) induces apoptosis in a dose-dependent manner. Conversely, subapoptotic doses of CDDO promote phagocytic activity and granulocytic-monocytic differentiation of HL60 cells through increased de novo synthesis of p42 CEBPA protein. CEBPA translational up-regulation is required for CDDO-induced granulocytic differentiation and depends on the integrity of the CEBPA upstream open reading frame (uORF). Moreover, CDDO increases the ratio of transcriptionally active p42 and the inactive p30 CEBPA isoform, which, in turn, leads to transcriptional activation of CEBPA-regulated genes (eg, GSCFR) and is associated with dephosphorylation of eIF2α and phosphorylation of eIF4E. In concordance with these results, CDDO induces a CEBPA ratio change and differentiation of primary blasts from patients with acute myeloid leukemia (AML). Because AML is characterized by arrested differentiation, our data suggest the inclusion of CDDO in the therapy of AML characterized by dysfunctional CEBPA expression.
Introduction
The triterpenoid 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO) and its derivatives, CDDO-methylester (CDDO-Me) and CDDO-imidazole (CDDO-Im), induce growth arrest and apoptosis of a variety of solid tumor and leukemic cell lines in vitro and in vivo.1,2 Different signaling pathways account for the proapoptotic and antiproliferative effects of CDDO. CDDO induces apoptosis through both caspase-independent and -dependent mechanisms, the latter involving caspase-8 activation, Bid cleavage, cytochrome c release, and caspase-3 activation.3-6 Furthermore, JNK, p38, and ERK pathways are involved in CDDO-induced apoptosis of tumor cell lines7-9 mediated by disrupted intracellular redox balance and involving decreased glutathione and increased reactive oxygen species9-12
CDDO-induced growth arrest of breast cancer cell lines correlates with transactivated PPARgamma and leads to up-regulation of p21cip1waf1, GADD153, CCAAT enhancer–binding proteins (CEBPs), and proteasome-regulatory factors, and to down-regulation of cyclin D1, PCNA, and IRS1.13 CDDO and CDDO-Im activate the TGFβ pathway through activation of Smad2/3,14,15 which is required for the repression of inflammatory molecules by CDDO.16
Interestingly, CDDO and its derivatives also induce differentiation of leukemic cells.1,2,7,17 Differentiation of normal hematopoietic stem cells into their mature progeny critically depends on a fine-tuned interplay of hematopoietic transcription factors.18 Among these, we have recently shown that increased CEBP beta (CEBPB) expression is critical for CDDO-Im–induced monocytic differentiation, and this was partially dependent upon ERK activation and TGFβ-mediated Smad activation.17 Granulocytic differentiation requires the presence of functional CEBPA, since mice with a targeted disruption of the CEBPA gene demonstrate a selective lack of granulocytes and an accumulation of immature myeloid cells.19 The expression and/or function of CEBPA is severely altered in a significant fraction of acute myeloid leukemia (AML) subtypes.20,21 CEBPA is mutated in 7% of all AML cases with normal cytogenetics, and this results in a balance shift from the transcriptionally active full-length isoform (p42) toward the dominant-negatively acting p30 isoform.22 The fusion protein AML1-ETO suppresses CEBPA transcription,23 and AML1-MDS1-EVI1 and CBFB-SMMHC oncogenes inhibit CEBPA translation through activation of the RNA-binding protein calreticulin.24,25 In these AML subtypes, re-expression of functional CEBPA restores granulocytic differentiation, suggesting that suppression of CEBPA is essential for the phenotype of AML blasts. Likewise, differentiation of myeloid progenitors in chronic myelogenous leukemia (CML) blast crisis is disturbed by BCR/ABL-induced suppression of CEBPA mRNA translation through the activation of the MAPK-hnRNP E2 pathway23,26 . Here, we provide evidence that CDDO potently induces granulocytic differentiation of leukemic cell lines and patient-derived primary AML blasts by translationally enhancing the expression and function of CEBPA through a mechanism that involves increase of p42 and the p42/p30 ratio. Moreover, we show that CDDO-induced CEBPA expression requires the integrity of the CEBPA uORF.
Patients, materials, and methods
IRB approval was obtained for both the assessment of the clinical samples (University Hospital of Muenster) and the phase 1 clinical study (M. D. Anderson Cancer Center). Informed consent was obtained in accordance with the Declaration of Helsinki.
Cells, transfection, and reagents
HL60, K562, 32Dcl3, WEHI-3, and HEK293A cells were obtained from ATCC (Manassas, VA) and were maintained in RPMI or DMEM (HEK293) medium, respectively, (Gibco/Invitrogen, Karlsruhe, Germany), with 10% fetal bovine serum (FBS) (Hyclone, Logan, UT), 1% penicillin-streptomycin (P/S), 1% glutamine (both from Sigma, Munich, Germany), and 10% WEHI-3 cell supernatant (32Dcl3). 6.15 (32D-BCR/ABL) cells have been previously described.26 Ficoll-separated bone marrow cells from patients with AML were used freshly or after freezing in liquid nitrogen and cultured in IMDM (Gibco) with 20% FBS, 1% P/S, and 1% glutamine. CDDO was synthesized by Dr Sporn, Dartmouth, NH, and was diluted in dimethyl sulfoxide (DMSO) to obtain working concentrations, and identical volumes of DMSO and CDDO (in DMSO) were added to the cultures. DMSO, all-trans retinoic acid (ATRA), cycloheximide, GW9662, calpain inhibitor I, 2-aminopurine (2-AP), calyculin A, transforming growth factor beta (TGFβ), and SB505124 were purchased from Sigma, Munich, Germany or Sigma, St Louis, MO.
For transient CEBPA expression, 8 × 105 HEK293A cells were seeded in 10-cm tissue culture dishes and transfected with 2.72 μg pSG5-rCEBPA-uORFwt or pSG5-rCEBPA-uORFopt (Dopt)27 DNA, using Fugene6 reagent (Roche Diagnostics, Basel, Switzerland). DMSO or 0.5 μM CDDO was added to the cultures for 2 hours, and protein lysates were prepared 24 hours after transfection using the TCA method.28
HL60 cell morphology and NBT assays
HL60 cells were cultured in the presence of CDDO, ATRA, or vehicle (DMSO) for up to 8 days, and cytospins were stained using Wright-Giemsa stain. HL60 cells (5 × 105) were incubated in phosphate-buffered saline (PBS), nitroblue tetrazolium (NBT), and 0.33 μM phorbol myristate acetate (Sigma, St Louis, MO) for 20 minutes at 37°C; the reaction was terminated by incubation on ice; and cytospins were prepared and counterstained with 0.5% safranin in 20% ethanol.
Phagocytosis assay
Phagocytosis was assessed using the Fluorescent Particles (E coli labeled with Alexa Fluor 488) and the Opsonization reagent (Molecular Probes/Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. After reconstitution and opsonization of the E coli bioparticles, HL60 cells were cultured for 2 days in the presence or absence of DMSO, ATRA, or CDDO. Seventy-five bioparticles per cell were added to 1 × 106 HL60 cells, and the mixture was incubated for 1 hour at 37°C. Cells were washed twice with PBS and subjected to flow cytometry for green fluorescence. For fluorescence microscopy, cytospun cells were fixed with Histochoice tissue fixative (Sigma) for 10 minutes; preparations were washed twice and mounted in ProlongFade (Molecular Probes). Light microscopy was performed with a Zeiss Axioplan microscope (Zeiss, Göttingen, Germany) using a 25× Plan-Neofluar 0.80, 63× Plan-Apochromat 1.4 oil, or 100× Plan-Neofluar 1.30 oil lens. Images were captured using Adobe Photoshop version 5.5 (Adobe Systems, San Jose, CA) and Microsoft Powerpoint 2000 (Microsoft, Redmond, WA).
Flow cytometry
Fluorescence-activated cell sorting (FACS) analysis was performed as described.29 Antibodies against the following antigens were used: CD4, CD11b, CD11c, CD14, CD15, CD16, HLA-DR, and isotype control (Becton Dickinson, Heidelberg, Germany). For analysis of apoptosis, the annexin V-FITC kit from Immunotech (Marseille, France) was used according to the manufacturer's instructions. Cells were assessed for annexin V/propidium iodide staining on day 2 of culture.
Northern blot and quantitative real-time RT-PCR analyses
Total RNA for Northern blot analysis was isolated using the AGPEP method (TriReagent; Molecular Research Center, Cincinnati, OH) from untreated and CDDO-treated HL60 cells. RNA (10 μg) was subjected to Northern blotting as described previously30 and hybridized to a 32P-labeled human granulocyte colony-stimulating factor receptor (G-CSFR; 0.72-kb SacII/NdeI fragment) or a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe (a 1.5-kb PstI fragment).30
For real-time reverse-transcription–polymerase chain reaction (RT-PCR), total RNA was isolated using the RNEasy Mini kit according to the manufacturer's recommendations (Qiagen, Hilden, Germany). Total RNA (1 μg) was used for reverse transcription with Superscript reverse transcriptase (Gibco). cDNA was diluted to 200 μL with ddH20, and 2.5 μL was used for each PCR reaction. Relative gene expression levels were calculated as follows: % of GAPDH expression = 100/(2CT[gene]−CT[GAPDH]). The primers and probes used are listed in Table S1, available on the Blood website; see the Supplemental Table link at the top of the online article. PCR reactions contained: 160 nM 5′ primer, 160 nM 3′ primer, 80 nM probe (for Taqman), 2.5 μL cDNA, 6.25 μL 2 × qPCR Mastermix (Plus QuickGoldStar for Taqman PCR and SYBR green I QuickGoldStar for SYBR green PCR, both from Eurogentec, Cologne, Germany), and filled to 12.5 μL with ddH2O. All primers and probes were purchased from Gibco/Invitrogen or Eurogentec. PCR conditions were as follows: 2 minutes at 50°C, 3 minutes at 94°C, followed by 40 cycles of 15 seconds at 94°C and 1 minute at 60°C.
Western blot analysis
Western blots were performed as described.28 Briefly, cells were washed with ice-cold phosphate-buffered saline (PBS) and lysed in radioimmunoprecipitation assay (RIPA) buffer or 10% trichloracetate. After pH adjustment, sample buffer was added immediately, and the samples were boiled for 10 minutes. The blots were probed with the following antibodies: rabbit anti-CEPBA, rabbit anti-HSP90, mouse anti–beta-tubulin, mouse anti–beta-actin, goat anti–rabbit or goat anti–mouse immunoglobulin G–horseradish peroxidase (IgG-HRP) (Santa Cruz Biotechnology, Santa Cruz, CA); rabbit anti-eIF2α, rabbit anti–phospho-eIF2α, rabbit anti-eIF4E, rabbit anti–phospho-eIF4E (Cell Signaling Technology, Danvers, MA); rabbit anticalreticulin (Sigma); and mouse anti-HA (Covance, Princeton, NJ); rabbit anti–hnRNP-E2 antibodies were a kind gift of Raul Andino (UCSF Comprehensive Cancer Center). Quantitative densitometric analysis was performed by scanning the autoradiographs with the Image J software (National Institutes of Health, http://rsb.info.nih.gov/ij/) (Table 1).
Densitometric analysis of CEBPA isoforms detected by Western blotting
| . | p42/p30 ratio . | Total CEBPA (p42+p30) . | p42 . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| DMSO . | CDDO . | DMSO/CDDO . | DMSO . | CDDO . | CDDO/DMSO . | DMSO . | CDDO . | CDDO/DMSO . | |
| Figure 3D | 0.54 | 1.53 | 2.85 | 1.89 | 1.77 | 0.93 | 0.66 | 1.07 | 1.62 |
| Figure 4A | 0.98 | 2.01 | 2.05 | 2.74 | 2.20 | 0.80 | 1.36 | 1.47 | 1.08 |
| Figure 4B | 0.83 | 1.95 | 2.36 | 1.45 | 1.87 | 1.29 | 0.66 | 1.24 | 1.89 |
| Figure 5A | 1.04 | 3.41 | 3.26 | 1.71 | 1.45 | 0.84 | 0.87 | 1.12 | 1.28 |
| Figure 5F | 0.47 | 1.36 | 2.91 | 1.13 | 1.59 | 1.42 | 0.36 | 0.92 | 2.56 |
| Figure 6A | 0.06 | 1.45 | 23.49 | 0.65 | 1.20 | 1.86 | 0.04 | 0.71 | 18.92 |
| Figure 6C | 1.33 | 2.34 | 1.76 | 1.54 | 1.11 | 0.72 | 0.88 | 0.78 | 0.89 |
| Figure 6D | 0.43 | 0.96 | 2.22 | 1.45 | 2.03 | 1.41 | 0.44 | 1.00 | 2.28 |
| Mean | 0.71 | 1.88 | 5.11 | 1.57 | 1.65 | 1.16 | 0.66 | 1.04 | 3.81 |
| SD | 0.41 | 0.75 | 7.44 | 0.61 | 0.39 | 0.40 | 0.40 | 0.25 | 6.13 |
| t test | — | 0.002 | — | — | 0.744 | — | — | 0.037 | — |
| . | p42/p30 ratio . | Total CEBPA (p42+p30) . | p42 . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| DMSO . | CDDO . | DMSO/CDDO . | DMSO . | CDDO . | CDDO/DMSO . | DMSO . | CDDO . | CDDO/DMSO . | |
| Figure 3D | 0.54 | 1.53 | 2.85 | 1.89 | 1.77 | 0.93 | 0.66 | 1.07 | 1.62 |
| Figure 4A | 0.98 | 2.01 | 2.05 | 2.74 | 2.20 | 0.80 | 1.36 | 1.47 | 1.08 |
| Figure 4B | 0.83 | 1.95 | 2.36 | 1.45 | 1.87 | 1.29 | 0.66 | 1.24 | 1.89 |
| Figure 5A | 1.04 | 3.41 | 3.26 | 1.71 | 1.45 | 0.84 | 0.87 | 1.12 | 1.28 |
| Figure 5F | 0.47 | 1.36 | 2.91 | 1.13 | 1.59 | 1.42 | 0.36 | 0.92 | 2.56 |
| Figure 6A | 0.06 | 1.45 | 23.49 | 0.65 | 1.20 | 1.86 | 0.04 | 0.71 | 18.92 |
| Figure 6C | 1.33 | 2.34 | 1.76 | 1.54 | 1.11 | 0.72 | 0.88 | 0.78 | 0.89 |
| Figure 6D | 0.43 | 0.96 | 2.22 | 1.45 | 2.03 | 1.41 | 0.44 | 1.00 | 2.28 |
| Mean | 0.71 | 1.88 | 5.11 | 1.57 | 1.65 | 1.16 | 0.66 | 1.04 | 3.81 |
| SD | 0.41 | 0.75 | 7.44 | 0.61 | 0.39 | 0.40 | 0.40 | 0.25 | 6.13 |
| t test | — | 0.002 | — | — | 0.744 | — | — | 0.037 | — |
This table shows the densitometric analysis of p42 and p30 CEBPA isoform and loading control bands as detected by Western blotting in Figures 3 through 6. DMSO and CDDO designate the type of stimulation of the HL60 cells, and the ratio of CDDO-induced changes is depicted as the DMSO/CDDO fraction. The p42/p30 ratio was calculated by dividing the densitometric values of p42 and p30. Total CEBPA (p42+p30) was calculated by addition of p42 and p30 bands and dividing this value by the densitometric value of the loading control band. p42 CEBPA was calculated by dividing the densitometric values of p42 and of the loading control band. Mean and standard deviation (SD) of all blots in the presented figures are shown, and DMSO- and CDDO-stimulated data were compared using a Student t test.
— indicates not applicable
EMSA
For electromobility shift assays (EMSAs), nuclear extracts were prepared as described.30 Briefly, 10 μg nuclear extracts from HL60 cells were incubated with double-stranded oligonucleotide derived from the G-CSF receptor promoter (bp −57 to −38): upper strand, 5′-AAGGTGTTGCAATCCCCAGC; lower strand, 5′-GCTGGGGATTGCAACACCTT. For supershift assays, 1 μL polyclonal anti-CEBPA antibody or monoclonal anti-CEBP beta antibodies (Santa Cruz Biotechnology) was added to the binding reaction. Binding reactions were resolved on a 4% polyacrylamide gel electrophoresis (PAGE)/1× TBE.30
Clinical trial of CDDO in patients with AML
Patients with refractory/relapsed AML were treated with CDDO (from 0.6 to 9.6 mg/m2 per hour × 5 days) in a phase 1 clinical trial, following informed consent according to the University of Texas M. D. Anderson Cancer Center guidelines (Table 2). Cells were collected from peripheral blood (PB) or bone marrow (BM) and assessed for expression of CD11b, CD14, and CD34 by flow cytometry at the indicated time points. In addition, routine blood counts and differentials were obtained daily and bone marrows were evaluated at baseline and on day 22 of treatment.
Patients with AML entered into the CDDO phase 1 clinical trial
| ID . | FAB . | Etiology . | Cytogenetics . | Dose level of RTA401 (CDDO), mg/m2 per h for 5 d . | Prior no. of Rx . |
|---|---|---|---|---|---|
| 301 | UNK | AHD | 46, XY, t(1;22)(p36.3;q11.2), del(5)(q13q33)[13], 48, XY, del(5)(q13q33),+22, +mar[7] | 0.6 | 1 |
| 302 | UNK | De novo | 46, XY, t(1;4)(p32;p16)[1], 46, XY[19] | 1.2 | 3 |
| 303 | RAEB-T | 2° | 45, X,−Y, der(3)ins(22;9)(q11.2;q34q34)t(3;22)(p23;q11.2), der(9)ins(22;9), der(22)t(3;22)[1], 44, X, −Y, der(3)ins(22;9)(q11.2;q34q34)t(3;22)(p23;q11.2), der(9)ins(22;9), −18, der(22)t(3;22)[8], 46, XX[11] | 2.4 | 2 |
| 304 | UNK | AHD | 46, XX[19] | 4.8 | 1 |
| 305 | M4 | AHD | NA | 9.6 | 3 |
| ID . | FAB . | Etiology . | Cytogenetics . | Dose level of RTA401 (CDDO), mg/m2 per h for 5 d . | Prior no. of Rx . |
|---|---|---|---|---|---|
| 301 | UNK | AHD | 46, XY, t(1;22)(p36.3;q11.2), del(5)(q13q33)[13], 48, XY, del(5)(q13q33),+22, +mar[7] | 0.6 | 1 |
| 302 | UNK | De novo | 46, XY, t(1;4)(p32;p16)[1], 46, XY[19] | 1.2 | 3 |
| 303 | RAEB-T | 2° | 45, X,−Y, der(3)ins(22;9)(q11.2;q34q34)t(3;22)(p23;q11.2), der(9)ins(22;9), der(22)t(3;22)[1], 44, X, −Y, der(3)ins(22;9)(q11.2;q34q34)t(3;22)(p23;q11.2), der(9)ins(22;9), −18, der(22)t(3;22)[8], 46, XX[11] | 2.4 | 2 |
| 304 | UNK | AHD | 46, XX[19] | 4.8 | 1 |
| 305 | M4 | AHD | NA | 9.6 | 3 |
All patients had a diagnosis of AML.
Rx indicates treatment; UKN, unknown; AHD, antecedent hematologic disease; 2°, secondary; and NA, not available.
Results
CDDO-induced granulocytic differentiation of HL60 acute myeloid leukemia cells is independent of apoptosis and correlates with enhanced expression of differentiation-regulated genes
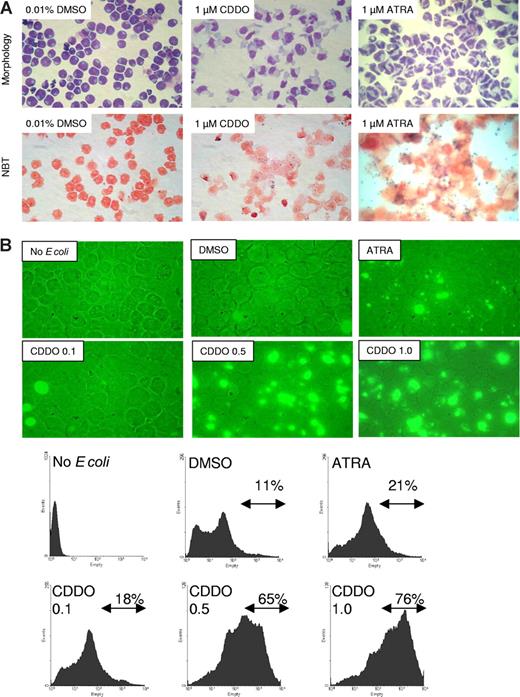
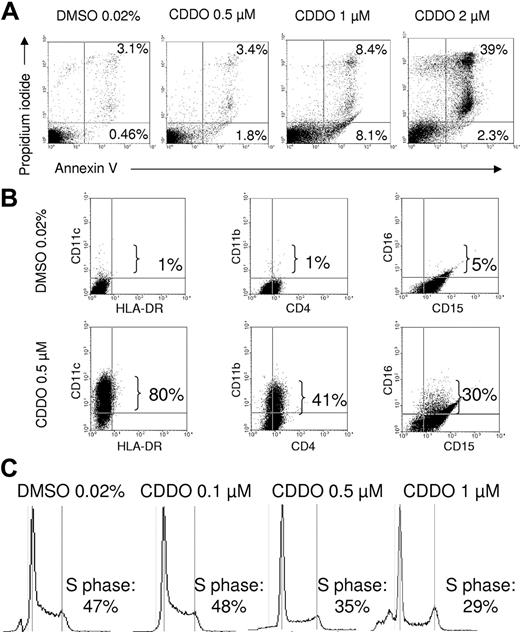
In HL60 acute myeloid leukemia (AML) cells, the proapoptotic and differentiation-inducing effects of CDDO are dose dependent. In fact, treatment with 1 μM CDDO for 4 to 5 days induced signs of both granulocytic and monocytic differentiation (increased cytoplasm-to-nucleus ratio, nuclear segmentation, and decreased cytoplasmic basophilia) without inducing cell death (Figure 1A top panel). Maturation toward both the monocytic and the granulocytic pathway was evident in CDDO-treated HL60 cells (Figure 1A), while, as expected,31 exposure to ATRA (1 μM) predominantly induced granulocytic differentiation of HL60 cells (Figure 1A). Accordingly, peroxidase function assessed by NBT assay was increased by CDDO, albeit to a lesser extent than by ATRA (Figure 1A lower panel). By contrast, phagocytosis of fluorescence-labeled bacteria was increased by treatment with CDDO and was more potent than with ATRA (Figure 1B). Interestingly, while higher CDDO doses (2 μM) induce apoptosis in approximately 40% of treated cells (Figure 2A), differentiation was already observed at subapoptotic CDDO concentrations (up to 1 μM). In fact, expression of the differentiation markers CD11b, CD11c, and CD16 was significantly and markedly increased in HL60 cells treated for 48 hours with 0.5 μM CDDO (Figure 2B). Notably, the percentage of CD14+ HL60 cells was also increased (data not shown). Consistent with the ability to promote differentiation, subapoptotic doses of CDDO induced cell cycle arrest as detected by a decrease of cells in S-phase (Figure 2C). Furthermore, consistent with the growth-inhibitory activity, a 75%, 81%, 56%, 50%, and 44% decrease of cell number was observed in HL60 cells cultured for 8 days with 0.05, 0.1, 0.25, 0.5, and 1 μM CDDO, respectively, but not with 0.01% DMSO used as vehicle (data not shown).
CDDO induces granulocytic-monocytic differentiation and phagocytic activity of HL60 cells. (A) Wright-Giemsa stain (top panel) and NBT assay (bottom panel) of HL60 cells after 4 to 5 days of culture. The cells were cultured in the presence of either 0.01% DMSO (vehicle), 1 μM ATRA, or 1 μM CDDO. (B) HL60 cells were cultured in the presence of either 0.01% DMSO, 1 μM ATRA, or 0.1 to 1 μM CDDO for 2 days prior to incubation with FITC-labeled E coli for 1 hour and analyzed for FITC positivity using fluorescence microscopy (top panel) or flow cytometry (bottom panel). The percentage of FITC-positive cells as detected by flow cytometry is indicated.
CDDO induces granulocytic-monocytic differentiation and phagocytic activity of HL60 cells. (A) Wright-Giemsa stain (top panel) and NBT assay (bottom panel) of HL60 cells after 4 to 5 days of culture. The cells were cultured in the presence of either 0.01% DMSO (vehicle), 1 μM ATRA, or 1 μM CDDO. (B) HL60 cells were cultured in the presence of either 0.01% DMSO, 1 μM ATRA, or 0.1 to 1 μM CDDO for 2 days prior to incubation with FITC-labeled E coli for 1 hour and analyzed for FITC positivity using fluorescence microscopy (top panel) or flow cytometry (bottom panel). The percentage of FITC-positive cells as detected by flow cytometry is indicated.
CDDO induces CD11b, CD11c, and CD16 expression and cell cycle arrest at subapoptotic doses. HL60 cells were cultured in the presence of 0.02% DMSO or different doses of CDDO for 2 days and analyzed for CD11c, HLA-DR, CD11c, CD4, CD16, and CD15 surface expression on day 5 of culture (A), or annexin V/propidium iodide positivity (B) and cell cycle phase distribution (C) using flow cytometry on day 2 of culture. Sub-G1 designates the fraction of cells that is undergoing apoptosis. The percentage of positive cells for each the designated fractions is indicated.
CDDO induces CD11b, CD11c, and CD16 expression and cell cycle arrest at subapoptotic doses. HL60 cells were cultured in the presence of 0.02% DMSO or different doses of CDDO for 2 days and analyzed for CD11c, HLA-DR, CD11c, CD4, CD16, and CD15 surface expression on day 5 of culture (A), or annexin V/propidium iodide positivity (B) and cell cycle phase distribution (C) using flow cytometry on day 2 of culture. Sub-G1 designates the fraction of cells that is undergoing apoptosis. The percentage of positive cells for each the designated fractions is indicated.
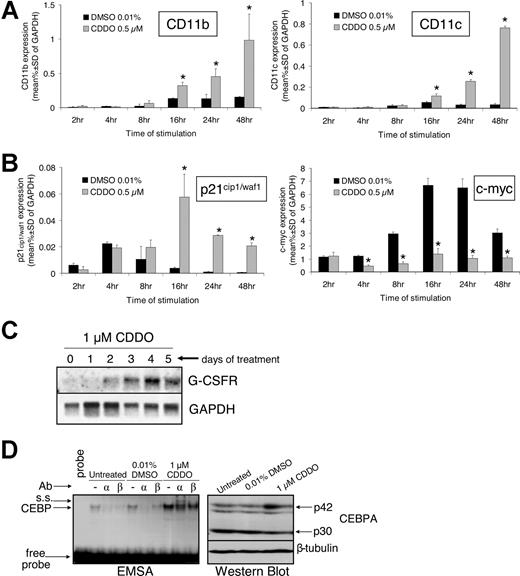
Quantitative RT-PCR showed significantly higher CD11b and CD11c mRNA levels within 16 hours of exposure to 0.5 μM CDDO than cells exposed to DMSO (Figure 3A). Likewise, p21cip1/waf1 mRNA expression was also induced by CDDO within 16 hours but subsequently decreased (Figure 3B). However, p21 levels in CDDO-treated cells remained significantly higher than in DMSO-treated control cells even 48 hours after treatment (Figure 3B left panel). Conversely, mRNA levels of c-myc, which has been described to antagonize granulocytic/monocytic differentiation of myeloid progenitors,32 were not induced but rather slightly inhibited by 4-hour exposure to 0.5 μM CDDO (Figure 3B right panel). Notably, while c-myc mRNA increased 7-fold in DMSO cultures most likely due to addition of fresh medium at the beginning of the experiment, c-myc mRNA expression in CDDO-treated cells increased only up to baseline levels and remained significantly lower than in DMSO cultures (Figure 3B right panel). Importantly, Northern blot analysis of CDDO-treated HL60 cells showed that mRNA expression of the CEBPA transcriptionally regulated G-CSF receptor (GCSFR)31 progressively increased within 5 days of exposure to 0.5 μM CDDO (Figure 3C). Moreover, real-time RT-PCR analysis showed that mRNA levels of the secondary granule genes, lysozyme, myeloperoxidase (MPO), and neutrophil elastase (NE) were 9-fold, 1.6-fold, and 3.5-fold, respectively, higher in cells treated for 48 hours with 0.5 μM CDDO than 0.01% DMSO (P = .001, P = .006, P = .027) (data not shown). Because GCSFR, NE, and MPO are transcriptional targets of CEBPA activity, it is likely that CDDO exerted its differentiation-promoting effects through induction of CEBPA expression. Notably, expression of the late differentiation-regulator CCAAT enhancer–binding protein epsilon (CEBPE) was not affected by CDDO (data not shown).
CDDO increases mRNA expression of CD11b, CD11c, p21cip1/waf1, and G-CSFR; induces CEBPA DNA binding; and suppresses the expression of c-myc. HL60 cells were exposed to 0.01% DMSO or 0.5 μM CDDO for the indicated times' RNA was extracted and DNAse-treated and retrotranscribed into cDNA. Subsequently, real-time PCR was performed as described in “Patients, materials, and methods.” The expression of CD11b and CD11c (A) as well as p21cip1/waf1 and c-myc (B) was assessed and is shown as the percentage of GAPDH mRNA expression. *P < .05 versus DMSO treatment (2-sided t test). (C) HL60 cells were cultured with 1 μM CDDO for the indicated time, RNA was extracted, and Northern blotting was performed using a probe for the human granulocyte colony-stimulating factor receptor (GCSFR). GAPDH mRNA expression served as a loading control. (D) Gel shift assay of CEBP protein DNA-binding activity in HL60 cells using a G-CSFR probe in the presence or absence of 1 μM CDDO (left panel). Supershift (s.s.) was analyzed using anti-CEBPA or anti-CEBPbeta antibodies (Ab). Total protein was comparable in the 3 lysates (right panel).
CDDO increases mRNA expression of CD11b, CD11c, p21cip1/waf1, and G-CSFR; induces CEBPA DNA binding; and suppresses the expression of c-myc. HL60 cells were exposed to 0.01% DMSO or 0.5 μM CDDO for the indicated times' RNA was extracted and DNAse-treated and retrotranscribed into cDNA. Subsequently, real-time PCR was performed as described in “Patients, materials, and methods.” The expression of CD11b and CD11c (A) as well as p21cip1/waf1 and c-myc (B) was assessed and is shown as the percentage of GAPDH mRNA expression. *P < .05 versus DMSO treatment (2-sided t test). (C) HL60 cells were cultured with 1 μM CDDO for the indicated time, RNA was extracted, and Northern blotting was performed using a probe for the human granulocyte colony-stimulating factor receptor (GCSFR). GAPDH mRNA expression served as a loading control. (D) Gel shift assay of CEBP protein DNA-binding activity in HL60 cells using a G-CSFR probe in the presence or absence of 1 μM CDDO (left panel). Supershift (s.s.) was analyzed using anti-CEBPA or anti-CEBPbeta antibodies (Ab). Total protein was comparable in the 3 lysates (right panel).
CDDO augments CEBPA activity in acute myeloid leukemia cells by translationally enhancing the p42/p30 CEBPA ratio in a CEBPA uORF-dependent manner
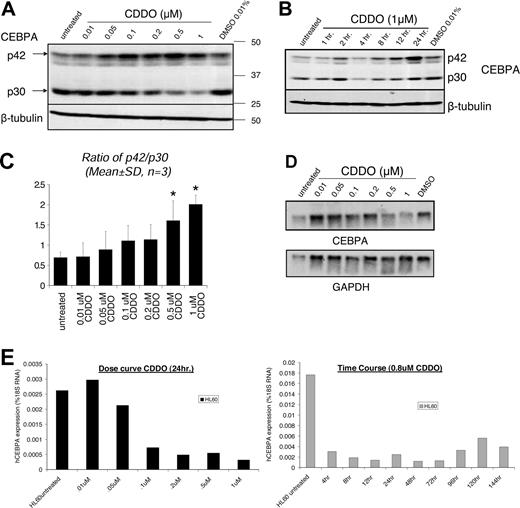
Because early steps of granulocytic and monocytic differentiation are critically dependent on the function of the CEBPA transcription factor, we investigated whether CDDO affects CEBPA expression and/or function in acute myeloid leukemia cells. In HL60 cells treated with CDDO, DNA binding of CEBP members CEBPA and CEBPB (and possibly other proteins involved in the complex) to a human GCSFR probe was increased compared with DMSO-treated cells (Figure 3D left panel), suggesting that CEBPA might be induced by CDDO treatment. Indeed, Western blot analysis revealed that 1 μM CDDO potently enhanced expression of p42 CEBPA in HL60 cells (Figure 3D right panel; Table 1). Moreover, CDDO dose-dependently increased p42 CEBPA protein within the first 24 hours of treatment (Figure 4A,B). When p42 and p30 isoforms were combined, no significant increase by CDDO was detected (Table 1), since CDDO induced a decrease of p30 expression (Figure 4A,B). Interestingly, the ratio (p42/p30) of the transcriptionally active 42-kDa and the transcriptionally inactive 30-kDa isoforms of CEBPA showed a significant dose-dependent increase up to 2.9-fold upon treatment with 1 μM CDDO (Figure 4C; Table 1). By contrast, CEPBA mRNA levels decreased upon exposure to CDDO (Figure 4D,E), suggesting that CDDO does not influence CEBPA transcription but that it may favor translation and/or posttranslationally stabilize p42 CEBPA.
CDDO increases the expression of p42/p30 CEBPA protein isoforms at a posttranscriptional level. (A) CEBPA protein expression after 24 hours in untreated, DMSO-treated, and CDDO-treated HL60 cells is shown by Western blotting using a polyclonal rabbit anti-CEBPA antibody that recognizes the C-terminal part of CEBPA (top panel). P42 and p30 designate the 2 prominent isoforms of CEBPA, and beta-tubulin served as a loading control. (B) Time course of HL60 cells cultured in the presence or absence of 0.01% DMSO or 1 μM CDDO. Protein lysates were obtained at the indicated time points, and Western blotting for CEBPA was performed. Beta-tubulin served as a loading control. (C) The ratio of p42/p30 as determined by densitometry is shown in the panel on the right. *P < .05 versus untreated control. (D) HL60 cells were cultured for 24 hours without treatment or treated with 0.01% DMSO-treated or CDDO at the indicated doses, RNA was extracted, and Northern blotting performed. CEBPA mRNA expression was detected using a probe against the 3′UTR of human CEBPA. GAPDH mRNA served as a loading control. (E) Real-time RT-PCR analysis of RNA extracted from HL60 cells that were treated or not treated with different doses of CDDO for 24 hours (left panel) or 0.8 μM CDDO for different periods of time (right panel), as indicated. Human CEBPA mRNA expression is shown as the percentage of 18S RNA.
CDDO increases the expression of p42/p30 CEBPA protein isoforms at a posttranscriptional level. (A) CEBPA protein expression after 24 hours in untreated, DMSO-treated, and CDDO-treated HL60 cells is shown by Western blotting using a polyclonal rabbit anti-CEBPA antibody that recognizes the C-terminal part of CEBPA (top panel). P42 and p30 designate the 2 prominent isoforms of CEBPA, and beta-tubulin served as a loading control. (B) Time course of HL60 cells cultured in the presence or absence of 0.01% DMSO or 1 μM CDDO. Protein lysates were obtained at the indicated time points, and Western blotting for CEBPA was performed. Beta-tubulin served as a loading control. (C) The ratio of p42/p30 as determined by densitometry is shown in the panel on the right. *P < .05 versus untreated control. (D) HL60 cells were cultured for 24 hours without treatment or treated with 0.01% DMSO-treated or CDDO at the indicated doses, RNA was extracted, and Northern blotting performed. CEBPA mRNA expression was detected using a probe against the 3′UTR of human CEBPA. GAPDH mRNA served as a loading control. (E) Real-time RT-PCR analysis of RNA extracted from HL60 cells that were treated or not treated with different doses of CDDO for 24 hours (left panel) or 0.8 μM CDDO for different periods of time (right panel), as indicated. Human CEBPA mRNA expression is shown as the percentage of 18S RNA.
To determine whether CDDO stabilized p42 protein, we cultured HL60 cells in the presence of DMSO or CDDO with or without the protein synthesis inhibitor cycloheximide (20 μg/mL) and evaluated CEBPA protein expression. CEBPA protein was reduced in the presence of cycloheximide regardless of the presence of CDDO (Figure 5A), suggesting that CDDO does not influence CEBPA protein turnover but that, most likely, it influences CEBPA mRNA translation. Reportedly, the ratio of CEBPA p42 and p30 isoforms is translationally controlled by a short upstream open reading frame (uORF) separated by a short C-rich spacer region from the main CEBPA p42 ATG, and deletion of this uORF results in an increased p42/p30 ratio.27 To determine whether CDDO-induced CEBPA expression requires the uORF translation regulatory element, we transiently transfected HEK293A cells with a CEPBA expression plasmid containing both the cDNA and the 5′-untranslated region (UTR). The 5′UTR contained either the wild-type uORF/spacer (uORFwt) or a mutated uORF/spacer region carrying a Kozak sequence around the uORF ATG (uORFopt). As shown, wild-type, but not mutated, uORF-driven CEBPA expression was sensitive to CDDO treatment (Figure 5B). In fact, CDDO increased the expression of p42 over p30 CEBPA only in cells transduced with wild-type uORF/spacer-containing construct (Figure 5C).
CDDO enhances granulocytic differentiation through CEBPA translation, and this involves the uORF of CEBPA. (A) HL60 cells were cultured for 22 hours with 0.01% DMSO or 0.5 μM CDDO after a 2-hour preincubation period with 20 μg/mL cycloheximide (CHX) and analyzed for CEBPA and beta-actin protein expression using Western blotting. (B) HEK293A cells were transiently transfected with an expression plasmid harboring the human CEBPA coding sequence and either a wild-type uORF sequence (uORFwt) or an uORF with an optimized Kozak sequence around the uORF ATG (uORFopt). Two hours after transfection, the cells were treated with 0.01% DMSO or 0.1 μM CDDO, and 24 hours after transfection, cells were lysed and subjected to Western blotting, using antibodies against CEBPA or beta-actin (left panel). (C) The ratio of p42/p30 was analyzed by densitometric analysis of panel B. (D) 32Dcl3, 6.15 + MigR1, and 6.15 + WT-uORF cells were cultured with G-CSF and treated with 0.01% DMSO or 0.5 μM CDDO for 48 hours, and Western blotting was performed using anti-HA, anti-HSP90, or anti–hnRNP-E2 antibodies. (E) May-Grünwald/Giemsa staining of 32Dcl3, 6.15 + MigR1, and 6.15 + WT-uORF cells cultured with G-CSF for 9 days in the presence or absence of 0.5 μM CDDO. (F) Western blot analysis of HL60 cells treated with DMSO or 0.5 μM CDDO. The blot was probed with CEBPA and hnRNP-E2 antibodies. HSP90 served as a loading control.
CDDO enhances granulocytic differentiation through CEBPA translation, and this involves the uORF of CEBPA. (A) HL60 cells were cultured for 22 hours with 0.01% DMSO or 0.5 μM CDDO after a 2-hour preincubation period with 20 μg/mL cycloheximide (CHX) and analyzed for CEBPA and beta-actin protein expression using Western blotting. (B) HEK293A cells were transiently transfected with an expression plasmid harboring the human CEBPA coding sequence and either a wild-type uORF sequence (uORFwt) or an uORF with an optimized Kozak sequence around the uORF ATG (uORFopt). Two hours after transfection, the cells were treated with 0.01% DMSO or 0.1 μM CDDO, and 24 hours after transfection, cells were lysed and subjected to Western blotting, using antibodies against CEBPA or beta-actin (left panel). (C) The ratio of p42/p30 was analyzed by densitometric analysis of panel B. (D) 32Dcl3, 6.15 + MigR1, and 6.15 + WT-uORF cells were cultured with G-CSF and treated with 0.01% DMSO or 0.5 μM CDDO for 48 hours, and Western blotting was performed using anti-HA, anti-HSP90, or anti–hnRNP-E2 antibodies. (E) May-Grünwald/Giemsa staining of 32Dcl3, 6.15 + MigR1, and 6.15 + WT-uORF cells cultured with G-CSF for 9 days in the presence or absence of 0.5 μM CDDO. (F) Western blot analysis of HL60 cells treated with DMSO or 0.5 μM CDDO. The blot was probed with CEBPA and hnRNP-E2 antibodies. HSP90 served as a loading control.
CDDO restores CEBPA expression in differentiation-arrested BCR/ABL-positive 6.15 myeloid precursor cells
The uORF/spacer-interacting RNA-binding protein hnRNP-E2 translationally suppresses CEBPA expression in CML blast crisis.26,33 To determine whether CDDO increases the p42/p30 CEBPA ratio by suppressing the translation-inhibitory function of hnRNP-E2, we stably transduced 6.15 cells, which (similarly to K562 cells) express BCR-ABL and aberrantly lack CEBPA mRNA expression,33 with a retroviral vector expressing rat CEBPA cDNA under the control of a wild-type uORF/spacer region (MigRI-WTuORF/spacer-CEBPA-HA) or with the empty MigRI vector and stimulated these cells with 1 μM CDDO in the presence of G-CSF for 7 to 9 days. As expected, anti-HA Western blot analysis showed that CDDO- and G-CSF–treated vector-transduced BCR/ABL-positive cells did not express CEBPA and did not show morphologic signs of granulocytic differentiation (Figure 5D,E). By contrast, wt-uORF-HA-CEBPA–transduced 6.15 cells showed weak expression of CEBPA compared with parental 32Dcl3 cells (Figure 5D lane 4 versus lane 1) and minimal morphologic signs of myeloid maturation (indented nuclei) (Figure 5E). Interestingly, CDDO treatment of wt-uORF-HA-CEBPA–transduced cells rescued p42 CEBPA (Figure 5D lane 5) and partially restored granulocytic maturation of differentiation-arrested BCR/ABL-expressing myeloid 32Dcl3 cells (30%-40% of postmitotic cells and 10%-20% of mature polymorph nuclear cells if compared with untreated uORF-expressing 6.15 cells) (Figure 5E), suggesting that CDDO enhances CEBPA expression and rescues differentiation by re-expression of CEBPA. CDDO treatment of BCR/ABL-positive K562 cells, which do not express CEBPA mRNA,30 failed to induce granulocytic differentiation, and flow cytometry for CD11b and CD11c and NBT assay was similar in untreated and treated K562 cells (data not shown), again suggesting that CDDO may induce granulocytic differentiation via CEBPA. hnRNP-E2 expression was inhibited by CDDO in 6.15 cells, suggesting that CDDO released CEBPA from translational inhibition by hnRNP-E2. Although p42 and the ratio of p42/p30 were increased in CDDO-treated HL60 cells (Figure 4A-C), neither changes of hnRNP-E2 protein expression nor altered hnRNP-E2 binding to the C-rich element present in the spacer region of the CEBPA mRNA was observed by Western blots and RNAelectromobility shift assays (REMSAs) in HL60 cells (Figure 5F, and data not shown). Likewise, in HL60 cells, expression of calreticulin, a RNA-binding protein that translationally inhibits CEBPA expression in t(3,21)(q26;q22) AML cells,24 was not affected by treatment with CDDO (not shown). Thus, in AML patient–derived cells, CDDO induces p42 and the p42/p30 ratio through a mechanism that is independent of translational suppression by hnRNP-E2 or calreticulin.
CDDO translationally alters the p42/p30 CEBPA ratio and modulates eIF2α and eIF4E phosphorylation in acute myeloid leukemia HL60 cells
Because CEBPA translation is influenced by eukaryotic translation initiation factors eIF2a and eIF4E,27 we studied whether expression and/or phosphorylation of these factors were affected in HL60 cells by treatment with CDDO. Reportedly, activation of eIF2α and eIF4E is associated with their dephosphorylation and phosphorylation, respectively.34 In HL60 cells, 16-hour treatment with 1 μM CDDO led to dephosphorylation of eIF2α but did not affect eIF2α expression (Figure 6A), and a close inverted temporal association of eIF2α dephosphorylation and increased p42/p30 ratio was observed in HL60 cells treated with 1 μM CDDO for 16 hours (Figure 6B). Accordingly, eIF4E phosphorylation was increased by CDDO with no changes in total eIF4E levels (Figure 6A). Moreover, de novo protein synthesis but not increased CEBPA stability was required for the CDDO-dependent induction and maintenance of CEBPA levels over at least a 16-hour period (Figure 6A). In fact, while p42 was increased in the presence of CDDO alone, this was not seen after addition of cycloheximide (Figure 6A). As expected, neither eIF2α dephosphorylation nor eIF4E phosphorylation was detectable in HL60 cells cotreated with CDDO and cycloheximide (Figure 6A), suggesting that CDDO does not stabilize CEBPA protein but possibly increases p42 CEBPA translation through an eiF-dependent mechanism.
CDDO-induced CEBPA isoform changes require de novo protein synthesis, are associated with activation of eIF2α and eIF4E, but are independent of TGFβ and PPARgamma pathways. (A) HL60 cells were cultured for the indicated times in the presence of 1 μM CDDO with or without cycloheximide (20 μg/mL, CHX), and Western blotting was performed using the anti-CEBPA antibody. Blots were reprobed with anti–P-eIF2α, anti-eIF2α, anti–P-eIF4E, or anti-eIF4E antibodies. (B) Densitometric analysis of CEBPA p42/p30 and P-eIF2α/eIF2α bands from panel A. (C) HL60 cells were cultured for 24 hours in the presence or absence of 1 μM CDDO, and Western blotting was performed using anti-CEBPA, anti–P-eIF2α, and anti-eIF2α antibodies. In addition, cells were incubated in the presence of either the PPARgamma antagonist GW9662, calpain I inhibitor, an inhibitor of the eIF2α kinase PKR (2-aminopurine), or an inhibitor of the eIF2α phosphatase PP1/PP2A (calyculin A). (D) HL60 cells were treated with 0.01% DMSO, 0.1 μM CDDO, or 0.5 μM CDDO for 22 hours after a preincubation time of 2 hours with either no inhibitor or 1 μM of the TGFβ pathway inhibitor, SB505124, and analyzed for CEBPA and beta-actin by Western blotting. (E) HL60 cells were preincubated with the TGFβ pathway inhibitor (TGFβ/ALK5 receptor inhibitor) SB505124 for 2 hours and then stimulated with 10 ng/mL TGFβ1 for 1 hour and subjected to Western blotting using phospho-SMAD2 (P-SMAD2) or β-actin antibodies.
CDDO-induced CEBPA isoform changes require de novo protein synthesis, are associated with activation of eIF2α and eIF4E, but are independent of TGFβ and PPARgamma pathways. (A) HL60 cells were cultured for the indicated times in the presence of 1 μM CDDO with or without cycloheximide (20 μg/mL, CHX), and Western blotting was performed using the anti-CEBPA antibody. Blots were reprobed with anti–P-eIF2α, anti-eIF2α, anti–P-eIF4E, or anti-eIF4E antibodies. (B) Densitometric analysis of CEBPA p42/p30 and P-eIF2α/eIF2α bands from panel A. (C) HL60 cells were cultured for 24 hours in the presence or absence of 1 μM CDDO, and Western blotting was performed using anti-CEBPA, anti–P-eIF2α, and anti-eIF2α antibodies. In addition, cells were incubated in the presence of either the PPARgamma antagonist GW9662, calpain I inhibitor, an inhibitor of the eIF2α kinase PKR (2-aminopurine), or an inhibitor of the eIF2α phosphatase PP1/PP2A (calyculin A). (D) HL60 cells were treated with 0.01% DMSO, 0.1 μM CDDO, or 0.5 μM CDDO for 22 hours after a preincubation time of 2 hours with either no inhibitor or 1 μM of the TGFβ pathway inhibitor, SB505124, and analyzed for CEBPA and beta-actin by Western blotting. (E) HL60 cells were preincubated with the TGFβ pathway inhibitor (TGFβ/ALK5 receptor inhibitor) SB505124 for 2 hours and then stimulated with 10 ng/mL TGFβ1 for 1 hour and subjected to Western blotting using phospho-SMAD2 (P-SMAD2) or β-actin antibodies.
To assess the molecular mechanism whereby CDDO influences eIF2α, we treated HL60 cells with different chemical inhibitors that alter mRNA translation by affecting the function of eIF2α. Treatment with 2-aminopurine (2-AP), which inhibits the RNA-dependent protein kinase PKR that, in turn, phosphorylates eIF2α,34 increased the p42/p30 CEBPA ratio (Figure 6C). However, there was no synergism between 2-AP and CDDO (Figure 6C), suggesting that both drugs may act through similar pathways. Moreover, enhanced eIF2α phosphorylation and loss of CEBPA expression were observed upon treatment of HL60 cells with the PP1/PP2A phosphatase inhibitor calyculin A, regardless of the presence and absence of CDDO (Figure 6C), suggesting that either PP1 or PP2A might be involved in the eIF2α-dependent regulation of CEBPA expression. We sought to study whether 2 pathways that had previously been linked to CDDO-induced apoptosis were involved in CDDO-induced CEBPA changes. These results showed that the CDDO-induced increase of p42/p30 CEBPA ratio does not involve PPARgamma as shown by treatment with the PPARgamma receptor antagonist, GW9662 (Figure 6C). Inhibition of the TGFβ pathway by the inhibitor SB505124,35 which, like SMAD7, blocks TGFβ receptor type I and downstream SMAD activity, mildly reduced the expression of p42 CEBPA in the absence of CDDO but did not alter the CDDO-induced ratio change of p42/p30 CEBPA (Figure 6D). SB505124 was used at 1 μM, which efficiently inhibited TGFβ pathway activation as demonstrated by suppression of TGFβ1-induced SMAD-2 phosphorylation (Figure 6E). Calpain inhibitor I treatment was performed to study whether p30 was a result of p42 cleavage. However, we found that calpain inhibitor I treatment did not lead to increased “uncleaved” p42 protein but, by itself, decreased p42 (Figure 6C). Therefore, cleavage of p42 by calpain is not a likely event in HL60 cells. Importantly, the effects of CDDO on the CEBPA isoform ratio were also independent of caspase activation, since preincubation of HL60 cells with the caspase-3 inhibitor Z-DEVD-fmk (25 μM) alone or in combination with CDDO did not influence the p42/p30 ratio (data not shown).
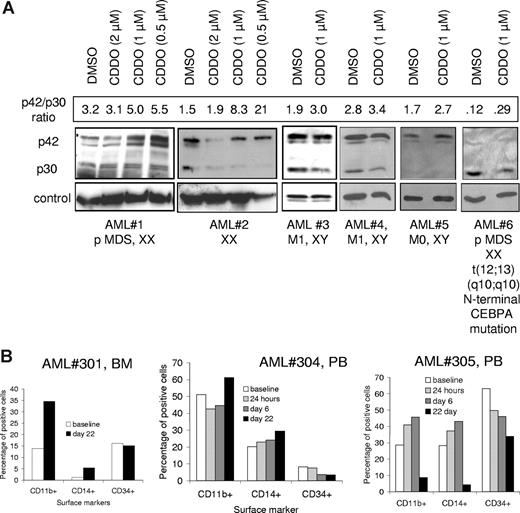
CDDO increases the ratio of p42/p30 CEBPA in a subset of patients with AML, and CDDO induces differentiation of AML blasts in vivo
CDDO has been described to induce granulocytic differentiation of primary blasts from patients with AML. We tested whether CDDO-mediated alteration of the ratio of CEBPA could be confirmed in primary blasts from patients with AML. Indeed, we could detect a CDDO-mediated increase of the p42/p30 ratio (either detected by an increase of p42 or a decrease of p30) in 6 of 13 patients who expressed detectable CEBPA protein (Figure 7A). One of these patients harbored a C68 deletion in the N-terminal CEBPA coding sequence. This resulted in a decrease of the p42 isoform as detected by Western blotting. Interestingly, the remaining p30 isoform was decreased when cells from this patient were exposed to CDDO (Figure 7A). To investigate the in vivo differentiating effects of CDDO, cells from 5 patients treated during a phase 1 clinical trial with increasing doses of CDDO were evaluated for the percentage of CD11b+, CD14+, and CD34+ cells before and after treatment. Differentiation antigen expression demonstrated an increase in CD11b+ and CD14+ cells and a concomitant reduction of immature cells expressing CD34 in 3 of 5 patients (Figure 7B). Clinically, patients did not fulfill response criteria and differential counts did not change significantly, except in patient no. 305 (who received the highest dose of CDDO) with AML FAB M4, whose bone marrow blasts and monocytes decreased from 68% to 53% on day 22. In this patient, who initially required hydroxyurea because of rising circulating blasts, white blood cells (WBCs) and circulating blasts remained low for 3 weeks without additional therapy.
CDDO-mediated increase of active CEBPA protein and signs of differentiation in primary blasts from patients with AML. (A) AML blasts were cultured for 24 hours in the presence or absence of CDDO at different concentrations as indicated and subjected to Western blotting using an anti-CEBPA antibody. Anti–β-actin (AML nos. 1,2), anti-eIF2α (AML no. 3), or anti–beta-tubulin (AML nos. 4-6) antibodies were used for loading controls. Information about the type of AML and the karyotype and/or a CEBPA mutation are given below the blots. The densitometric units of p42/p30 are indicated as numbers above the Western blots. (B) Patients with refractory or relapsed AML were treated with CDDO (RTA401) during a phase 1 clinical trial, and cells were collected from the peripheral blood (PB) or bone marrow (BM) and assessed for expression of surface markers CD11b, CD14, and CD34 by flow cytometry at the indicated times (see also Table 2). Three of 5 patients (patients nos. 301, 304, 305) showed alterations of these parameters during the observed period. PB baseline percentages are not available from patient no. 301, therefore, BM percentages are provided.
CDDO-mediated increase of active CEBPA protein and signs of differentiation in primary blasts from patients with AML. (A) AML blasts were cultured for 24 hours in the presence or absence of CDDO at different concentrations as indicated and subjected to Western blotting using an anti-CEBPA antibody. Anti–β-actin (AML nos. 1,2), anti-eIF2α (AML no. 3), or anti–beta-tubulin (AML nos. 4-6) antibodies were used for loading controls. Information about the type of AML and the karyotype and/or a CEBPA mutation are given below the blots. The densitometric units of p42/p30 are indicated as numbers above the Western blots. (B) Patients with refractory or relapsed AML were treated with CDDO (RTA401) during a phase 1 clinical trial, and cells were collected from the peripheral blood (PB) or bone marrow (BM) and assessed for expression of surface markers CD11b, CD14, and CD34 by flow cytometry at the indicated times (see also Table 2). Three of 5 patients (patients nos. 301, 304, 305) showed alterations of these parameters during the observed period. PB baseline percentages are not available from patient no. 301, therefore, BM percentages are provided.
Discussion
In this report, we show that CDDO induces phagocytic activity and granulocytic-monocytic differentiation via translational up-regulation of CEBPA expression. Our data are in line with previous data describing CDDO-induced differentiation of AML cell lines.1,2,7,17 Since phagocytic activity of AML cells has been shown to be decreased compared with healthy controls,36,37 CDDO may be useful to enhance phagocytic function in this patient population.
CDDO, CDDO-Im, and CDDO-Me have been shown to induce growth arrest in a variety of cancer cell lines in vitro and in vivo.2,13,38,39 Cell cycle arrest was associated with down-regulation of c-myc, and up-regulation of p21cip1/waf1.13,38,40 Induction of p21 and down-regulation of cyclin D1 were essential for CDDO-induced growth arrest in breast cancer cell lines.13 In HL60 cells, c-myc down-regulation was first seen 4 hours after exposure to CDDO (Figure 3B). Interestingly, the first signs of CEBPA protein up-regulation occurred within 5 hours of CDDO treatment (Figure 6A). Negative regulation of the c-myc promoter by CEBPA has been described,32 and it will be interesting to study whether these 2 processes are linked. p21 was induced approximately 8 hours after CDDO treatment, and the p21 promoter has been described to be induced by CEBPA in rat hepatoma cells upon dexamethasone treatment.41
CCAAT enhancer–binding proteins (CEBPs) are critical for the differentiation of granulocytes19,42 and monocytes.42,43 Recently, CEBP beta (CEBPB) protein was described to be up-regulated during CDDO-Im–induced monocytic differentiation of HL60 cells as early as 30 minutes after treatment, and monocytic differentiation was partially dependent upon ERK activation and TGFβ-mediated Smad activation.17 While CEBPB DNA-binding activity was enhanced in our CDDO-treated cells, no significant up-regulation of CEBPB protein was found within 16 hours of treatment (data not shown). Differences between CDDO and CDDO-Im have been described earlier.1,44,45 In a previous report, CDDO-Im but not CDDO induced the monocytic marker CD36,1,17 suggesting that the CDDO derivatives may induce qualitatively different patterns of myelomonocytic differentiation. In addition, while CDDO acted as a partial agonist for the PPARgamma receptor, another close relative, CDDO-Me, which binds to PPARg with similar affinity, is an antagonist.44 These differences were attributed to differences in their capacity to recruit or displace cofactors of transcriptional regulation as CDDO releases the nuclear receptor corepressor, NCoR, from PPARg, while CDDO-Me does not.44 Another study by Chintharlapalli et al found that CDDO-Im showed higher activity to induce PPARgamma interactions with the corepressor SMRT than CDDO.45
CDDO induced the transcriptionally active isoform of CEBPA (p42), increased the p42/p30 ratio, and enhanced CEBPA DNA-binding activity (Figures 3D and 4A-C). Importantly, CDDO partially restored granulocytic maturation of differentiation-arrested BCR/ABL-expressing cells and rescued p42 CEBPA expression from a construct bearing the uORF/spacer region of CEBPA 5′UTR. As CDDO was unable to induce granulocytic differentiation in cells that do not express CEBPA mRNA (K562 cells, 6.15 cells), our data indicate that CDDO may release CEBPA from the translation-inhibitory effects of RNA-binding proteins such as hnRNP-E2. The requirement of CEBPA expression for the differentiation-promoting effects of CDDO is further supported by the lack of differentiation in CDDO-treated Kasumi-6 cells that harbor a mutation in the 3′ end of the CEBPA coding sequence. These cells express a C-terminally mutated CEBPA protein that has lost its DNA-binding activity46 and fail to show any signs of granulocytic differentiation (morphology, NBT assay, G-CSFR RNA) when exposed to CDDO, although (mutant) CEBPA p42 and p30 isoforms were expressed (not shown).
Interestingly, while hnRNP-E2 was a target of CDDO in 6.15 cells, CDDO did not suppress hnRNP-E2 protein expression or hnRNP-E2 RNA-binding activity in HL60 cells (data not shown). In addition, the expression of calreticulin, another RNA-binding protein that has been described to affect CEBPA translation,24,25,47 was unaltered by CDDO treatment in HL60 cells. However, it remains possible, if not likely, that other RNA-binding proteins may play a role in CDDO-induced CEBPA translation and granulocytic differentiation of HL60 cells. The eukaryotic translation initiation factors, eIF2α and eIF4E, have been implicated in controlling the ratio of truncated isoforms of CEBPA and CEBPB.27 During the final stage of adipocytic differentiation of 3T3-L1 fibroblasts, translation of p42 was decreased upon activation of eIF2α or eFI4E, while the translation of p30 was increased.27 Conversely, CDDO-induced p42/p30 was associated with a decrease of eIF2α and an increase of eIF4E activity in HL60 cells, and the inhibitor of eIF2α phosphorylation, 2-AP, increased the p42/p30 ratio to a similar degree as CDDO, arguing for cell type–specific effects of CEBPA translational regulation.
Since both hnRNP-E2 and eIFs require the presence of the uORF to regulate CEBPA translation,27 we asked whether the effect of CDDO might also work through the uORF. Deletion of the uORF results in an enhanced p42/p30 ratio. Therefore, we used a mutant that contains an optimized Kozak sequence at the ATG of the uORF. Using this mutant that has been shown to increase p30 at the expense of p42,27 we found that CDDO was no longer able to increase p42 or decrease p30 and that a wild-type uORF was required for the CDDO-induced p42/p30 ratio change. It is tempting to speculate that alteration of eFI2alpha activity was involved in this regulation, but future studies are needed to show whether these CDDO-induced effects are linked. To exclude the possibility of mutations in the CEBPA gene, we have sequenced the entire CEBPA gene including the uORF in HL60 cells but found no mutations (data not shown).
The CDDO-induced CEBPA isoform ratio change was independent of other pathways previously linked to CDDO, such as TGFβ (Figure 6D) and PPARgamma signaling pathways (Figure 6C), and the latter has also been described for CDDO-Im–induced differentiation of U937 cells.1
CDDO and its derivatives induce dose-dependent apoptosis in cancer cell lines.3,5,9,10,13,48-50 However, the CDDO-induced effects on differentiation, gene expression, and the CEBPA p42/p30 ratio that are described in the present report were independent from induction of apoptosis (Figure 2). The first clinical trials of CDDO and CDDO-Me in patients with AML are ongoing, and since CDDO causes differentiation even at subapoptotic doses, patients may already benefit from low doses of the drug, as has been described for novel demethylating substances such as decitabine.51 In addition, combination therapy with differentiation-inducing drugs may prove useful in the future, as it has been shown that CDDO-Im synergizes with vitamin D3 to induce monocytic differentiation17 and sensitized PML-RARA–positive cells to the differentiation inducer, ATRA.48 Given our own results of CDDO-induced G-CSF receptor expression (Figure 3C), G-CSF would also be an interesting partner for combination therapy. We were able to confirm CDDO-induced changes of CEBPA expression and induction of differentiation in primary blasts from patients with AML (Figure 7). Interestingly, CDDO decreased the p30 isoform in blasts from a patient with an N-terminal CEBPA mutation (Figure 7A). This suggests that patients with heterozygous CEBPA mutations may benefit from CDDO-induced decrease of the described dominant-negative effect of the remaining p30 isoform,22 thereby increasing the amount of transcriptionally active CEBPA protein.
In summary, we report that CDDO enhances p42 CEBPA protein at the level of translation. This makes it an attractive drug to target blasts from patients with AML or MDS that are defective in granulocytic-monocytic differentiation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants of the Deutsche Forschungsgemeinschaft (DFG) (KO2155/1–1, KO2155/2–1) and the IZKF [Interdisciplinary Center for Clinical Research] Münster to S.K., C.M.-T., and H.S. This study was supported in part by research funding from (REATA [Reata Pharmaceuticals]) to M.A. and by NCI [National Cancer Institute] (1 P50 CA100632, P01 CA55164, R01 CA89346, 2P30-CA16672 to M.A.), NCI grant CA095512 to D.P., and LLSA [Leukemia and Lymphoma Society of America] (R6149–07 01 to MK). D.P. is a Scholar of the Leukemia and Lymphoma Society of America. E.L. has been supported by grants from FAMRI [Flight Attendant Medical Research Institute] and IASLC [International Society for the Study of Lung Cancer].
We thank Dr Cornelis Calkhoven (Jena, Germany) for providing CEBPA uORF expression plasmids; Dr Raul Andino (University of California, San Francisco) for anti–hnRNP-E2 antibody; Linda Kamp, Emiliano Fabiani, and Francesco Guidi for excellent technical assistance; and Prof Giuseppe Leone for his support.
Authorship
Contribution: S. Koschmieder, F.D., H.R., M.A., D.P., M.B.S., W.E.B., and H.S. designed research; S. Koschmieder, F.D., H.R., C.S., J.S.C., M.K., R.S., B.S., and A.D.R. performed research and collected the data; M.A., D.P., M.B.S., C.M.-T., and H.S. contributed vital reagents; M.A., M.K., H.K., and J.C.W. designed and conducted the clinical protocol and analyzed the clinical samples; S.Koschmieder., F.D., S. Kobayashi, E.L., N.S., M.T.V., and D.P. analyzed data; S. Koschmieder, F.D., W.E.B., C.M.-T., H.S., D.P., and D.G.T. wrote the paper. S. Koschmieder and F.D. contributed equally to this work.
Conflict-of-interest disclosure: Two of the authors (M.A. and M.K.) have declared a financial interest in a company whose (potential) product was studied in the present work. Several of the authors (M.B.S., N.S., M.A., and M.K.) hold a patent related to the work that is described in the present study. All other authors declare no competing financial interests.
Correspondence: Steffen Koschmieder, Department of Medicine/Hematology and Oncology, University of Münster, 48149 Münster, Germany; e-mail:steffen.koschmieder@ukmuenster.de.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal