The proteasome has been successfully targeted for the treatment of multiple myeloma and mantle cell lymphoma; however, in other hematologic malignancies, bortezomib has been less effective as a single agent. Here, we describe effects of NPI-0052, a novel proteasome inhibitor, in leukemia model systems. In cell lines, NPI-0052 inhibits all 3 proteolytic activities associated with the proteasome: chymotrypsin-, trypsin-, and caspase-like. NPI-0052 also induces DNA fragmentation in leukemia lines and in mononuclear cells from a Ph + acute lymphoblastic leukemia (ALL) patient. Caspase-3 activation by NPI-0052 was seen in wild-type Jurkat cells, but was significantly lessened in Fas-associated death domain (FADD)–deficient or caspase-8–deficient counterparts. NPI-0052–induced apoptosis was further probed using caspase-8 inhibitors, which were more protective than caspase-9 inhibitors. N-acetyl cysteine (NAC) also conferred protection against NPI-0052–induced apoptosis, indicating a role for oxidative stress by NPI-0052. In support of the drug's in vitro activities, biweekly treatment with NPI-0052 lessened total white blood cell (WBC) burden over 35 days in leukemic mice. Interestingly, combining NPI-0052 with either MS-275 or valproic acid (VPA) induced greater levels of cell death than the combination of bortezomib with these histone deacetylase inhibitors (HDACi). These effects of NPI-0052, alone and in combination with HDACi, warrant further testing to determine the compound's clinical efficacy in leukemia.
Introduction
The 26S proteasome, composed of 19S and 20S components, is a multicatalytic complex responsible for degrading most intracellular proteins in eukaryotes.1,2 Ubiquitinated proteins are recognized by 19S regulatory caps and are digested into small peptides by the 20S proteasome. Three distinguishable proteolytic activities are localized to beta subunits present in the 20S proteasome and are classified as chymotrypsin-like, caspase-like, and trypsin-like.3 Of these activities, the chymotrypsin-like activity is rate limiting4 and has been targeted by bortezomib (Velcade, PS-341), which is currently the only proteasome inhibitor approved by the Food and Drug Administration.
The rationale to target the proteasome in cancer cells stems from data indicating that malignant cells accumulate more misfolded/mutated/damaged proteins, which are disposed of by the proteasome: therefore, they are more dependent on proteasome activity.5 Validation of this rationale has been provided by bortezomib, a boronic acid dipeptide proteasome inhibitor, currently in use for the treatment of refractory multiple myeloma and mantle cell lymphoma.6,–8 Furthermore, inhibition of the proteasome induces apoptosis and has antitumor effects in several types of cancer cell lines and xenograft models, such as prostate,9 pancreatic,10 lymphoma,11 head and neck,12 melanoma,13 lung,14 breast,15 and leukemia.16 However, the efficacy of bortezomib as a single agent in clinical trials for these malignancies has been restricted to specific disease subtypes.17 Thus, a proteasome inhibitor with a different mode of action may be useful in tumor types where bortezomib did not demonstrate a therapeutic advantage.
This study focuses on NPI-0052, which is currently in clinical trials at the University of Texas M. D. Anderson Cancer Center. NPI-0052 (salinosporamide A)18 is an orally active, nonpeptide small molecule proteasome inhibitor derived from marine bacteria19 . NPI-0052 structurally resembles omuralide, the active form of lactacystin, a potent and specific bacterially derived proteasome inhibitor that is not suitable for in vivo application in humans.20 Besides structural differences,21 NPI-0052 possesses several other features that distinguish it from bortezomib. Unlike bortezomib, which was developed to inhibit the chymotrypsin-like activity of the proteasome, NPI-0052 can inhibit the chymotrypsin-like, caspase-like, and trypsin-like activities of purified human erythrocyte 20S proteasomes. Subsequent studies have shown that bortezomib also inhibits all 3 activities, but NPI-0052 appears to more effectively inhibit the chymotrypsin-like and trypsin-like activities in vivo and in vitro.21 Another distinction between bortezomib and NPI-0052 is the latter's ability to irreversibly bind to the 20S proteasome.22
Cell death resulting from proteasome inhibition requires caspase activation in most cases23 and has been linked to increased levels of reactive oxygen species (ROS).11,16,24 Not surprisingly, bortezomib is well documented to activate several caspases, including caspase-9, -8, -3, and -4.25,26 The relative contributions of caspase-8 and caspase-9 to bortezomib-induced cell death appear similar.25 In contrast, data from multiple myeloma cells21 and our data presented herein from leukemia systems suggest that NPI-0052 relies more heavily on a caspase-8–dependent pathway. We further show that caspase-8 participation is critical for synergy observed when NPI-0052 is combined with the HDACi (histone deacetylase inhibitors), MS-275, and valproic acid (VPA). This reliance on caspase-8 outlines yet another mechanism by which NPI-0052 may exert unique effects in leukemia cells.
Materials and methods
Cells
Jurkat, K562, ML-1, and I2.1 (Fas-associated death domain [FADD]-deficient Jurkat) human leukemia cell lines were purchased from American Type Culture Collection (Rockville, MD). Caspase-8–deficient Jurkat cells, I9.2, were kindly provided by Dr Michael Andreeff (University of Texas M. D. Anderson Cancer Center, Houston, TX). All cells were cultured in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Hyclone, Logan, UT), L-glutamate, and penicillin/streptomycin (Sigma, St Louis, MO). Cells were maintained at 37°C with 5% CO2. Peripheral blood from a Philadelphia-positive (Ph +) acute lymphoblastic leukemia (ALL) patient was obtained after consent under the aegis of a protocol reviewed by the Institutional Review Board of M. D. Anderson Cancer Center. Mononuclear cells were isolated using double-density Ficoll-Hypaque gradients composed of Histopaque 1077 and 1119 (Sigma) as previously described.27
Reagents
NPI-0052 was provided by Nereus Pharmaceuticals (San Diego, CA). Bortezomib (Millennium Pharmaceuticals, Cambridge, MA) was obtained from the M. D. Anderson Cancer Center pharmacy. HDACi's, MS-275, and VPA were obtained from Calbiochem and Sigma (San Diego, CA and St Louis, MO, respectively). Fluorogenic substrates, suc-LLVY-amc and z-LLE-amc, were from AG Scientific, Inc (San Diego, CA) and boc-LLR-amc from Bachem (King of Prussia, PA). N-acetyl cysteine (NAC) and staurosporine were purchased from Sigma. The dyes 6-carboxy-2′,7′-dichlorodihydrofluorescein (H2DCF-DA), dihydroethidium (HEt), and tetramethylrhodamine ethyl ester (TMRE) were obtained from Molecular Probes (Eugene, OR). Antibodies were purchased for caspase-8, caspase-9, caspase-3, FADD, and Bid (Cell Signaling, Beverly, MA); CH-11 (MBL International, Woburn, MA); p27 (Transduction Laboratories, San Diego, CA); cytochrome c (BD PharMingen, San Diego, CA); and actin (Sigma). The caspase inhibitors zVAD-fmk, IETD-fmk, and LEHD-fmk were purchased from Calbiochem. The caspase-3 substrate, DEVD-amc, was obtained from Biomol International, LP (Plymouth Meeting, PA). Annexin V–fluorescein isothiocyanate (Annexin V-FITC) was purchased from BD Bioscience (Franklin Lakes, NJ).
20S proteasome activity assay
The chymotrypsin-like, trypsin-like, and caspase-like activity of the 20S proteasome of leukemia cells was determined by measurement of fluorescence generated from the cleavage of the fluorogenic substrates suc-LLVY-amc, boc-LRR-amc, and z-LLE-amc, respectively.28 Cells were incubated for 1 hour in the presence of diluent or 1 μM NPI-0052, washed with phosphate buffered saline (PBS) and resuspended in 300 μL of a solution containing 20 mM Tris (tris(hydroxymethyl)aminomethane), pH 7.5, 0.1 mM EDTA (ethylenediaminetetraacetic acid), pH 8.0, 20% glycerol, 0.05% Nonidet-P40, 1 mM 2-β mercaptoethanol, 1 mM adenosine triphosphate (ATP) and lysed by freezing and thawing 3 times on dry ice. After centrifugation, supernatants were combined with substrate buffer (50 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], pH 7.5, 5 mM EGTA (ethylene glycol tetraacetic) acid pH 7) and the specific fluorogenic substrate in a 96-well plate and analyzed on a spectrofluorometer (SpectraMax Gemini EM, Molecular Devices, Sunnyvale, CA), using an excitation of 380 nm and an emission of 460 nm.
Western blotting
Cells (5-10 × 106 ) were incubated with indicated concentrations of NPI-0052 and washed with PBS followed by lysing as previously described.29 Aliquots of protein lysates (50 μg) were run on sodium dodecyl sulfate (SDS)–polyacrylamide gels. Protein was transferred to nitrocellulose membranes and blocked either for 1 hour at room temperature or overnight at 4°C with 5% nonfat dry milk. Membranes were incubated with a 1:1000 dilution of primary antibodies in 5% milk/Tris-buffered saline with 0.05% Tween-20 (TBST), followed by corresponding secondary antibodies (1:1000 dilution with 5% milk/TBST). Bound antibodies were detected using enhanced chemiluminescence (ECL plus Western blotting detection system, Amersham Biosciences UK limited, Little Chalfont Buckinghamshire, England).
Assessment of DNA fragmentation
Apoptosis was assessed by propidium iodide (PI) staining followed by fluorescence-activated cell sorting (FACS) analysis as described previously.27 Following incubation with different doses of NPI-0052 for 24 hours, cells were pelleted by centrifugation and resuspended with PBS containing 50 μg/mL PI, 0.1% Triton-X-100, and 0.1% sodium citrate. Samples were stored at 4°C for 24 hours and vortexed before analysis on the FL-3 channel of a flow cytometer (FACSCalibur; Becton Dickinson; Franklin Lakes, NJ). Data were analyzed using Cell Quest Software (BD Bioscience).
Annexin V staining
Externalization of phosphatidylserine (PS) was measured by Annexin V-FITC staining according to the manufacturer's protocol. Briefly, 1 × 106 cells were treated with indicated doses of NPI-0052 for 6 hours, washed twice in cold PBS, and resuspended in 1× binding buffer (0.01 M HEPES, pH 7.4; 0.14 M NaCl; 2.5 mM CaCl 2) and incubated for 30 minutes in the dark at room temperature with 5 μL Annexin V-FITC and 10 μL of 50 μg/mL PI. Samples were analyzed by flow cytometry.
In vivo studies
Eight- to twelve-week-old male CB.17/severe combined immunodeficiency disease (SCID) mice were purchased from Taconic Farms (Hudson, NY). Animal experiments were approved by the M. D. Anderson Institutional Animal Care and Use Committee (IACUC). SCID mice were injected intravenously (tail vein) with 2 × 107 ML-1 cells in 0.2 mL of PBS. Viability of injected cells was greater than 90% as assessed by trypan blue exclusion. Mice were separated into control and treatment groups (n = 6/group), and treatment with NPI-0052 (0.15 mg/kg) or vehicle alone (1% dimethyl sulfoxide [DMSO] in PBS) was given intraperitoneally twice a week for 5 weeks. Blood samples were collected on days 17, 21, 23, 32, and 35. To avoid tail necrosis, the same mouse was not bled at every time point, accounting for 9 bleeds from each group (control versus treated) over the course of 35 days. Complete cell blood counts were performed using a minimum of 50 μL of peripheral blood using a Cell-Dyn (Abbott Diagnostics, Abbott Park, IL) counter at the Department of Veterinary Medicine and Surgery, Section of Veterinary Laboratory Medicine at M. D. Anderson Cancer Center. Post-hoc power analysis indicated that with 9 observations per group (control or NPI-0052), we had 26% power to see a statistically significant (P > .05) difference in slopes of the regression lines for white blood cell (WBC) count (tumor burden) over time. With 36 observations per group, we would have 80% power to see a statistically significant difference between the slopes of the regression lines for WBC count over time.
Transient transfection
Full-length FADD EGFP-N1 plasmid,30 kindly provided by Dr Scott Kaufmann (Mayo Clinic, Rochester, MN), was transfected into FADD-deficient I2.1 cells using Nucleofector Kit V (Amaxa Biosystems, Cologne, Germany) according to the manufacturer's protocol. After 24 hours, the brightest 25% green fluorescent protein (GFP)–positive cells of the total population were sorted by FACS. These GFP-positive cells were treated with 200 nM NPI-0052 for 6 hours and analyzed for caspase-3 activity.
Caspase-3 activity assays
Cells were pelleted, resuspended in 150 μL PBS, and lysed by freezing and thawing. Fifty μL was loaded in triplicate on a 96-well plate. To each well, 150 μL of 50 μM DEVD-amc in DEVD buffer (10% sucrose, 0.001% IGEPAL, 0.1% 3-[(3-cholamidopropyl)dimethylamonio]-1-propyl sulfonate [CHAPS], 5 mM HEPES, pH 7.25) was added. Release of fluorescence (amc) was measured using a spectrofluorometer using an excitation of 355 nm and an emission of 460 nm.
Detection of intracellular peroxides and superoxide
Measurements of intracellular ROS were determined by using cell-permeable dyes as previously described.31 Pelleted cells were resuspended in 1 mL of RPMI medium containing either 10 μM CM-H2DCF-DA or 10 μM HEt (measuring intracellular peroxide and superoxide levels, respectively) and incubated at 37°C for 30 minutes in the dark. Fluorescence intensity was read by flow cytometry on the FL-1 (DCF) or FL-3 (HEt) channel.
Measurement of changes in mitochondrial membrane potential
Cells were pelleted, washed with PBS, and incubated with 25 nM TMRE in 10 mM HEPES, 150 mM NaCl, 5 mM KCl, 1 mM MgCl, and 1.8 mM CaCl, pH 7.4 in a volume of 1 mL for 30 minutes at 37°C in the dark. Cells were washed in PBS, and fluorescence intensity was analyzed on the FL-2 channel of a flow cytometer.
Statistical analyses
Values represent the mean 1 standard deviation (SD) around the mean from 3 separate studies performed in triplicate. Differences in groups were assessed by using paired Student t test and were considered statistically significant at P < .05. For the experiments combining proteasome inhibitors and HDACi, synergism was determined using isobologram analysis based on the Chou and Talalay method with Calcusyn (Biosoft, Ferguson, MO) software program.32 A combination index (CI) value higher than 1.0 indicates synergism: from 0.1 to 0.3 indicates strong synergism, and from 0.3 to 0.7 synergism. A CI = 1.0 indicates additive effects. In vivo data were analyzed with linear regression to estimate the rate of change (ie, the slope of the regression line) of the WBC count over time for the mice treated with diluent or NPI-0052.
Results
NPI-0052 inhibits the 20S proteolytic activities in leukemia cells
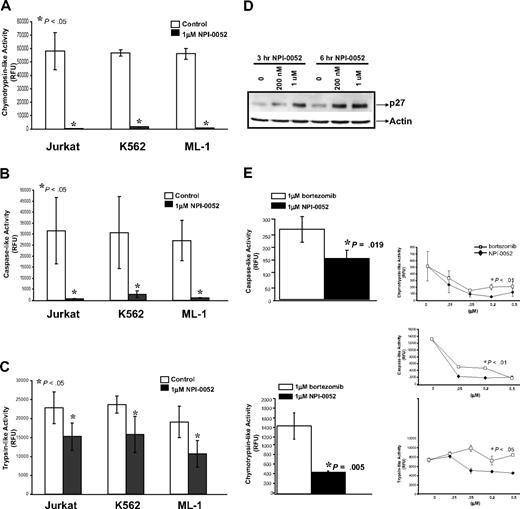
To measure NPI-0052's effects on the proteolytic activities of the 20S proteasome in leukemia cells, we used cell lines representative of chronic myeloid leukemia (CML), ALL, and acute myeloid leukemia (AML) (K562, Jurkat, and ML-1, respectively). Cells were incubated with 1 μM NPI-0052 for 1 hour, and the chymotrypsin-like, caspase-like, and trypsin-like activities were measured using distinct fluorogenic peptides. Figures 1A and 1B show that 1 μM NPI-0052 inhibited the chymotrypsin-like and caspase-like activity by greater than 90% compared with diluent in the 3 leukemia cells lines, whereas the trypsin-like activity was inhibited to a lesser extent (Figure 1C). The accumulation of p27 (Figure 1D) and p21 (data not shown), both known proteasome protein substrates,33,34 in cells treated with NPI-0052 served as functional confirmation of proteasome inhibition.
NPI-0052 inhibits the proteolytic activities of the 20S proteasome in leukemia cell lines. (A) NPI-0052 inhibits the chymotrypsin-like activity of the 20S proteasome in Jurkat, K562, and ML-1 cells. Cells were incubated for 1 hour in the presence (black bars) or absence (white bars) of 1 μM NPI-0052. The chymotrypsin-like activity of the 20S proteasome of leukemia cells was determined by measurement of fluorescence generated from the cleavage of the fluorogenic substrate suc-LLVY-amc. Release of fluorescence (amc) was measured using a spectrofluorometer using an excitation of 380nm and an emission of 460nm. Proteasome activity was evaluated in relative fluorescence units (RFU). (B) Measurement of caspase-like activity in Jurkat, K562, and ML-1 cells treated with NPI-0052. Caspase-like activity of the 20S proteasome detected with fluorogenic peptide z-LLE-amc. (C) Effects on the trypsin-like activity by NPI-0052 in Jurkat, K562, and ML-1 cells. Trypsin-like activity of 20S proteasome measured with boc-LRR-amc fluorogenic substrate. For panels A, B, and C, the data presented are from 3 independent experiments with similar results, mean ± SD (P < .05, significantly different from control). (D) NPI-0052–induced accumulation of proteasomal substrate, p27. Lysates from Jurkat cells treated with NPI-0052 for 3 hours and 6 hours were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted for p27 and actin. (E) Comparison between NPI-0052 and bortezomib inhibition of proteolytic activities of the 20S proteasomes. 1 μM NPI-0052 (black bars) inhibits the caspase-like activity (P = .019) and chymotrypsin-like activity more effectively (P = .005) than 1 μM bortezomib (white bars) in Jurkat cells. The RFU for control was 58081.5 (data not shown). Dose response curves for NPI-0052 (black rhombus) and bortezomib (white squares), measuring the chymotrypsin-, caspase-, and trypsin-like activities (P < .05 for 200 nM dose). The data presented are the mean ± SD from 3 independent experiments performed in triplicate with similar results.
NPI-0052 inhibits the proteolytic activities of the 20S proteasome in leukemia cell lines. (A) NPI-0052 inhibits the chymotrypsin-like activity of the 20S proteasome in Jurkat, K562, and ML-1 cells. Cells were incubated for 1 hour in the presence (black bars) or absence (white bars) of 1 μM NPI-0052. The chymotrypsin-like activity of the 20S proteasome of leukemia cells was determined by measurement of fluorescence generated from the cleavage of the fluorogenic substrate suc-LLVY-amc. Release of fluorescence (amc) was measured using a spectrofluorometer using an excitation of 380nm and an emission of 460nm. Proteasome activity was evaluated in relative fluorescence units (RFU). (B) Measurement of caspase-like activity in Jurkat, K562, and ML-1 cells treated with NPI-0052. Caspase-like activity of the 20S proteasome detected with fluorogenic peptide z-LLE-amc. (C) Effects on the trypsin-like activity by NPI-0052 in Jurkat, K562, and ML-1 cells. Trypsin-like activity of 20S proteasome measured with boc-LRR-amc fluorogenic substrate. For panels A, B, and C, the data presented are from 3 independent experiments with similar results, mean ± SD (P < .05, significantly different from control). (D) NPI-0052–induced accumulation of proteasomal substrate, p27. Lysates from Jurkat cells treated with NPI-0052 for 3 hours and 6 hours were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted for p27 and actin. (E) Comparison between NPI-0052 and bortezomib inhibition of proteolytic activities of the 20S proteasomes. 1 μM NPI-0052 (black bars) inhibits the caspase-like activity (P = .019) and chymotrypsin-like activity more effectively (P = .005) than 1 μM bortezomib (white bars) in Jurkat cells. The RFU for control was 58081.5 (data not shown). Dose response curves for NPI-0052 (black rhombus) and bortezomib (white squares), measuring the chymotrypsin-, caspase-, and trypsin-like activities (P < .05 for 200 nM dose). The data presented are the mean ± SD from 3 independent experiments performed in triplicate with similar results.
Next, we compared the effects of NPI-0052 and bortezomib on the chymotrypsin-like and caspase-like activities. At the 1 μM dose, NPI-0052 inhibited both the chymotrypsin-like and caspase-like activities more effectively than bortezomib (P = .005 and P = .019) in Jurkat cells (Figure 1E). To further characterize the ability of NPI-0052 to inhibit proteasome activities in comparison to bortezomib, dose-dependent effects of the 2 inhibitors were assessed (Figure 1E, inset). At the 200 nM dose, NPI-0052 was more effective than bortezomib at inhibiting the chymotrypsin-like, caspase-like, and trypsin-like activities in Jurkat cells.
NPI-0052 induces apoptotic cell death in vitro and in vivo
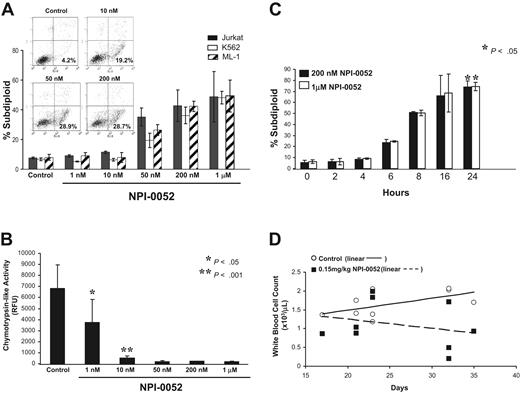
We next examined the cytotoxic effects of NPI-0052 in K562, Jurkat, and ML-1 cells. These cell lines were exposed to 1 nM-1 μM NPI-0052 for 24 hours. NPI-0052 induced DNA fragmentation, which peaked at 200 nM, in a dose-dependent manner as measured by PI staining in the 3 leukemia cell lines (Figure 2A). An increase of Annexin V-positive and PI-negative cells were detected after 6 hours of treatment with various doses of NPI-0052 (19.2% for 10 nM, 28.9% for 50 nM, and 28.7% for 200 nM) (Figure 2A, inset). Cells also were collected after 1 hour of exposure with NPI-0052 and analyzed for inhibition of chymotrypsin-like activity using these same doses. NPI-0052 doses that induced apoptosis also effectively inhibited the chymotrypsin-like activity in Jurkat cells (Figure 2B). Proteasome inhibition by NPI-0052 triggered apoptosis in a time-dependent fashion, with maximum DNA fragmentation occurring at 24 hours (Figure 2C). At a lower dose (10 nM), NPI-0052 achieved significant proteasome inhibition but did not cause DNA fragmentation. Analysis of phosphatidylserine exposure, an early apoptotic event, at the 10 nM dose, however, revealed that 19.2% of cells were Annexin V-positive. These findings indicate that higher doses of NPI-0052 are required to induce apoptosis than those needed to inhibit proteasome activity.
In vitro and in vivo effects of NPI-0052. (A) NPI-0052 treatment causes DNA fragmentation in Jurkat, K562, and ML-1 cells. Cells were treated with increasing doses (0–1 μM) of NPI-0052 for 24 hours and stained with PI. DNA fragmentation was assessed by flow cytometry using the FL-3 channel of a Becton Dickinson FACS Calibur. Inset: Jurkat cells were treated with 0, 10, 50, or 200 nM NPI-0052 for 6 hours, and early apoptotic cells were detected by monitoring FITC-conjugated Annexin V binding to the cell surface (x-axis) and PI staining (y-axis) by flow cytometry. Shown are representative dot plots of 3 separate experiments. Percentages indicate cells that were Annexin V positive and PI negative. (B) NPI-0052 inhibits the chymotrypsin-like activity at the same doses it induces DNA fragmentation. Cells treated for DNA fragmentation with NPI-0052 in Figure 2A also were harvested after only 1 hour of exposure and assessed for chymotrypsin-like activity as described in Figure 1 in triplicate. P < .05 for 1 nM dose and P < .001 for 10 nM dose, compared with control. (C) NPI-0052 triggers DNA fragmentation in a time-dependent manner. Jurkat cells were exposed to 200 nM or 1 μM NPI-0052 for indicated times. DNA fragmentation was determined by flow cytometric analysis of PI staining (P < .05 for 24 hours compared with 0 hours). (D) Total white blood cell counts in NPI-0052–treated mice. CB.17/SCID mice (n = 6/group) were inoculated with 2 × 107 ML-1 cells by tail-vein injection. Mice were administered 0.15 mg/kg NPI-0052 or vehicle alone, IP twice weekly, starting on day 2. Blood samples were collected by tail-vein bleeds at days 17, 21, 23, 32, and 35 and analyzed for complete cell blood counts. Shown are white blood cell counts in 103 /μL over time and linear regression analysis of control (solid line) versus treatment (dotted line). Each data point represents data obtained from one mouse at the indicated time.
In vitro and in vivo effects of NPI-0052. (A) NPI-0052 treatment causes DNA fragmentation in Jurkat, K562, and ML-1 cells. Cells were treated with increasing doses (0–1 μM) of NPI-0052 for 24 hours and stained with PI. DNA fragmentation was assessed by flow cytometry using the FL-3 channel of a Becton Dickinson FACS Calibur. Inset: Jurkat cells were treated with 0, 10, 50, or 200 nM NPI-0052 for 6 hours, and early apoptotic cells were detected by monitoring FITC-conjugated Annexin V binding to the cell surface (x-axis) and PI staining (y-axis) by flow cytometry. Shown are representative dot plots of 3 separate experiments. Percentages indicate cells that were Annexin V positive and PI negative. (B) NPI-0052 inhibits the chymotrypsin-like activity at the same doses it induces DNA fragmentation. Cells treated for DNA fragmentation with NPI-0052 in Figure 2A also were harvested after only 1 hour of exposure and assessed for chymotrypsin-like activity as described in Figure 1 in triplicate. P < .05 for 1 nM dose and P < .001 for 10 nM dose, compared with control. (C) NPI-0052 triggers DNA fragmentation in a time-dependent manner. Jurkat cells were exposed to 200 nM or 1 μM NPI-0052 for indicated times. DNA fragmentation was determined by flow cytometric analysis of PI staining (P < .05 for 24 hours compared with 0 hours). (D) Total white blood cell counts in NPI-0052–treated mice. CB.17/SCID mice (n = 6/group) were inoculated with 2 × 107 ML-1 cells by tail-vein injection. Mice were administered 0.15 mg/kg NPI-0052 or vehicle alone, IP twice weekly, starting on day 2. Blood samples were collected by tail-vein bleeds at days 17, 21, 23, 32, and 35 and analyzed for complete cell blood counts. Shown are white blood cell counts in 103 /μL over time and linear regression analysis of control (solid line) versus treatment (dotted line). Each data point represents data obtained from one mouse at the indicated time.
The ability of NPI-0052 to exert effects in vivo was examined by reconstituting SCID mice with ML-1 cells. Biweekly administration of 0.15 mg/kg NPI-0052 decreased tumor burden, as evidenced by lower total WBCs (Figure 2D) over the course of 5 weeks. Linear regression modeling of WBC over time found a difference in the y-intercepts of the regression lines (P = .06), comparing mice administered NPI-0052 (n = 6 with 9 observations) versus diluent (n = 6 with 9 observations). It is worth noting that the slope of the regression line for the NPI-0052 group was negative (−0.0244) and the slope of the regression line for the control group was positive (0.0317), and that the 2 lines intersect on the first day of treatment (day 2).
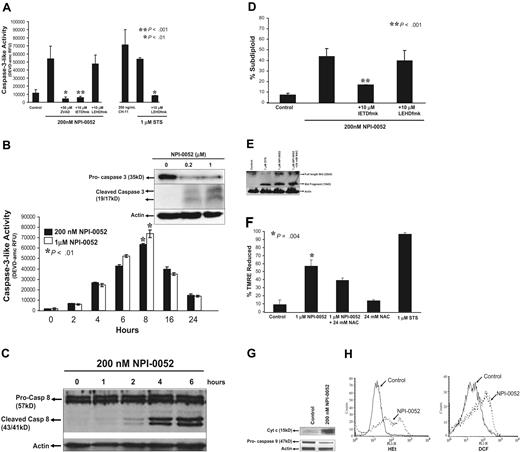
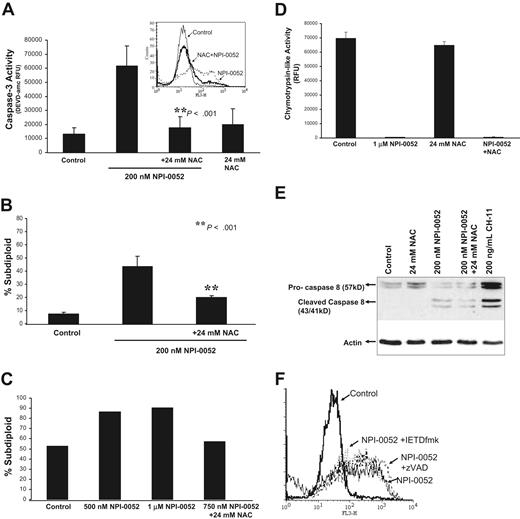
Since DNA fragmentation is a consequence of caspase-3 activation,35 we examined caspase-3 activity in response to NPI-0052 (Figure 3A). Treatment with anti-Fas antibody (CH-11) served as a positive control for caspase-3 activation. As shown in Figure 3A, 200 nM NPI-0052 increased caspase-3 activity by 4-fold as compared with control. Inhibition of caspases by a pan-caspase inhibitor, zVAD-fmk, attenuated the NPI-0052–induced caspase-3 activity. Furthermore, 10 μM IETD-fmk, a caspase-8 inhibitor, abrogated caspase-3 activity by NPI-0052 (P < .001), whereas an equimolar dose of caspase-9 inhibitor, LEHD-fmk, did not. The combination of LEHD-fmk and staurosporine was used as a positive control to demonstrate the effectiveness of the caspase-9 inhibitor (Figure 3A). NPI-0052 induced caspase-3 activity in a time-dependent manner, beginning at 4 hours and plateauing at 8 hours (Figure 3B). Detection of 19-kDa and 17-kDa cleaved products by Western blot analysis (Figure 3B inset) further confirmed caspase-3 activation by NPI-0052.
NPI-0052 induced apoptosis through caspase activation and mitochondrial perturbations. (A) A caspase-8 inhibitor blocks NPI-0052–induced caspase-3 activation. Jurkat cells were treated with 200 nM NPI-0052 and 10 μM IETD-fmk, 10 μM LEHD-fmk, or 50 μM zVAD-fmk for 6 hours. Cells were lysed and caspase-3 activity was measured by the cleavage of DEVD-amc fluorescent peptide as described in “Materials and methods”. ** P < .001, significantly different from NPI-0052–treated cells. (B) NPI-0052 induces activation and cleavage of caspase-3. Cells were treated with 200 nM or 1 μM NPI-0052 over a period of 24 hours. Cells were lysed at indicated time points, and caspase-3 activity was measured as previously described. *P > 0.01 for 8 hour time point compared with 0 hour time point. (B) Inset: Jurkat cell lysates from cells treated with 200 nM or 1 μM NPI-0052 for 8 hours were subjected to SDS-PAGE. Membranes were probed with caspase-3–specific antibody to detect pro–caspase-3 and/or cleaved products or actin-specific antibody. (C) NPI-0052 induces cleavage of caspase-8 in Jurkat cells. Cells were stimulated with 200 nM NPI-0052 for indicated times and subjected to SDS-PAGE. Immunoblotting was performed using caspase-8–specific antibody that detects full and cleaved caspase-8 or actin-specific antibody. (D) A caspase-8 inhibitor protects from NPI-0052–induced apoptosis. DNA fragmentation by PI staining was assessed after exposure of Jurkat cells for 24 hours to 200 nM NPI-0052 and 10 μM caspase-8 inhibitor (IETD-fmk), or 10 μM caspase-9 inhibitor (LEHD-fmk). * P > .001. (E) NPI-0052 induces cleavage of Bid. Jurkat cells were preincubated with 24 mM NAC for 15 minutes followed by treatment with 1 μM NPI-0052 or treated with1 μM NPI-0052 alone or 1 μM staurosporine (STS) as a positive control for 8 hours. Western blot detected full-length and cleaved Bid. (F) NPI-0052 triggers mitochondrial injury. Drops in mitochondrial membrane potential (ΔΨm) were assessed by TMRE staining of Jurkat cells treated with NPI-0052 alone or with pretreatment with 24 mM NAC for 15 minutes prior to NPI-0052 exposure. Samples were analyzed by flow cytometry. *P = .004, compared with control. (G) NPI-0052 induces cytochrome c release and caspase-9 activation. After exposure to 200 nM NPI-0052, Western blots detected cytochrome c in cytosolic fractions after 4 hours and reduction of pro–caspase-9 after 6 hours in lysates from Jurkat cells. (H) NPI-0052 increases intracellular superoxide and hydrogen peroxide levels. Jurkat cells were exposed to 200 nM NPI-0052 for 14 hours and stained with HEt or CM-H2DCF-DA followed by FACS analysis. Experiments A, B, D, and F were from 3 independent experiments and presented as mean ± SD.
NPI-0052 induced apoptosis through caspase activation and mitochondrial perturbations. (A) A caspase-8 inhibitor blocks NPI-0052–induced caspase-3 activation. Jurkat cells were treated with 200 nM NPI-0052 and 10 μM IETD-fmk, 10 μM LEHD-fmk, or 50 μM zVAD-fmk for 6 hours. Cells were lysed and caspase-3 activity was measured by the cleavage of DEVD-amc fluorescent peptide as described in “Materials and methods”. ** P < .001, significantly different from NPI-0052–treated cells. (B) NPI-0052 induces activation and cleavage of caspase-3. Cells were treated with 200 nM or 1 μM NPI-0052 over a period of 24 hours. Cells were lysed at indicated time points, and caspase-3 activity was measured as previously described. *P > 0.01 for 8 hour time point compared with 0 hour time point. (B) Inset: Jurkat cell lysates from cells treated with 200 nM or 1 μM NPI-0052 for 8 hours were subjected to SDS-PAGE. Membranes were probed with caspase-3–specific antibody to detect pro–caspase-3 and/or cleaved products or actin-specific antibody. (C) NPI-0052 induces cleavage of caspase-8 in Jurkat cells. Cells were stimulated with 200 nM NPI-0052 for indicated times and subjected to SDS-PAGE. Immunoblotting was performed using caspase-8–specific antibody that detects full and cleaved caspase-8 or actin-specific antibody. (D) A caspase-8 inhibitor protects from NPI-0052–induced apoptosis. DNA fragmentation by PI staining was assessed after exposure of Jurkat cells for 24 hours to 200 nM NPI-0052 and 10 μM caspase-8 inhibitor (IETD-fmk), or 10 μM caspase-9 inhibitor (LEHD-fmk). * P > .001. (E) NPI-0052 induces cleavage of Bid. Jurkat cells were preincubated with 24 mM NAC for 15 minutes followed by treatment with 1 μM NPI-0052 or treated with1 μM NPI-0052 alone or 1 μM staurosporine (STS) as a positive control for 8 hours. Western blot detected full-length and cleaved Bid. (F) NPI-0052 triggers mitochondrial injury. Drops in mitochondrial membrane potential (ΔΨm) were assessed by TMRE staining of Jurkat cells treated with NPI-0052 alone or with pretreatment with 24 mM NAC for 15 minutes prior to NPI-0052 exposure. Samples were analyzed by flow cytometry. *P = .004, compared with control. (G) NPI-0052 induces cytochrome c release and caspase-9 activation. After exposure to 200 nM NPI-0052, Western blots detected cytochrome c in cytosolic fractions after 4 hours and reduction of pro–caspase-9 after 6 hours in lysates from Jurkat cells. (H) NPI-0052 increases intracellular superoxide and hydrogen peroxide levels. Jurkat cells were exposed to 200 nM NPI-0052 for 14 hours and stained with HEt or CM-H2DCF-DA followed by FACS analysis. Experiments A, B, D, and F were from 3 independent experiments and presented as mean ± SD.
Caspase-8 activation by NPI-0052
To verify a role for caspase-8 activation as an early event in NPI-0052–induced cell death, we measured cleavage of caspase-8 in Jurkat cells (Figure 3C) over a 6 hour time period by immunoblotting. Exposure of Jurkat cells to 200 nM NPI-0052 caused activation of caspase-8 starting at 2 hours, indicated by the appearance of the 43-kDa and 41-kDa cleavage fragments.
The relative contributions of caspase-8 and caspase-9 on NPI-0052–induced apoptosis were assessed using peptide inhibitors. Jurkat cells were treated with 200 nM NPI-0052 alone or in combination with caspase-8 and caspase-9 inhibitors for 24 hours, and DNA fragmentation was assessed. When pretreated with a caspase-8 inhibitor, Jurkat cells were protected against NPI-0052–induced apoptosis in a statistically significant manner (P < .001), whereas a caspase-9 inhibitor did not confer protection (Figure 3D).
Bid is a proapoptotic BH-3 domain–containing member of the Bcl-2 family and is a substrate for caspase-8.36 Exposure to 1 μM NPI-0052 for 8 hours induced the cleavage of Bid (Figure 3E), generating a 15-kDa fragment, tBid, which can translocate to mitochondria. Thus, NPI-0052 is likely exerting its cytotoxic effects through a caspase-8-tBid-mitochondria–dependent pathway.
Mitochondrial injury by NPI-0052 was evaluated by detecting drops in mitochondrial membrane potential (ΔΨm). Reduction in TMRE fluorescence (indicative of loss of ΔΨm) by 56.74% was seen in Jurkat cells treated with 1 μM NPI-0052 for 6 hours (Figure 3F). This drop in potential was not abrogated by an antioxidant, NAC. Staurosporine, a positive control, caused a 96.8% reduction in ΔΨm. These results support a model in which NPI-0052 causes caspase-8 activation, leading to Bid cleavage and mitochondrial perturbations.
Loss of mitochondrial membrane potential generally precedes cytochrome c release from mitochondria, which leads to caspase-9 activation in the cytosol.37 Figure 3G shows the presence of cytochrome c in cytosol after 4 hours of NPI-0052 treatment but not in diluent-treated cells. Accordingly, levels of pro–caspase-9 were decreased in NPI-0052–treated cells, indicating activation of the zymogen (Figure 3G).
Several reports indicate that proteasome inhibition causes increased ROS levels.12,16,24 To determine if ROS levels are heightened by NPI-0052, Jurkat cells were treated with 200 nM NPI-0052 and stained with HEt or dichlorofluorescein (DCF). Monitoring ROS over time revealed strong increases in intracellular superoxide and hydrogen peroxide after 14 hours of exposure (Figure 3H).
Requirement for caspase-8 in NPI-0052–induced apoptosis in leukemia cells
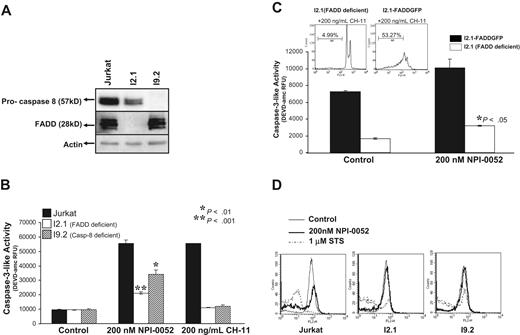
To address if caspase-8 is required for NPI-0052–induced apoptosis, we used Jurkat cells lacking caspase-8 (I9.2) and cells lacking the caspase-8 adaptor molecule FADD (I2.1). The absence of caspase-8 and FADD was confirmed by Western blotting (Figure 4A). Both of these cell lines showed significant inhibition of chymotrypsin-like proteasome activity after exposure to 1 μM NPI-0052 (data not shown), indicating that caspase-8 and FADD are not required for NPI-0052 to inhibit the proteasome.
The role of caspase-8 and FADD in NPI-0052–induced apoptosis. (A) Caspase-8 and FADD expression. Jurkat, I2.1 and I9.2 protein lysates were assessed for expression of caspase-8 and FADD by immunoblotting. (B) NPI-0052–induced caspase-3 activity is mediated by caspase-8 and FADD. Jurkat, I2.1 (FADD deficient) and I9.2 (caspase-8 deficient) cells were incubated in the presence of 200 nM NPI-0052 for 6 hours, and caspase-3 activity was monitored by cleavage of DEVD-amc. Anti-Fas antibody (CH-11) was used as positive control. Values represent the mean ± SD from 3 separate studies performed in triplicate (P > .05). (C) Re-expression of FADD in I2.1 cells induces caspase-3 activation. I2.1 cells and I2.1 transfected cells with full-length FADD (I2.1-FADD GFP) were treated with NPI-0052 for 6 hours, and caspase-3 activity was monitored. *P > .05. Inset: PI staining assessed DNA fragmentation in I2.1 and I2.1-FADD GFP cells exposed to 200 ng/mL CH-11 for 12 hours. (D) FADD and caspase-8 are required for NPI-0052–induced mitochondrial injury. Following 4 hours of 200 nM NPI-0052 exposure, TMRE staining, followed by FACS analysis, was employed to monitor Jurkat, I2.1 and I9.2 cells for mitochondrial injury. Histograms shown are representative of 3 separate experiments.
The role of caspase-8 and FADD in NPI-0052–induced apoptosis. (A) Caspase-8 and FADD expression. Jurkat, I2.1 and I9.2 protein lysates were assessed for expression of caspase-8 and FADD by immunoblotting. (B) NPI-0052–induced caspase-3 activity is mediated by caspase-8 and FADD. Jurkat, I2.1 (FADD deficient) and I9.2 (caspase-8 deficient) cells were incubated in the presence of 200 nM NPI-0052 for 6 hours, and caspase-3 activity was monitored by cleavage of DEVD-amc. Anti-Fas antibody (CH-11) was used as positive control. Values represent the mean ± SD from 3 separate studies performed in triplicate (P > .05). (C) Re-expression of FADD in I2.1 cells induces caspase-3 activation. I2.1 cells and I2.1 transfected cells with full-length FADD (I2.1-FADD GFP) were treated with NPI-0052 for 6 hours, and caspase-3 activity was monitored. *P > .05. Inset: PI staining assessed DNA fragmentation in I2.1 and I2.1-FADD GFP cells exposed to 200 ng/mL CH-11 for 12 hours. (D) FADD and caspase-8 are required for NPI-0052–induced mitochondrial injury. Following 4 hours of 200 nM NPI-0052 exposure, TMRE staining, followed by FACS analysis, was employed to monitor Jurkat, I2.1 and I9.2 cells for mitochondrial injury. Histograms shown are representative of 3 separate experiments.
Caspase-3 activity was assessed after NPI-0052 exposure in I2.1 and I9.2 cells. Caspase-8– and FADD-deficient cell lines moderately increased caspase-3 activity 3-fold, whereas wild-type Jurkat cells exhibited a 6-fold increase as compared with control (Figure 4B). Thus, caspase-8 and FADD mediate NPI-0052–induced caspase-3 activity.
The specific requirement for FADD in NPI-0052–induced apoptosis was further tested by transfection of wild-type FADD-GFP into the FADD-deficient I2.1 cells and subsequent treatment with 200 nM NPI-0052 (Figure 4C). In comparison to parental I2.1 cells, the FADD transfectants displayed higher levels of caspase-3 activity, and the difference was statistically significant (P < .05). As a positive control, we show that 200 ng/mL CH-11 induced DNA fragmentation in I2.1 FADD-GFP transfectants (53.27%) but not in the I2.1 cell line (4.99%) (Figure 4C, inset).
To place caspase-8 activation in the context of other biochemical apoptotic events, we examined alterations of ΔΨm in parental, caspase-8–deficient, and FADD-deficient Jurkat cells. Jurkat cells exposed to 200 nM NPI-0052 displayed a loss of ΔΨm as detected by TMRE staining and indicated by a shift of the histogram (Figure 4D). The caspase-8– and FADD-deficient, I9.2 and I2.1, cells did not display a drop in ΔΨm in the presence of NPI-0052. These data indicate that caspase-8 and FADD are required for NPI-0052 to cause mitochondrial perturbations.
Requirement for NPI-0052–induced ROS in cytotoxicity and proteasome inhibition
To test if increased levels of ROS by NPI-0052 contribute to apoptosis, Jurkat cells were treated with 200 nM NPI-0052 alone or in combination with the antioxidant, NAC. Figure 5A shows that an 8 hour exposure to NPI-0052 caused an increase in caspase-3 activity that was attenuated 3-fold in the presence of NAC. Superoxide levels generated by 100 nM NPI-0052 were blunted in the presence of NAC (Figure 5A, inset). Furthermore, the antioxidant conferred significant protection (P < .001) against NPI-0052–induced apoptosis as measured by DNA fragmentation (Figure 5B). Similarly, in mononuclear cells isolated from a Ph + ALL patient, NAC also protected against cell death by NPI-0052 (Figure 5C).
The free radical scavenger, NAC, protects from NPI-0052–induced apoptosis. (A) NAC attenuates NPI-0052 activation of caspase-3. Jurkat cells pretreated with 24 mM NAC for 15 minutes prior to exposure of 200 nM NPI-0052 for 6 hours. Caspase-3 activity was assessed by measuring the cleavage of DEVD-amc. A significant reduction (P > .001) in caspase-3 activity was observed in the presence of NAC. Inset: Pretreatment of Jurkat cells with 24 mM NAC for 15 minutes prior to 100 nM NPI-0052 exposure reduces superoxide levels as measured by HEt staining and flow cytometric analysis on the FL3 channel. (B) NAC confers protection from NPI-0052–induced apoptosis. DNA fragmentation was assessed by PI staining of Jurkat cells treated with 200 nM NPI-0052 alone or in combination with 24 mM NAC (P > .001) for 24 hours. (C) NPI-0052 induces oxidant-dependent apoptosis in a Ph + ALL patient sample. Lymphocytes from patient material were separated by centrifugation on a double-density Histopaque gradient. Cells were incubated in the presence/absence of 24 mM NAC and 500 nM, 1 μM or 750 nM NPI-0052. After 24 hours, apoptosis was assessed by PI staining and subsequent flow cytometric analysis. (D) Effects of combination treatment with NPI-0052 and NAC on the chymotrypsin-like activity. Chymotrypsin-like activity was determined in Jurkat cells exposed to 1 μM NPI-0052 and/or 24 mM NAC after 1 hour. (E) NAC does not protect from NPI-0052 activation of caspase-8. Cell lysates from Jurkats treated with 24 mM NAC, 200 nM NPI-0052, and 200 ng/mL CH-11 (positive control) for 6 hours were subjected to immunoblotting for caspase-8 and actin. (F) Neither a pan-caspase nor caspase-8 inhibitor protects from NPI-0052–induced ROS generation. HEt staining was performed in Jurkat cells treated with 200 nM NPI-0052 and/or 50 μM zVAD-fmk or 50 μM IETD-fmk for 14 hours to detect intracellular O2 levels.
The free radical scavenger, NAC, protects from NPI-0052–induced apoptosis. (A) NAC attenuates NPI-0052 activation of caspase-3. Jurkat cells pretreated with 24 mM NAC for 15 minutes prior to exposure of 200 nM NPI-0052 for 6 hours. Caspase-3 activity was assessed by measuring the cleavage of DEVD-amc. A significant reduction (P > .001) in caspase-3 activity was observed in the presence of NAC. Inset: Pretreatment of Jurkat cells with 24 mM NAC for 15 minutes prior to 100 nM NPI-0052 exposure reduces superoxide levels as measured by HEt staining and flow cytometric analysis on the FL3 channel. (B) NAC confers protection from NPI-0052–induced apoptosis. DNA fragmentation was assessed by PI staining of Jurkat cells treated with 200 nM NPI-0052 alone or in combination with 24 mM NAC (P > .001) for 24 hours. (C) NPI-0052 induces oxidant-dependent apoptosis in a Ph + ALL patient sample. Lymphocytes from patient material were separated by centrifugation on a double-density Histopaque gradient. Cells were incubated in the presence/absence of 24 mM NAC and 500 nM, 1 μM or 750 nM NPI-0052. After 24 hours, apoptosis was assessed by PI staining and subsequent flow cytometric analysis. (D) Effects of combination treatment with NPI-0052 and NAC on the chymotrypsin-like activity. Chymotrypsin-like activity was determined in Jurkat cells exposed to 1 μM NPI-0052 and/or 24 mM NAC after 1 hour. (E) NAC does not protect from NPI-0052 activation of caspase-8. Cell lysates from Jurkats treated with 24 mM NAC, 200 nM NPI-0052, and 200 ng/mL CH-11 (positive control) for 6 hours were subjected to immunoblotting for caspase-8 and actin. (F) Neither a pan-caspase nor caspase-8 inhibitor protects from NPI-0052–induced ROS generation. HEt staining was performed in Jurkat cells treated with 200 nM NPI-0052 and/or 50 μM zVAD-fmk or 50 μM IETD-fmk for 14 hours to detect intracellular O2 levels.
Examination of proteasome activity in Jurkat cells revealed that protection by NAC was not due to interference with NPI-0052 effects on proteasome function (Figure 5D). In addition, Western blot analysis results demonstrate that NAC did not prevent caspase-8 or Bid cleavage in Jurkat cells treated with NPI-0052 (Figure 5E and 3E). Interestingly, neither pan-caspase inhibitors nor specific caspase-8 inhibitors inhibited superoxide production by NPI-0052 (Figure 5F). We also observed that NAC does not protect from alterations to mitochondrial membrane potential (Figure 3F). These results indicate that inhibition of the proteasome and increases in ROS by NPI-0052 are independent of caspase-8 and Bid activation and mitochondrial perturbations.
NPI-0052 interacts with HDACi to induce synergistic apoptosis
HDACi have been previously reported to synergistically interact with proteasome inhibitors to induce apoptosis.38,39 Therefore, we examined whether the HDACi, MS-275, and VPA can be combined with NPI-0052 to enhance apoptosis in leukemia cells. Jurkat cells were treated with increasing doses of MS-275 (1-5 μM) or VPA (1-5 mM) and low doses of NPI-0052 (10 nM or 5 nM) for 24 hours. The combination of 2.5 μM or 5 μM MS-275 and 10 nM NPI-0052 significantly increases DNA fragmentation (P < .05) when compared with cells exposed to a single agent (Figure 6A). Furthermore, cells treated with 10 nM NPI-0052 and MS-275 had a higher percent increase in the subdiploid population (Figure 6C; P < .05) and displayed greater synergism (CI = 0.27 for 2.5 μM MS-275 and CI = 0.21 for 5 μM MS-275) compared with cells treated with 10 nM bortezomib and MS-275 (CI = 0.48 for 2.5 μM MS-275 and CI = 0.47 for 5 μM MS-275). To explore the dependence upon caspase-8 of this observed synergy, we compared parental Jurkat cells to caspase-8–deficient counterparts (I9.2). As predicted by the subdiploid data, caspase-3 activation was potentiated in parental Jurkat cells treated with the combination of NPI-0052 and MS-275 (Figure 6B; P < .001). However, caspase-8–deficient cells did not display additive or synergistic effects. Combined treatment of NPI-0052 and MS-275 resulted in an increase of intracellular superoxide levels (Figure 6B, inset). Exposure of Jurkat cells to low doses of VPA (1 or 2.5 mM) and NPI-0052 (5 nM) also displayed significantly enhanced apoptosis (Figure 6D; P < .05). As shown in Figure 6D (inset), treatment with 5 nM NPI-0052 and 2.5 mM VPA resulted in a greater increase in the subdiploid population (P < .05) as compared with 5 nM bortezomib and VPA. Finally, combination index values indicated synergism between 5 nM NPI-0052 and 2.5 mM VPA (CI = 0.70), while 5 nM bortezomib and 2.5 mM VPA displayed additive effects (CI = 1.06). These findings demonstrate for the first time that combination of HDACi (either MS-275 or VPA) with low doses of NPI-0052 results in synergistic induction of apoptosis, and these effects are more potent than those seen when bortezomib is combined with the same agents.
Low doses of NPI-0052 and HDAC inhibitors induce synergistic apoptosis. (A) NPI-0052 synergizes with MS-275 to induce cytotoxicity. Jurkat cells were exposed to increasing doses of MS-275 (1-5 μM) and 10 nM NPI-0052 for 24 hours. Cells were assessed for apoptosis by measuring DNA fragmentation as previously described. The combination of 2.5 mM or 5 mM MS-275 with 10 nM NPI-0052 was statistically significant, P > .05, when compared with either single agent alone. (B) ROS and caspase-8 involvement in NPI-0052– and MS-275–induced apoptosis. Caspase-3 activity in wild-type Jurkat and their caspase-8–deficient counterparts exposed to 10 nM NPI-0052 and 5 μM MS-275 for 6 hours. *P > .001 comparing NPI-0052 treatment versus combination treatment in Jurkat. **P > .001 comparing Jurkat treated with NPI-0052 and MS-275 versus I9.2 cells also treated with both agents. Inset: Superoxide levels measured by HEt staining in Jurkat cells incubated with MS-275 and NPI-0052. Shown is a representative histogram of 3 separate experiments and mean fluorescent (MF) values. (C) NPI-0052 synergizes with MS-275 more effectively than bortezomib. Cells were treated with indicated doses of MS-275 and 10 nM NPI-0052 or 10 nM bortezomib for 24 hours. PI staining determined the percentage of cells undergoing DNA fragmentation. Shown is the percent increase in the subdiploid population; *P > .05. Isobologram analysis determined the combination index (CI) values, calculated by Calcusyn software. CI > 0.1 indicates synergism, where a CI range from 0.1-0.3 indicates strong synergism, and from 0.3-0.7 synergism. CI values for 10 nM bortezomib and MS-275 were CI = 0.48 for 2.5 μM MS-275 and CI = 0.47 for 5 μM MS-275. CI values for 10 nM NPI-0052 and MS-275 were CI = 0.27 for 2.5 μM MS-275 and CI = 0.21 for 5 μM MS-275. (D) Low-dose NPI-0052 and Valproic acid synergistically enhance apoptosis. DNA fragmentation was assessed in cells treated with 1 mM or 2.5 mM VPA and 5 nM NPI-0052 for 24 hours. *P > .05. Inset: Jurkat cells were incubated with 2.5 mM VPA and 10 nM NPI-0052 or 10 nM bortezomib. After 24 hours, cells were stained with PI to determine DNA fragmentation. Shown is the percent increase in the subdiploid population. Analysis of synergism by isobologram determined the CI values. CI > 0.1 indicates synergism. CI = 1.0 indicates additive effects.
Low doses of NPI-0052 and HDAC inhibitors induce synergistic apoptosis. (A) NPI-0052 synergizes with MS-275 to induce cytotoxicity. Jurkat cells were exposed to increasing doses of MS-275 (1-5 μM) and 10 nM NPI-0052 for 24 hours. Cells were assessed for apoptosis by measuring DNA fragmentation as previously described. The combination of 2.5 mM or 5 mM MS-275 with 10 nM NPI-0052 was statistically significant, P > .05, when compared with either single agent alone. (B) ROS and caspase-8 involvement in NPI-0052– and MS-275–induced apoptosis. Caspase-3 activity in wild-type Jurkat and their caspase-8–deficient counterparts exposed to 10 nM NPI-0052 and 5 μM MS-275 for 6 hours. *P > .001 comparing NPI-0052 treatment versus combination treatment in Jurkat. **P > .001 comparing Jurkat treated with NPI-0052 and MS-275 versus I9.2 cells also treated with both agents. Inset: Superoxide levels measured by HEt staining in Jurkat cells incubated with MS-275 and NPI-0052. Shown is a representative histogram of 3 separate experiments and mean fluorescent (MF) values. (C) NPI-0052 synergizes with MS-275 more effectively than bortezomib. Cells were treated with indicated doses of MS-275 and 10 nM NPI-0052 or 10 nM bortezomib for 24 hours. PI staining determined the percentage of cells undergoing DNA fragmentation. Shown is the percent increase in the subdiploid population; *P > .05. Isobologram analysis determined the combination index (CI) values, calculated by Calcusyn software. CI > 0.1 indicates synergism, where a CI range from 0.1-0.3 indicates strong synergism, and from 0.3-0.7 synergism. CI values for 10 nM bortezomib and MS-275 were CI = 0.48 for 2.5 μM MS-275 and CI = 0.47 for 5 μM MS-275. CI values for 10 nM NPI-0052 and MS-275 were CI = 0.27 for 2.5 μM MS-275 and CI = 0.21 for 5 μM MS-275. (D) Low-dose NPI-0052 and Valproic acid synergistically enhance apoptosis. DNA fragmentation was assessed in cells treated with 1 mM or 2.5 mM VPA and 5 nM NPI-0052 for 24 hours. *P > .05. Inset: Jurkat cells were incubated with 2.5 mM VPA and 10 nM NPI-0052 or 10 nM bortezomib. After 24 hours, cells were stained with PI to determine DNA fragmentation. Shown is the percent increase in the subdiploid population. Analysis of synergism by isobologram determined the CI values. CI > 0.1 indicates synergism. CI = 1.0 indicates additive effects.
Discussion
Recent studies have described the effects of NPI-0052 in myeloma cell lines and animal models,21 and in primary chronic lymphocytic leukemia (CLL) cells.40 Our work details NPI-0052's effects on proteasome activity and the apoptotic machinery in other hematologic malignancies, with a focus on ALL and AML model systems. In the current study, we show that NPI-0052 inhibits all 3 activities associated with the 20S proteasome in leukemia cells (Figure 1) and induces apoptosis in a variety of leukemia cells. Cells representative of ALL, CML, AML, as well as mononuclear cells from a Ph + ALL patient, are sensitive to NPI-0052 (Figure 2A and 5C). In vivo administration of NPI-0052 also decreases tumor burden in leukemia-bearing mice (Figure 2E). Our data are in agreement with findings from recent studies in multiple myeloma21 and CLL40 and demonstrate a spectrum of activity that extends to numerous hematologic malignancies.
Several unique features of NPI-0052 hint at the potential for efficacy in diseases where bortezomib has been less successful. Firstly, NPI-0052 inhibits the 20S proteolytic activities in leukemic cells to different degrees, blocking the chymotrypsin-like and caspase-like activities more effectively than the trypsin-like activity (Figure 1A-C). In addition, we found NPI-0052 to be more potent than bortezomib in inhibiting the rate-limiting activity of the proteasome (Figure 1E). Since others report that proteasome activities are allosterically regulated4 and that inhibition of multiple sites of the proteasome is necessary to block significant protein degradation,41 NPI-0052's pattern of inhibition may impact proteasome function.
The mode of apoptosis induction by NPI-0052 also may be influenced by this unique differential inhibition of proteasome activities. Caspase-8 inhibition appears to be critical for NPI-0052's cytotoxicity, whereas caspase-9 inhibitors did not significantly protect against DNA fragmentation or caspase-3 activation (Figure 3A,D). Nevertheless, pro-enzyme caspase-9 disappearance is detected by Western blot (Figure 3G), indicating activation of caspase-9 by NPI-0052. Since upstream caspases (caspase-8 in our case) can cause activation of other caspases, it is likely that the disappearance of pro–caspase-9 is via this mechanism. Our results further suggest that caspase-8 is activating caspase-9, since experiments using caspase-8 inhibitors, or ALL cell lines lacking caspase-8 or FADD, showed diminished mitochondrial perturbations, caspase-3 activity, and DNA fragmentation (Figures 3A,D and 4B,D). These results place caspase-8 at the apex of the apoptotic cascade triggered by NPI-0052 (Figure 7). Our use of lymphocyte models with total caspase-8 and FADD deficiency extends and confirms data from multiple myeloma cell lines transfected with dominant-negative caspase-8, caspase-9, or FADD constructs.21 Furthermore, our studies show Bid cleavage and mitochondrial perturbations by NPI-0052 in wild-type Jurkat cells, whereas caspase-8– and FADD-deficient Jurkat cells do not undergo mitochondrial potential drops (Figure 4D). These results clearly link NPI-0052 to caspase-8, causing cleavage of Bid, loss of mitochondrial membrane potential, cytochrome c release, caspase-9 activation, caspase-3 activation, and DNA fragmentation (Figure 7).
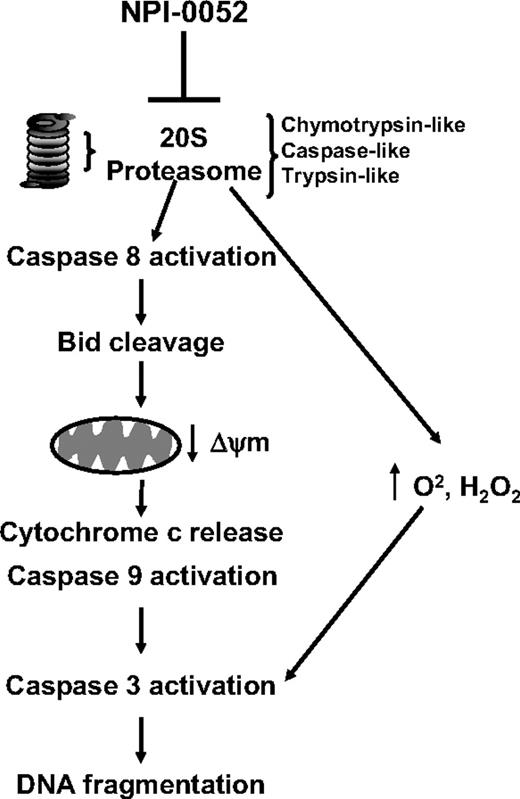
Schematic representation of the mechanism of action of NPI-0052. Inhibition of the chymotrypsin-like, caspase-like, and trypsin-like activities of the 20S proteasome in leukemia cells by NPI-0052 results in activation of caspase-8. This leads to Bid cleavage and drops of mitochondrial membrane potential (ΔΨm). Release of cytochrome c and activation of caspase-9, likely consequences of mitochondrial perturbations, follow. Activation of effector caspase-3 and consequent DNA fragmentation then are observed. Increased levels of intracellular superoxide and peroxide are highest after 14 hours of exposure to NPI-0052. However, caspase-8 inhibitors do not prevent this increase, suggesting that a parallel pathway contributes to cell death by NPI-0052, since the antioxidant NAC prevents the drug's cytotoxicity.
Schematic representation of the mechanism of action of NPI-0052. Inhibition of the chymotrypsin-like, caspase-like, and trypsin-like activities of the 20S proteasome in leukemia cells by NPI-0052 results in activation of caspase-8. This leads to Bid cleavage and drops of mitochondrial membrane potential (ΔΨm). Release of cytochrome c and activation of caspase-9, likely consequences of mitochondrial perturbations, follow. Activation of effector caspase-3 and consequent DNA fragmentation then are observed. Increased levels of intracellular superoxide and peroxide are highest after 14 hours of exposure to NPI-0052. However, caspase-8 inhibitors do not prevent this increase, suggesting that a parallel pathway contributes to cell death by NPI-0052, since the antioxidant NAC prevents the drug's cytotoxicity.
The mechanism by which proteasome inhibition leads to caspase-8 activation is unclear. Cells lacking caspase-8 or FADD still exhibit proteasome inhibition after exposure to NPI-0052 (data not shown), ruling out the possibility that proteasome function may be modulated by caspase-8. In experiments conducted with other proteasome inhibitors, up-regulation of the death receptors, DR4 and DR5, was observed.42,–44 Decreased levels of cellular FLICE-inhibitory protein (cFLIP), which competes with and blocks caspase-8 activation, also have been documented in bortezomib-treated cells45 and could allow caspase-8 activation to proceed unchecked. Although these experiments provide potential insight into our data, further studies need to be done to address activation of caspase-8 as a consequence of proteasome inhibition by NPI-0052. Recent work shows that bortezomib causes endoplasmic reticulum (ER) stress and apoptosis mediated by caspase-4.26 NPI-0052 similarly causes caspase-4 activation in CLL cells.40 However, a hierarchical ordering of activation of other caspases relative to caspase-4 was not conducted in that study, and our results indicate that caspase-4 inhibition does not affect NPI-0052 cytotoxicity in ALL cells (data not shown), pointing to cell type–dependent activities of proteasome inhibition.
The antioxidant NAC conferred protection against NPI-0052–induced caspase-3 activity and apoptosis (Figure 5A,B). Furthermore, increased peroxide and superoxide levels are detectable after exposure to NPI-0052 (Figure 3H). Since caspase-8 activation proceeds in the presence of NAC (Figure 5E) and caspase-8 inhibitors do not affect NPI-0052's ability to raise intracellular superoxide levels (Figure 5F), these data place ROS alterations in a parallel pathway. Consistent with this model, NAC does not alter proteasome activity (Figure 5D). Several structurally dissimilar proteasome inhibitors are reported to cause elevated levels of intracellular superoxide and intracellular peroxides.11,12,24,46 The source of this oxidant production is of interest, since inhibition of ROS by NAC prevents cytotoxicity in numerous models.
Our results show for the first time that HDACi and low doses of novel proteasome inhibitor, NPI-0052, synergistically induce apoptosis in leukemic cells. This is in accordance with previous reports that show HDACi synergize with other proteasome inhibitors, namely bortezomib and MG132, to induce cell death.39,47,48 We observed synergistic interactions in leukemia cells exposed to low doses of NPI-0052 and MS-275 or VPA (Figure 6), whereas in cells treated with bortezomib and HDACi, less synergistic and additive effects were observed with MS-275 and VPA, respectively (Figure 6C,D, inset). The mechanism by which NPI-0052 synergizes with HDACi still remains to be resolved. However, HDACi have been shown to raise intracellular ROS levels,49 and our results indicate that the combination of NPI-0052 and MS-275 cause a further increase in superoxide than seen with either agent alone (Figure 6B, inset). Thus, it is conceivable that this greater oxidative challenge may contribute to the synergistic effects. Our results also implicate caspase-8 in the synergistic apoptosis induced by NPI-0052 and MS-275 (Figure 6B). Our data reveal the potential of administering NPI-0052 at low doses (nontoxic) with HDACi, for clinical benefit. This is a promising finding, given the recent FDA approval of the HDACi vorinostat (Zolinza).
Taken together, our data suggest that NPI-0052 is a potent proteasome inhibitor with potential therapeutic value in several hematologic malignancies. With NPI-0052 currently in clinical trials for refractory solid tumors and lymphoma, the present findings warrant consideration of testing efficacy in leukemia.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
C.M. is funded by National Institutes of Health grant F31CA123645–01. One of the authors (M.P.) is employed by a company (Nereus Pharmaceuticals) whose (potential) product was studied in the present work.
J.C. received support from the Leukemia SPORE Career Development Award to M. D. Anderson from NCI (P50 CA100632–04).
National Institutes of Health (NIH)
Authorship
Contribution: C.M. performed research, analyzed data, and cowrote the paper. K.B. and M.D. performed research. D.M. aided in designing the research. M.M. analyzed data, M.P. contributed vital reagents. J.C. designed the research, performed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Joya Chandra, PhD, Box 853, Pediatrics Research, University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: jchandra@mdanderson.org.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal