Abstract
Minor histocompatibility antigens (mHags) play an important role in both graft-versus-tumor effects and graft-versus-host disease (GVHD) after allogeneic stem cell transplantation. We applied biochemical techniques and mass spectrometry to identify the peptide recognized by a dominant tumor-reactive donor T-cell reactivity isolated from a patient with relapsed multiple myeloma who underwent transplantation and entered complete remission after donor lymphocyte infusion. A frequently occurring single nucleotide polymorphism in the human ATP-dependent interferon-responsive (ADIR) gene was found to encode the epitope we designated LB-ADIR-1F. Although gene expression could be found in cells from hematopoietic as well as nonhematopoietic tissues, the patient suffered from only mild acute GVHD despite high percentages of circulating LB-ADIR-1F–specific T cells. Differential recognition of nonhematopoietic cell types and resting hematopoietic cells as compared with activated B cells, T cells, and tumor cells was demonstrated, illustrating variable LB-ADIR-1F expression depending on the cellular activation state. In conclusion, the novel mHag LB-ADIR-1F may be a suitable target for cellular immunotherapy when applied under controlled circumstances.
Introduction
Allogeneic stem cell transplantation (SCT) is a curative treatment in patients with hematologic cancers. In addition to the antitumor effects of chemotherapy, antibody treatment and/or irradiation administered as the conditioning regimen prior to transplantation, an allogeneic graft-versus-tumor (GVT) immunoreactivity significantly contributes to the curative potential of this therapy.1–5 This GVT reactivity following HLA-matched SCT has been demonstrated to be mediated primarily by T cells from the donor. All reactive T cells from donor origin may not only mediate the beneficial GVT effect but are also responsible for the development of graft-versus-host disease (GVHD), which is the major detrimental complication after allogeneic SCT.6,7 T-cell depletion of the stem cell graft removes both GVHD and GVT effect.8–10 The antitumor reactivity can be reintroduced in case of persistent or relapsed hematologic malignancies after transplantation by donor lymphocyte infusion (DLI).11–15 Whereas profound antitumor effects are frequently associated with GVHD in patients responding to DLI, postponed administration of DLI has been associated with a decreased risk of severe GVHD, and clinical observations indicate that more subtle antitumor reactivities can also be observed in the absence of GVHD.16–18
The main targets of both GVHD and GVT reactivity after HLA-matched allogeneic SCT are minor histocompatibility antigens (mHags).19–21 mHags are peptides differentially expressed by donor and recipient that can be recognized in the context of self-HLA molecules. mHags may arise from differential processing of peptides due to polymorphisms in the gene encoding the protein or by direct polymorphisms in the peptide sequence that is presented in the HLA molecules.22,23
The clinical manifestation of immune responses against mHags is likely to be determined by the specific tissue expression of the proteins encoding these antigens. Whereas mHags constitutively expressed in many tissues are likely to be targets for combined alloreactive GVHD and GVT responses, T-cell responses directed against antigens that are restricted to the hematopoietic cell lineages including the malignant T cells of hematopoietic origin are likely to mediate a GVT reactivity without severe GVHD.21,24–31 However, antigens with variable expression in tissues may also be targets for relatively specific GVT responses because these proteins may not be expressed or recognized on normal tissues under steady state conditions.19,32,33 By specific stimuli like inflammatory reactions they may be induced in target tissues of GVHD, leading to a detrimental local immune response.7,34,35 Characterization of mHags from patients responding to cellular immunotherapeutic interventions following allogeneic SCT in the presence or absence of GVHD will lead to a better understanding of the pathogenesis of GVHD and GVT and may lead to the development of specific antitumor T-cell therapy.
Recently, we studied in detail several patients treated for relapsed hematologic malignancies after allogeneic SCT with DLI. During the clinical GVT response, we isolated tumor-reactive T cells based on their ability to produce IFN-γ in response to specific activation by bone marrow containing the malignant cells.28,36 From one of the patients who were treated with DLI for relapsed multiple myeloma (MM) after transplantation with DLI and IFN-α we isolated a dominant cytotoxic T lymphocyte (CTL) clone capable of recognizing the malignant MM cells from the patient. At the time of the clinical response the patient suffered from mild acute GVHD of skin and liver (grade II), which resolved after discontinuation of IFN-α and short-term treatment with prednisone and cyclosporine. The patient entered a complete remission and is now, 6 years later in persistent complete remission without GVHD.
In the present study we identified the mHags recognized by this CTL clone to be encoded by the ATP-dependent interferon-responsive (ADIR) gene.37 This gene was found to be highly expressed in the MM cells, in other hematopoietic tumors, as well as nonhematopoietic tumor cell lines. Recognition of normal nonmalignant cells appears to be minor under steady state conditions, but activation of the target cell populations resulted in enhanced recognition by the CTL clone. We hypothesize that T-cell responses against mHags encoded by the ADIR gene may lead to a strong GVT reactivity, potentially coincident with GVHD depending on the activation state of the target tissues.
Materials and methods
CTL generation and culture
The HLA-A2–restricted mHag-specific CTL clone RDR2 was previously isolated using the IFN-γ secretion assay from a peripheral blood (PB) sample of a patient at the time of clinical response to DLI as treatment for relapsed MM after SCT.36 CTL clone RDR2 and the allo–HLA-A2 control clone MBM13 were expanded by stimulation with irradiated (50 Gy) allogeneic mononuclear cells (MNCs) and patient-derived Epstein-Barr virus–lymphoblastoid cell line (EBV-LCL) in IMDM (Cambrex, Verviers, Belgium) supplemented with penicillin-streptomycin (Cambrex), 3 mM l-glutamine (Cambrex), 5% fetal bovine serum (FBS) (Cambrex), 5% pooled human serum, 100 U/mL IL-2 (Chiron, Amsterdam, The Netherlands), and 0.8 μg/mL PHA (Remel, Dartford, United Kingdom).
Target-cell populations
After informed consent was obtained in accordance with the Declaration of Helsinki, with approval from Leiden University Medical Center's Institutional Review Board, BM and PB samples were collected from patients and donors, and MNCs were isolated by Ficoll-Isopaque separation. Recognition of target cells was measured in cytotoxicity assays, and stimulation of responder cells was measured using INF-γ secretion. Lineage-specific hematopoietic cell populations were isolated from PBMCs by magnetic-activated cell sorting using magnetic microbeads coupled to CD4, CD8, CD14, and CD19 monoclonal antibody (mAb) (Mylteni Biotec, Bergisch Gladbach, Germany) according to manufacturer's instructions. PHA blasts were generated from PBMCs by stimulation with 0.8 μg/mL PHA and subsequent culturing in IMDM supplemented with 100 IU/mL IL-2 and 10% FBS. EBV-LCLs were cultured in IMDM supplemented with 10% FBS. The HLA-A2–positive lymphoblastoid-processing defective cell line T238 was cultured in IMDM supplemented with 10% FBS. Hela/A2 was generated by retroviral transduction of HLA-A*0201 in Hela Tk− cells and cultured in IMDM supplemented with 10% FBS. Mesenchymal stem cells (MSCs) were generated from bone marrow cells by culturing adherent cells in low-glucose DMEM (Invitrogen, Paisley, Scotland) supplemented with 10% FBS.39 Commercially obtained biliary epithelial cells (BECs) (Sciencell, San Diego, CA) were grown in DMEM F12 (Cambrex) supplemented with 5% FBS, 2 × 10−9 M T3 hormone, 8.2 × 10−7 M hydrocortisone, 2 ng/mL epidermal growth factor, and 5 μg/mL insulin and were retrovirally transduced with HLA-A*0201. IFN-α modulation of stimulator cells was performed by addition of 1000 IU/mL IFN-α-2a (Roche, Woerden, The Netherlands). Adherent solid tumor cell lines from thyroid carcinoma (TT), breast adenocarcinoma (MCF7), and cervical carcinoma (CaSki) from the American Type Culture Collection (ATCC, Manassas, VA) and a melanoma cell line (Brown) were cultured in RPMI supplemented with 10% FBS.
Cytotoxicity assays
CTL-induced specific cytotoxicity of defined cell types in a heterogeneous target cell population was determined in CFSE-based cellular cytotoxicity assays as described before.40 Briefly, target cells were labeled with 2.5 μM CFSE (Molecular Probes, Leiden, The Netherlands) and incubated with target cells at a 1:1 ratio. After 4 and 24 hours, specific cell populations were counterstained with PE- or APC-labeled CD138, CD3, CD4, CD8, CD14, or CD19 mAb (Becton Dickinson, Erembodegem-Aalst, Belgium), and propidium iodide was added to exclude dead cells. To allow quantification of the surviving cell numbers in each sample, 104 Flow-count Fluorospheres (Coulter, Miami, FL) were added immediately before flow cytometric analysis. Cytotoxicity of CTL clones in standard 51Cr release assays was performed as described previously.41 HPLC-purified natural peptides or diluted synthetic peptides were tested for reactivity by loading 51Cr-labeled T2 cells for 1 hour at 37°C and 5% CO2 prior to addition of CTLs.
IFN-γ secretion assays
Quantification of CTL stimulation was performed by IFN-γ secretion assays. CTLs were cocultivated with various freshly isolated PBMC cell populations, EBV-LCLs, PHA blasts, transfected Hela/A2 cells, MSCs, or BECs. Stimulator cells and CTLs were diluted in IMDM supplemented with 10% FBS and cultured in 96-well microtiterplates for 24 hours at various responder-stimulator ratios. In each well 5 × 103 CTLs were stimulated with 2 × 104 PBMCs or Hela/A2, 5 × 103 BECs, or 1 × 103 MSCs. Supernatant was harvested, and IFN-γ was measured by standard enzyme-linked immunosorbent assay (ELISA) (Sanquin, Amsterdam, The Netherlands).
Peptide isolation, purification, and characterization
Purification of peptides from 8 × 1010 EBV-LCLs was performed as described before.24,42 Briefly, frozen cell pellets were lysed and affinity purified. Peptides were separated from proteins by size-exclusion centrifugation, and peptide concentrates were injected on a Smart System (Amersham Biosciences, Freiburg, Germany) and subjected to reverse-phase high-performance liquid chromatography (RP-HPLC) on a 10 cm × 2.1 mm C2/C18 3 μm particle column at 0.2 mL/min. A gradient from 20% to 50% organic phase containing 0.1% TFA was run while 0.1 mL fractions were collected in siliconized vials and stored at −80°C. Isopropanol or acetonitrile was used as organic phase. Reactive fractions were injected on a 15 cm × 75 um C18-Pepmap nano column (LC Packings/Dionex, Breda, The Netherlands), which was directly coupled to a QTOF1 mass spectrometer (Micromass Waters, Manchester, United Kingdom). The flow was delivered by a conventional JASCO binary gradient system (JASCO, Maarssen, The Netherlands), which was reduced to approximately 300 nL/min by an in-house–constructed flow splitter. Injections were performed with a FAMOS (LC Packings/Dionex). On-line HPLC tandem mass spectrometry analysis on the HCTplus (Bruker Daltonics, Bremen, Germany) was performed in a similar fashion. In this case the flow was delivered by an Ultimate (LC Packings/Dionex).
Sequence analysis of the ADIR gene
Trizol reagent (Invitrogen) was added to patient and donor cell pellets, RNA was isolated and purified, and 4 μg was reverse transcribed into cDNA for 1 hour at 37°C using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Invitrogen) in accordance with the manufacturer's instructions. Polymerase chain reactions (PCRs) of nucleotides 1 to 327 of the ADIR gene were performed in 50 μL GeneAmpII PCR buffer containing 1.5 mM MgCl2, 250 μM dNTPs, 800 nM forward primer (nucleotides 1 to 18), 800 nM reverse primer (nucleotides 309 to 327), 2% DMSO, and 1.5 units of AmpliTaq DNA polymerase. Amplification on an Applied Biosystems GeneAmp PCR system 2400 was achieved in 35 cycles of 15 seconds at 95°C and 30 seconds at 58°C. Sequence reactions were performed on 1 μL of purified PCR product using the Big Dye Terminator v3.1 sequencing kit (Applied Biosystems, Foster City, CA) and 1 μM reverse primer in 25 cycles of 10 seconds at 96°C, 5 seconds at 58°C, and 4 minutes at 60°C. After DNA purification, sequencing was performed using an ABI310 sequencer.
Transfection of constructs containing the LB-ADIR-1F epitope
Constructs from donor and patient cDNA containing the ADIR gene were generated using 3 different forward primers and 1 reverse primer and ligated in pCR3.1 expression vector (Invitrogen) as described previously.31 A mixture of 100 ng plasmid and 0.8 μL Lipofectamine (Invitrogen) was incubated for 30 minutes at room temperature and used to transiently transfect 2 × 104 Hela/A2 cells. After 5 hours of incubation at 37°C, 50 μL IMDM/10% FBS was added. After 24 hours, 104 RDR2 cells were added and again after 24 hours 50 μL of supernatant was harvested and tested for IFN-γ secretion in ELISA.
Ex vivo detection of LB-ADIR-1F–specific T cells and TCRBV analysis
Recombinant biotinylated HLA-A*0201 monomers were folded with both ADIR peptide variants. Streptavidin-APC tetramers were produced as described previously.43 Tetramer-positive events in thawed patient samples were sorted by fluorescence-activated cell sorting (FACS) to single cells per well, expanded, and tested for cytotoxicity as described previously.21 T-cell receptor V-B expression of cytotoxic clones and tetramer-positive T cells in patient material was determined by staining with FITC-conjugated mAb to TCRBV7 (Beckman Coulter, Mijdrecht, The Netherlands). Sequences of the TCRBV were determined as described previously,36 and TCR chains were named in accordance with the nomenclature described by Arden et al.44
Quantitative real-time-PCR to analyze gene expression
Quantitative real-time PCR analysis was performed as described previously.45 To normalize for variations in the procedures for RNA isolation and cDNA synthesis, 1% mouse spleen cells were added to each cell sample. Quantitative real-time PCR was performed on an ABI/PRISM 7700 Sequence Detector System (Applied Biosystems) using qPCR-Core Kit (Eurogentec, Seraing, Belgium). Human ADIR and PBGD results were normalized using the murine GAPDH expression. For each sample, relative gene expression was calculated using the results obtained from a serial diluted EBV-LCL. EBV-LCL expression was set to 100. Primers for human ADIR were designed spanning exons 1 to 3: forward 5′-GACGACTGTGACGAGGACGA-3′, reverse 5′-CAAATGCTGGCCATGCAG-3′, and probe 5′-(TET)-CTGGGCTGGCGCCTTCCTCTGT-(TAMRA)-3′. Primers and probe for human PBGD were as follows: forward 5′-GCAATGCGGCTGCAA-3′, reverse 5′-GGGTACCCACGCGAATG-3′, and probe 5′-(TET)-CTCATCTTTGGGCTGTTTTCTTCCGCC-(TAMRA)-3′. Primers and probe for murine GAPDH were as follows: forward 5′-GGGCTCATGACCACAGTCCA-3′, reverse 5′-ATACTTGGCAGGTTTCTCCAGG-3′, and probe 5′-(TET)-TCCTACCCCCAATGTGTCCGTCGT-(TAMRA)-3′.
Results
Isolation of the HLA-A2–restricted CD8+ CTL clone RDR2 recognizing a frequently expressed mHags
We previously described the isolation of various CTL clones from a female patient who was successfully treated with DLI after relapsed MM.36 CTL clones were generated by direct cloning of IFN-γ–producing cells upon stimulation by irradiated bone marrow cells harvested from the patient prior to SCT. Panel studies using unrelated EBV-LCLs and blocking studies with HLA allele–specific mAb showed that recognition by the most dominant CTL clones was restricted by HLA-A2. Extensive panel studies using PHA blasts and EBV-LCLs of unrelated sibling pairs demonstrated that most HLA-A2–restricted CTL clones designated RDR2 displayed an identical recognition pattern and lysed 57% of targets from all HLA-A2 individuals tested. All RDR2 clones were found to express an identical TCR-BV7S1, N region, and BJ1S4, illustrating that they were derived from the same clonal T cell.36
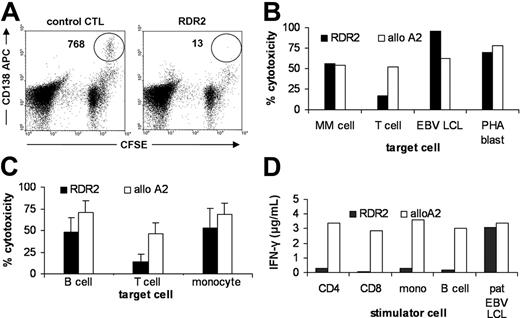
Because RDR2 was isolated from an MM patient, we investigated the sensitivity of her MM cells to lysis by the T-cell clone. The CFSE-based cytotoxicity assay was performed to allow quantitative measurement of lysis of MM cells that were present in relatively low frequencies within the heterogeneous bone marrow samples.40 Figure 1A shows dot plots of a quantitative CFSE-based cytotoxicity assay performed on thawed bone marrow cells from an mHag-positive patient containing 10% MM cells. Lysis was measured after incubation with a control CMV-pp65-A2–specific CTL clone or with CTL RDR2. CD138 was used as a marker for the malignant MM cells. A strong decrease of the number of CD138+ cells was observed. In a sample containing 1% MM cells that was taken from the patient from which CTL RDR2 was isolated, lysis was measured of bone marrow cells using CD138 as a marker for the malignant MM cells and CD3 as a marker for nonmalignant patient-derived T cells. Figure 1B illustrates that MM cells from the patient were strongly lysed whereas lysis of normal unstimulated T cells was low. Activated T cells (PHA blasts) and EBV-LCLs from the patient were strongly recognized. Donor-derived PHA blasts and EBV-LCLs were not recognized (data not shown). To further study susceptibility of normal hematopoietic cells to lysis by RDR2, PBMC subpopulations from HLA-A2–positive mHag-positive donors were tested for recognition by RDR2 and an allo-A2 clone. Whereas similar recognition of PHA blasts and EBV-LCLs by the allo-A2 clone and RDR2 was observed, RDR2-mediated lysis of normal B cells and T cells was lower as compared with allo-A2–mediated lysis (Figure 1C). In addition, PBMCs were separated by magnetic bead cell sorting into CD4+ T cells, CD8+ T cells, monocytes, and B cells and were used to stimulate RDR2 and allo-A2 CTL in IFN-γ release assays. Whereas all stimulator cell subpopulations were equally able to induce IFN-γ secretion by allo-A2 CTL, stimulation of RDR2 by normal hematopoietic cells was more than 10-fold lower as compared with stimulation by EBV-LCLs (Figure 1D).
Recognition pattern of CTL clone RDR2 on MM cells and normal hematopoietic cells. Recognition of MM cells by RDR2 in a heterogeneous bone marrow sample of an mHag-positive patient was determined in CFSE-based cytotoxicity assays after incubation with a control CMV-pp65-A2–specific CTL clone (left dot plot) or with CTL RDR2 (right dot plot) in 1:1 effector-target (E/T) ratios. CFSE-negative cells represent the CTL clones. Strong specific recognition of CFSE-positive CD138 APC-counterstained MM cells was observed (A). Recognition by RDR2 (▪) and control allo-A2 CTL (□) was tested in CFSE-based cytotoxicity assays and by IFN-γ secretion. Heterogeneous cell samples were incubated with CTLs at a 1:1 E/T ratio for 4 hours. Patient bone marrow cells were counterstained with CD138 mAb for detection of MM cells or with CD3 mAb for detection of T cells. Patient-derived MM cells were strongly lysed by RDR2 whereas T cells were weakly recognized. Both EBV-LCLs and PHA blasts were strongly lysed (B). PBMCs from 3 healthy mHag-positive donors were counterstained with different lineage-specific markers. Lysis by RDR2 was significantly diminished in both B cells (P = .02) and T cells (P < .001) as compared with lysis by allo-A2 CTL (shown as mean value ± SD) (C). Stimulation of CTLs was measured by INF-γ release after 24 hours of coculture. RDR2 stimulation by resting PBMC subpopulations was low as compared with allo-A2 CTL stimulation whereas activated B cells (EBV-LCL) induced similar IFN-γ release in both CTLs (D).
Recognition pattern of CTL clone RDR2 on MM cells and normal hematopoietic cells. Recognition of MM cells by RDR2 in a heterogeneous bone marrow sample of an mHag-positive patient was determined in CFSE-based cytotoxicity assays after incubation with a control CMV-pp65-A2–specific CTL clone (left dot plot) or with CTL RDR2 (right dot plot) in 1:1 effector-target (E/T) ratios. CFSE-negative cells represent the CTL clones. Strong specific recognition of CFSE-positive CD138 APC-counterstained MM cells was observed (A). Recognition by RDR2 (▪) and control allo-A2 CTL (□) was tested in CFSE-based cytotoxicity assays and by IFN-γ secretion. Heterogeneous cell samples were incubated with CTLs at a 1:1 E/T ratio for 4 hours. Patient bone marrow cells were counterstained with CD138 mAb for detection of MM cells or with CD3 mAb for detection of T cells. Patient-derived MM cells were strongly lysed by RDR2 whereas T cells were weakly recognized. Both EBV-LCLs and PHA blasts were strongly lysed (B). PBMCs from 3 healthy mHag-positive donors were counterstained with different lineage-specific markers. Lysis by RDR2 was significantly diminished in both B cells (P = .02) and T cells (P < .001) as compared with lysis by allo-A2 CTL (shown as mean value ± SD) (C). Stimulation of CTLs was measured by INF-γ release after 24 hours of coculture. RDR2 stimulation by resting PBMC subpopulations was low as compared with allo-A2 CTL stimulation whereas activated B cells (EBV-LCL) induced similar IFN-γ release in both CTLs (D).
In conclusion, RDR2 recognized an HLA-A2–restricted epitope causing strong lysis of MM cells and activated T cells and B cells. In contrast, reactivity with normal nonactivated hematopoietic cells was relatively low as measured both by direct cytotoxicity and by IFN-γ secretion.
Purification and mass spectrometric identification of the peptide
To identify the epitope that was recognized by CTL clone RDR2, EBV-LCL cells expressing the antigen were lysed, and peptide-HLA complexes were affinity purified using HLA-A2–specific BB7.2 mAb. After elution of peptides, size centrifugation was performed to separate peptides from HLA monomers and β2-microglobulin. After freeze drying, the peptide mixture was subjected to RP HPLC using isopropanol as organic solvent and fractions were collected. 51Cr-labeled T2 cells were loaded with a small sample of each fraction. RDR2 was added, and a single positive fraction could be detected. This fraction was subsequently subjected to RP HPLC with acetonitrile as organic solvent and fractionated. Fractions were tested for reactivity, and again a positive fraction was found. To determine the most abundant masses present in this fraction, part of the fraction was injected on a nano–liquid chromatography system directly coupled to a Q-TOF1 mass spectrometer. Abundantly present masses were fragmented by collision-activated dissociation on an HCTplus mass spectrometer. Analysis of obtained fragmentation patterns led to the sequence of several candidate peptides that were subsequently synthesized. RDR2 recognized an [M+2H]++ candidate peptide with mass/charge of 528.8 and sequence SVAPALALFPA at levels as low as 10 pM (data not shown). Furthermore, the fragmentation patterns of the synthetic peptide and the natural eluted peptide were identical (data not shown).
Identification of a polymorphic gene responsible for RDR2 recognition
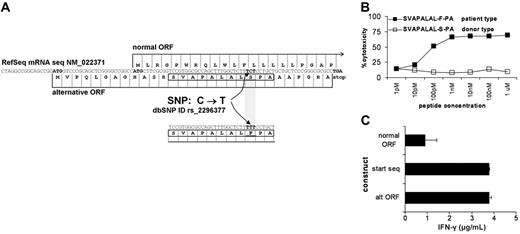
A basic local alignment sequence tool (BLAST) search of sequence SVAPALALFPA against a 6-frame translation of the European Molecular Biology Laboratory (EMBL) nucleotide database revealed 100% identity to amino acids 13 to 23 from an alternative open reading frame (ORF) of the ADIR gene, also known as TOR3A37 (National Center for Biotechnology Information [NCBI] mRNA Reference Sequence [RefSeq] NM_022371). A known single nucleotide polymorphism (SNP) in ADIR at nucleotide 78 from C>T (NCBI dbSNP cluster ID rs2296377) results in an amino acid change in an alternative transcript from serine (S) to phenylalanine (F) at position 21, corresponding to position 9 of the eluted peptide (Figure 2A). Both peptides were synthesized and loaded on T2 cells. Patient-type peptide SVAPALAL-F-PA but not donor-type peptide SVAPALAL-S-PA was recognized by RDR2 (Figure 2B). Sequence analysis of the ADIR PCR product flanking the SNP revealed that the donor was CC homozygous and the patient CT heterozygous. To demonstrate that patient-type T but not donor-type C of this gene was responsible for recognition by RDR2, constructs were generated from both patient and donor cDNA. Because RDR2 recognized a peptide arising from an alterative ORF controlled by a start codon 5′ upstream from the normal start codon, 3 different forward primers were composed. The first primer was chosen at the normal start codon, thus lacking the alternative start codon. The second primer was chosen at the start of the transcript, thus providing both start codons. The third primer was chosen at the alternative start codon and also contained the normal start codon. The constructs were transiently transfected into Hela/A2 cells. Only patient-derived constructs induced IFN-γ release by RDR2 CTL. Transfection of constructs containing only the normal ORF start codon and lacking the alternative ORF start codon showed a strong decrease of CTL recognition (Figure 2C). All donor-derived constructs failed to induce IFN-γ by RDR2 (data not shown). Next, a panel of 74 unrelated HLA-A2–positive individuals was analyzed by sequencing for determination of the polymorphism and susceptibility to lysis of PHA blasts by RDR2. A 100% correlation between presence of this specific SNP and CTL reactivity proved that this SNP from C>T in the ADIR gene generates the mHag epitope SVAPALALFPA that is recognized by RDR2 (Table 1). The mHag was designated LB-ADIR-1F.
Identification of ADIR as the polymorphic gene responsible for RDR2 recognition. BLAST searching of SVAPALALFPA against a translated EMBL database revealed 100% identity to amino acids 13 to 23 from an alternative ORF of the ADIR gene. A known SNP at nucleotide 78 results in an amino acid change from S>F. (A). Both patient-type peptide SVAPALAL-F-PA (▪) and donor-type peptide SVAPALAL-S-PA (□) were synthesized and tested for RDR2 reactivity on T2 cells in a 51Cr release assay. Only cells loaded with the patient-type peptide but not cells loaded with the donor-type peptide were lysed (B). Constructs containing patient-derived DNA were generated. The start of each construct was varied to obtain translation at the start of the transcript, resulting in both the normal and the alternative ORF or specific translation of only the normal or alternative ORF. Constructs were transiently transfected into Hela/A2 cells. RDR2 was cocultured for 24 hours, and IFN-γ release in supernatants was measured by ELISA. RDR2 stimulation was observed in all cases. Stimulation by constructs containing only the normal ORF start codon and lacking the alternative ORF start codon showed strongly diminished CTL recognition (C). Similar constructs containing the donor-derived DNA were not recognized by RDR2 (data not shown).
Identification of ADIR as the polymorphic gene responsible for RDR2 recognition. BLAST searching of SVAPALALFPA against a translated EMBL database revealed 100% identity to amino acids 13 to 23 from an alternative ORF of the ADIR gene. A known SNP at nucleotide 78 results in an amino acid change from S>F. (A). Both patient-type peptide SVAPALAL-F-PA (▪) and donor-type peptide SVAPALAL-S-PA (□) were synthesized and tested for RDR2 reactivity on T2 cells in a 51Cr release assay. Only cells loaded with the patient-type peptide but not cells loaded with the donor-type peptide were lysed (B). Constructs containing patient-derived DNA were generated. The start of each construct was varied to obtain translation at the start of the transcript, resulting in both the normal and the alternative ORF or specific translation of only the normal or alternative ORF. Constructs were transiently transfected into Hela/A2 cells. RDR2 was cocultured for 24 hours, and IFN-γ release in supernatants was measured by ELISA. RDR2 stimulation was observed in all cases. Stimulation by constructs containing only the normal ORF start codon and lacking the alternative ORF start codon showed strongly diminished CTL recognition (C). Similar constructs containing the donor-derived DNA were not recognized by RDR2 (data not shown).
Correlation of SNP and CTL reactivity in 76 individuals
| ADIR nucleotide 78* . | No. of individuals . | No. of lysed PHA blasts . | Frequency, % . |
|---|---|---|---|
| CC | 33 | 0 | 43 |
| CT | 36 | 36 | 47 |
| TT | 7 | 7 | 9 |
| ADIR nucleotide 78* . | No. of individuals . | No. of lysed PHA blasts . | Frequency, % . |
|---|---|---|---|
| CC | 33 | 0 | 43 |
| CT | 36 | 36 | 47 |
| TT | 7 | 7 | 9 |
Sequence analysis of nucleotides 1 to 327 of the ADIR gene in PBMCs. Nucleotide 78 represents the SNP.
Tetramer staining and FACS of LB-ADIR-1F–specific CTLs
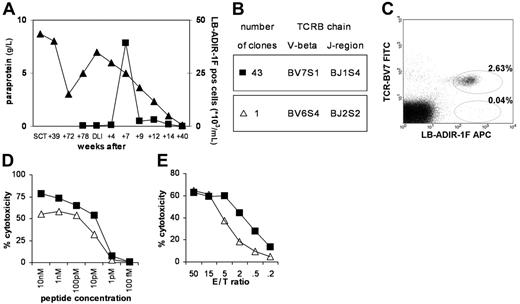
Both peptide LB-ADIR-1F and peptide SVAPALAL-S-PA were able to bind to recombinant HLA-A*0201 molecules, and tetrameric complexes were produced. RDR2 specifically bound the LB-ADIR-1F tetramer whereas SVAPALAL-S-PA tetramers and irrelevant HA-1 tetramers were negative (data not shown). LB-ADIR-1F tetramers were used to analyze a series of peripheral blood samples that were taken from the patient before and after DLI. Serum paraprotein levels were analyzed as a marker for disease activity. Whereas LB-ADIR-1F–specific T cells were not detectable prior to DLI, at 7 weeks after DLI high numbers of LB-ADIR-1F–specific CD8+ T cells could be detected (Figure 3A). The appearance of LB-ADIR-1F–specific T cells coincided with development of acute GVHD grade II and resulted in complete remission. GVHD was treated successfully with a short course of 1 mg prednisone per kilogram of body weight and cyclosporin. Tetramer-positive T cells were clonally isolated by FACS and expanded. All tetramer-positive CTL clones were able to lyse both patient EBV-LCLs and LB-ADIR-1F–pulsed donor EBV-LCLs (data not shown). TCR characterization of RDR2 showed usage of V-beta BV7S1, J-region BJ1S4, and identical CDR3 regions in 43 of 44 growing clones analyzed. One clone, however, expressed TCRBV6S4 (Figure 3B). Analysis of the patient sample at 7 weeks after DLI revealed that a low percentage of LB-ADIR-1F–positive T cells did not stain with mAb directed against TCR-BV7 (Figure 3C). Functional comparison of the original RDR2, newly isolated identical TCR-BV7S1 clones, and TCR-BV6S4–expressing clones was performed. Whereas cytotoxicity of the original RDR2 and newly isolated BV7S1 clones was similar (data not shown), the BV6S4 clone showed diminished recognition of peptide-loaded T2 cells (Figure 3D). When HLA-A2–positive and LB-ADIR-1F–expressing EBV-LCL cells were used as target cells, again the BV6S4 clone displayed lower cytotoxicity (Figures 3E). Similar results were obtained using HLA-A2–positive and LB-ADIR-1F–expressing PHA blasts (data not shown). To further investigate differential recognition, reactivity of both TCR BV7S1 and TCR BV6S4 was tested on EBV-LCL–expressing HLA-A*0205 and HLA-A*0220 subtypes. Lysis of LB-ADIR-1F–positive HLA-A*0205 EBV-LCLs was absent whereas identical strong lysis between both TCR types was observed of HLA-A*0220 EBV-LCLs. Pulsing of HLA-A*0205 EBV-LCLs with synthetic peptide could only partially restore the recognition to identical levels by both TCR types (data not shown).
Tetramer staining and clonal analysis of LB-ADIR-1F–specific CTLs in the patient. PBMCs from the patient taken at several time points after SCT and DLI were stained with LB-ADIR-1F tetramers (▪), and serum paraprotein levels were measured (▴) (A). LB-ADIR-1F tetramer-positive cells present at week 7 after DLI were single-well sorted and expanded. TCRBV sequence analysis was performed on 44 reactive clones revealing TCRBV7S1 in 43 clones and TCRBV6S4 in 1 clone (B). Reanalysis of the patient sample was performed using counterstaining with TCRBV7 confirming a low percentage of TCRBV7-negative cells in the LB-ADIR-1F–positive population (C). Reactivity of TCRBV7S1- (▪) and TCRBV6S4- (▵) expressing clones was determined using 51Cr release assays on peptide-pulsed T2 cells (D) and EBV-LCL cells (E), demonstrating that TCRBV6S4-expressing T cells displayed lower cytotoxicity.
Tetramer staining and clonal analysis of LB-ADIR-1F–specific CTLs in the patient. PBMCs from the patient taken at several time points after SCT and DLI were stained with LB-ADIR-1F tetramers (▪), and serum paraprotein levels were measured (▴) (A). LB-ADIR-1F tetramer-positive cells present at week 7 after DLI were single-well sorted and expanded. TCRBV sequence analysis was performed on 44 reactive clones revealing TCRBV7S1 in 43 clones and TCRBV6S4 in 1 clone (B). Reanalysis of the patient sample was performed using counterstaining with TCRBV7 confirming a low percentage of TCRBV7-negative cells in the LB-ADIR-1F–positive population (C). Reactivity of TCRBV7S1- (▪) and TCRBV6S4- (▵) expressing clones was determined using 51Cr release assays on peptide-pulsed T2 cells (D) and EBV-LCL cells (E), demonstrating that TCRBV6S4-expressing T cells displayed lower cytotoxicity.
Modulation of recognition by IFN-α and ADIR gene expression
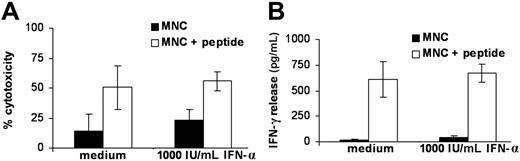
Because previous studies on the ADIR gene indicated that IFN-α could enhance gene expression37 and because the patient was also treated with IFN-α, we studied the effect of IFN-α on LB-ADIR-1F recognition by RDR2 using MNCs of LB-ADIR-1F–expressing donors. MNCs were precultured for 48 hours in the absence or presence of 1000 IU/mL IFN-α prior to addition of CTL RDR2 at a 1:1 ratio. Maximal recognition was determined by testing MNCs pulsed with saturating concentrations of synthetic peptide. Cytotoxicity was measured in a 4-hour CFSE assay (Figure 4A), and IFN-γ release was measured after 24 hours (Figure 4B). A nonsignificant increase of recognition was observed in all cases, whereas peptide-pulsed MNCs were recognized at levels comparable to EBV-LCL recognition (data not shown). Because recognition of LB-ADIR-1F–positive cells significantly differed between freshly isolated MNCs and activated cells such as EBV-LCLs and PHA blasts, we measured gene expression by performing quantitative real-time PCR on freshly isolated MNCs and MNCs cultured for 24 and 48 hours in medium containing 10% FBS alone or supplemented with either 1000 IU/mL IFN-α or 0.8 μg/mL PHA. To each cell sample a fixed percentage of 1% murine spleen cells was added prior to RNA isolation and cDNA synthesis. Each sample was assayed for expression of ADIR, PBGD, and murine GAPDH. To exclude variation in mRNA isolation and cDNA synthesis, both ADIR and PBGD expression levels were normalized to the murine GAPDH expression level, which maximally resulted in 2-fold higher or lower outcome. Whereas expression of the household gene PBGD was only influenced by PHA, ADIR expression already increased upon culturing of the cells, irrespective of the presence of IFN-α. High ADIR expression levels in EBV-LCLs and in PHA-stimulated MNCs were observed (Table 2).
Modulation of LB-ADIR-1F–specific recognition by IFN-α. Recognition of MNCs from 3 different LB-ADIR-1F–positive donors was measured by direct cytotoxicity in 4-hour CFSE assays (A) and by 24-hour IFN-γ release (B) following preincubation in medium alone or in medium containing 1000 IU/mL IFN-α for 48 hours (shown as mean value ± SD). Recognition of MNCs (▪) and MNCs exogeneously pulsed with saturating concentrations of synthetic peptide (□) was measured at an E/T ratio of 1:1. Only minimal up-regulation of recognition in IFN-α precultured MNCs was observed.
Modulation of LB-ADIR-1F–specific recognition by IFN-α. Recognition of MNCs from 3 different LB-ADIR-1F–positive donors was measured by direct cytotoxicity in 4-hour CFSE assays (A) and by 24-hour IFN-γ release (B) following preincubation in medium alone or in medium containing 1000 IU/mL IFN-α for 48 hours (shown as mean value ± SD). Recognition of MNCs (▪) and MNCs exogeneously pulsed with saturating concentrations of synthetic peptide (□) was measured at an E/T ratio of 1:1. Only minimal up-regulation of recognition in IFN-α precultured MNCs was observed.
Modulation of ADIR relative gene expression in PBMCs
| Incubation time, h . | ADIR . | PBGD . | ||||
|---|---|---|---|---|---|---|
| Control . | IFN . | PHA . | Control . | IFN . | PHA . | |
| 0 | 9 ± 3 | 9 ± 3 | 9 ± 3 | 5 ± 1 | 5 ± 1 | 5 ± 1 |
| 24 | 24 ± 4 | 19 ± 6 | 48 ± 22 | 5 ± 2 | 3 ± 1 | 60 ± 53 |
| 48 | 36 ± 17 | 23 ± 10 | 238 ± 91 | 8 ± 2 | 5 ± 2 | 178 ± 153 |
| Incubation time, h . | ADIR . | PBGD . | ||||
|---|---|---|---|---|---|---|
| Control . | IFN . | PHA . | Control . | IFN . | PHA . | |
| 0 | 9 ± 3 | 9 ± 3 | 9 ± 3 | 5 ± 1 | 5 ± 1 | 5 ± 1 |
| 24 | 24 ± 4 | 19 ± 6 | 48 ± 22 | 5 ± 2 | 3 ± 1 | 60 ± 53 |
| 48 | 36 ± 17 | 23 ± 10 | 238 ± 91 | 8 ± 2 | 5 ± 2 | 178 ± 153 |
PBMCs were incubated for the indicated times in the absence or presence of 1000 IU/mL INF-α or 0.8 μg/mL PHA. Prior to mRNA isolation and cDNA synthesis, 1% mouse spleen cells were added to each sample. Data were normalized to murine GAPDH expression. Expression of EBV-LCLs was set to 100.
Recognition of normal nonhematopoietic cells by LB-ADIR-1F–specific CTLs
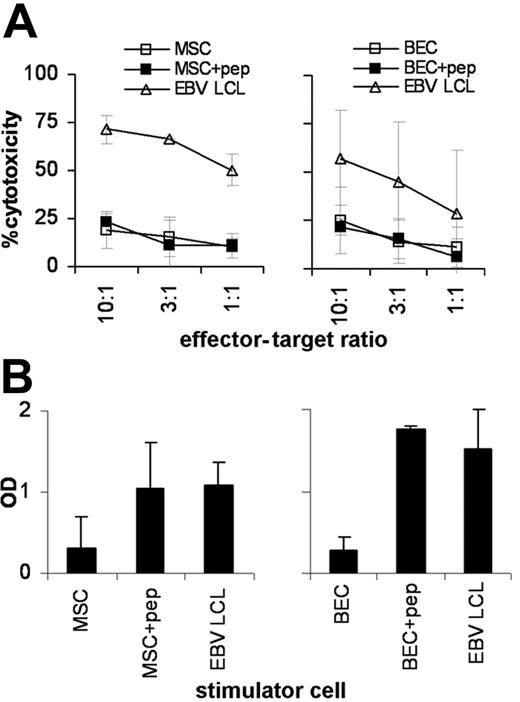
Recognition of nonhematopoietic normal tissues was studied using BECs and MSCs expressing the LB-ADIR-1F SNP variant in both direct cytotoxicity assays and stimulation assays. Whereas direct cytotoxicity against patient EBV-LCLs was strong, lysis of MSCs (Figure 5A) and BECs (Figure 5B) was low. LB-ADIR-1F peptide pulsing of MSCs and BECs did not significantly increase lysis, suggesting overall low susceptibility of these target cells to direct cytotoxicity. When MSCs (Figure 5C) and BECs (Figure 5D) were used as stimulator cells in IFN-γ release assays again, low stimulation by both nonhematopoietic cell types was observed. LB-ADIR-1F peptide pulsing of the stimulator cells increased stimulatory capacity of both MSCs and BECs to levels comparable to stimulation by patient EBV-LCLs.
Recognition of normal nonhematopoietic cells by LB-ADIR-1F–specific CTLs. Recognition of LB-ADIR-1F–expressing HLA-A2–positive MSCs (A,C) and BECs (B,D) by CTL RDR2 was studied in 3 separate 4-hour 51Cr release assays and in 3 separate IFN-γ secretion assays. Lysis of untreated target cells (□) and target cells exogeneously pulsed with saturating concentrations of synthetic peptide (▪) was compared with lysis of patient EBV-LCLs (▵) (A,B). IFN-γ secretion by CTL RDR2 was measured after 24 hours of cocultivating CTL RDR2 at a 3:1 responder-stimulator ratio with untreated cells, peptide-pulsed cells, and patient EBV-LCLs (C,D). Data are shown as mean values ± SD.
Recognition of normal nonhematopoietic cells by LB-ADIR-1F–specific CTLs. Recognition of LB-ADIR-1F–expressing HLA-A2–positive MSCs (A,C) and BECs (B,D) by CTL RDR2 was studied in 3 separate 4-hour 51Cr release assays and in 3 separate IFN-γ secretion assays. Lysis of untreated target cells (□) and target cells exogeneously pulsed with saturating concentrations of synthetic peptide (▪) was compared with lysis of patient EBV-LCLs (▵) (A,B). IFN-γ secretion by CTL RDR2 was measured after 24 hours of cocultivating CTL RDR2 at a 3:1 responder-stimulator ratio with untreated cells, peptide-pulsed cells, and patient EBV-LCLs (C,D). Data are shown as mean values ± SD.
Recognition of malignant hematopoietic cells and solid tumor cell lines
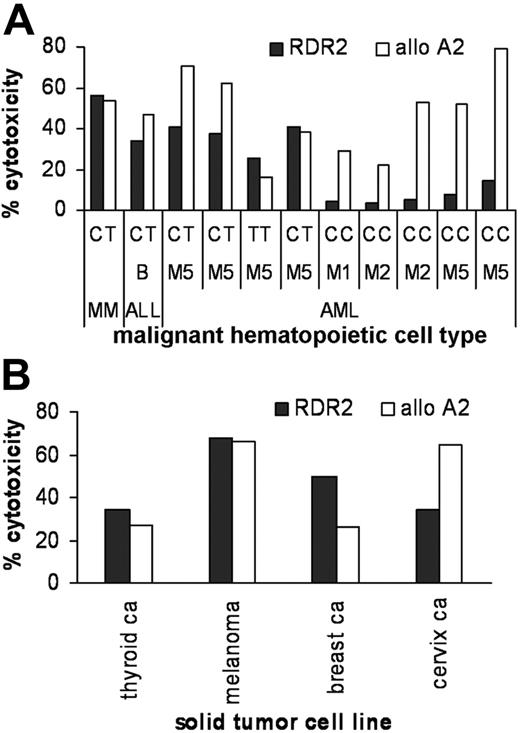
To investigate susceptibility to lysis by LB-ADIR-1F–specific CTLs, a panel of HLA-A2–positive malignant hematopoietic cells was subjected to sequence analysis on the LB-ADIR-1F polymorphism and lysis by CTL RDR2 and allo-A2 CTL. Prominent recognition was found of MM cells. Additionally, leukemic cells expressing the LB-ADIR-1F polymorphism were susceptible to lysis by RDR2 (Figure 6A). Furthermore, when a panel of LB-ADIR-1F–positive HLA-A2–expressing solid tumor lines was tested for recognition by RDR2, lysis at levels comparable to lysis by the allo-A2 CTL was observed (Figure 6B).
Recognition of malignant hematopoietic cells and solid tumor cell lines. Lysis of LB-ADIR-1F–expressing MM cells in heterogeneous bone marrow samples was measured using the CFSE assay at a 1:1 E/T ratio and leukemic blast cell populations using 4-hour 51Cr release assays at an E/T ratio of 20:1. Recognition by RDR2 is shown by ▪ and allo-A2 control CTLs by □. On the x-axis the malignant cell type and the SNP at nucleotide 78 of the ADIR gene are depicted. MM and leukemic cells expressing the LB-ADIR-1F epitope (CT or TT) were recognized by both CTLs whereas LB-ADIR-1F–negative (CC) targets were only lysed by control allo-A2 CTLs (A). A panel of HLA-A2–positive LB-ADIR-1F–expressing solid tumor cell lines was tested for lysis by RDR2 (▪) and control allo-A2 CTL (□) using 4-hour 51Cr release assays at an E/T ratio of 20:1. All tumor cell lines were lysed by LB-ADIR-1F–specific CTLs (B).
Recognition of malignant hematopoietic cells and solid tumor cell lines. Lysis of LB-ADIR-1F–expressing MM cells in heterogeneous bone marrow samples was measured using the CFSE assay at a 1:1 E/T ratio and leukemic blast cell populations using 4-hour 51Cr release assays at an E/T ratio of 20:1. Recognition by RDR2 is shown by ▪ and allo-A2 control CTLs by □. On the x-axis the malignant cell type and the SNP at nucleotide 78 of the ADIR gene are depicted. MM and leukemic cells expressing the LB-ADIR-1F epitope (CT or TT) were recognized by both CTLs whereas LB-ADIR-1F–negative (CC) targets were only lysed by control allo-A2 CTLs (A). A panel of HLA-A2–positive LB-ADIR-1F–expressing solid tumor cell lines was tested for lysis by RDR2 (▪) and control allo-A2 CTL (□) using 4-hour 51Cr release assays at an E/T ratio of 20:1. All tumor cell lines were lysed by LB-ADIR-1F–specific CTLs (B).
Discussion
In this study, we identified the ATP-dependent interferon-responsive gene to encode an activation-induced minor histocompatibility antigen that was recognized by MM-reactive T cells. A dominant HLA-A2–restricted CD8+ CTL clone designated RDR2 was isolated from a patient with relapsed MM after allogeneic SCT responding to DLI, resulting in a long-lasting complete remission. Biochemical analysis of peptides eluted from HLA molecules revealed that this CTL clone recognized the peptide SVAPALALFPA, which was encoded by an alternative ORF of the human ADIR gene. This mHag, designated LB-ADIR-1F, differed only by an S>F substitution from the donor-type allelic counterpart. A population study with 76 HLA-A2 individuals revealed a 100% correlation between SNP occurrence and recognition of target cells, confirming that the CTL clone was directed against this ADIR-derived epitope, which had a population frequency of 57%.
The function of the ADIR gene is unknown. When T-cell recognition of target cells and expression levels of ADIR mRNA were analyzed in various cell types, we demonstrated a relatively low expression of the gene in resting cells corresponding with low recognition. When proliferation was induced in PB T cells and B cells, strong gene up-regulation was observed, resulting in high recognition by the LB-ADIR-1F–specific T cells. Although concurrent expression of normally and alternatively transcribed ADIR proteins cannot be directly determined in the cell, the strong correlation between the ADIR SNP and susceptibility to lysis illustrates the absence of specific expression regulation of the alternatively translated protein.
Because the patient was treated with IFN-α and it has previously been described that ADIR gene expression could be up-regulated by interferons,37 we examined gene expression and LB-ADIR-1F–specific recognition of PBMCs that were preincubated with IFN-α. A minor increase in ADIR gene expression was found after in vitro culture in serum irrespective of IFN-α preincubation. In addition, in the presence of IFN-α only a minimal increase in susceptibility to lysis and stimulatory capacity was observed.
Not only MM cells but also other hematologic malignancies and nonhematopoietic solid tumor cell lines were recognized by LB-ADIR-1F–specific T cells. MSCs and BECs, which may represent possible target tissues of GVHD, displayed low recognition by the ADIR-specific T cells. Our results illustrate broad antitumor reactivity of LB-ADIR-1F–specific T cells asshown by the lysis of malignant hematopoietic cells as well as solid tumor cell lines combined with limited recognition of nonactivated tissues.
Kinetic studies of PB samples isolated from the patient before and after DLI using tetramers recognizing various mHags differentially expressed in donor and recipient illustrated a strong correlation between the clinical response and the occurrence of not only a high frequency of LB-ADIR-1F–specific T cells but also of HA-1 and LB-ECGF-1H–specific T cells as studied previously.21,28,31,36 The LB-ADIR-1F–specific T cells were the most dominant, comprising almost 3% of the CD8+ T cells. Approximately 1% LB-ECGF-1H–specific T cells and 0.9% HA1-specific T cells were found with similar kinetics. These in vivo circulating mHag-specific T cells represent 80% of activated PB CD8+ T cells as identified by coexpression of HLA-DR. Furthermore, because HA-1, ECGF, and ADIR proteins are highly expressed in MM cells and their relevant polymorphism can be recognized by T cells, we hypothesize that the 3 combined strong reactivities are likely to have caused the clinical course of the major response to DLI resulting in a complete remission lasting now for more than 6 years.
In conclusion, we characterized a novel mHag encoded by a frequently occurring SNP in the ADIR gene that is recognized by tumor-reactive CTLs. We hypothesize that the balance between GVT responses and GVHD is not only determined by the specificity of T cells but also by the activation state of GVHD target tissues.33–35,46 Up-regulated tissue susceptibility to mHag-specific T cells may be caused by the cytokine storm shortly after stem celltransplantation due to conditioning regimens resulting in tissue damage and the presence of high numbers of recipient-originated antigen-presenting cells. Homeostatic proliferation of these T cells during the lymphopenic phase may further amplify the magnitude of such responses. This state may result in severe GVHD if these T cells are administered as part of the stem cell graft. Immunotherapy of tumor cells by postponed adoptive transfer of LB-ADIR-1F–specific T cells following initially T-cell–depleted transplantation may result in a strong GVT response with limited risk of GVHD.
Authorship
Contribution: C.A.M.v.B., M.G.D.K., P.A.v.V., R.W., and J.H.F.F. designed the study; C.A.M.v.B., M.G.D.K., I.J., M.H.M.H., S.A.P.v.L.H., F.M.K., A.H.d.R., P.A.v.V., and J.H.F.F. performed research and analyzed data; W.A.F.M. and M.R.S. provided and analyzed the patient's materials; and C.A.M.v.B., R.W. and J.H.F.F. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
P.A.v.V. and J.H.F.F. contributed equally to this paper.
Correspondence: Cornelis A. M. van Bergen, Laboratory of Experimental Hematology, Department of Hematology, Leiden University Medical Center, PO Box 9600 2300 RC Leiden, The Netherlands; e-mail: c.a.m.van_bergen@lumc.nl.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Centre for Medical Systems Biology, a center of excellence approved by the Netherlands Genomic Initiative/Netherlands Organisation for Scientific Research (A.H.d.R. and P.A.v.V.), and AlloStem (EU grant 503319). The authors thank E. G. A. Lurvink and A. B. Kruisselbrink for their expert technical assistance with EBV-LCL cell culture and BEC cell culture and R. van der Linden for expert technical assistance with flow cytometric isolation.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal